SUMMARY
Both splicing factors and microRNAs are important regulatory molecules that play key roles in post-transcriptional gene regulation. By miRNA deep sequencing, we identified 40 miRNAs that are differentially expressed upon ectopic overexpression of the splicing factor SF2/ASF. Here we show that SF2/ASF and one of its upregulated microRNAs (miR-7) can form a negative feedback loop: SF2/ASF promotes miR-7 maturation, and mature miR-7 in turn targets the 3′UTR of SF2/ASF to repress its translation. Enhanced microRNA expression is mediated by direct interaction between SF2/ASF and the primary miR-7 transcript to facilitate Drosha cleavage and is independent of SF2/ASF’s function in splicing. Other miRNAs, including miR-221 and miR-222, may also be regulated by SF2/ASF through a similar mechanism. These results underscore a function of SF2/ASF in pri-miRNA processing and highlight the potential coordination between splicing control and miRNA-mediated gene repression in gene regulatory networks.
INTRODUCTION
MicroRNAs (or miRNAs) are small noncoding RNAs of great importance in posttranscriptional gene regulation. With few known exceptions (Vasudevan et al., 2007), miRNAs typically bind to the 3′ untranslated regions (UTRs) of protein-coding genes and repress their expression by translational inhibition and/or promoting mRNA degradation. It is estimated that each miRNA can have hundreds of potential functional targets (Bartel, 2004); thus, miRNAs are broadly involved in diverse cellular processes to establish and/or maintain cell identity (Alvarez-Garcia and Miska, 2005). Alterations in miRNA expression often lead to severe pathological consequences and are frequently observed in human diseases (Esquela-Kerscher and Slack, 2006; Lee and Dutta, 2009; He et al., 2007).
MiRNAs are primarily transcribed by RNA polymerase II as long primary transcripts called pri-miRNAs. Pri-miRNAs are first processed in the nucleus by an RNase III-like enzyme, Drosha, to liberate ~70 nt miRNA precursors (pre-miRNAs). Pre-miRNAs are then exported to the cytoplasm, where they are further cleaved by Dicer to produce mature miRNAs, which are subsequently loaded into an RNA-induced-silencing complex (RISC) to guide downstream gene repression (Bartel, 2004). Emerging evidence suggests that miRNA processing is a highly regulated event. For instance, multiple pri-miRNAs are detected in tumor cells but are not processed to precursor or mature miRNAs (Thomson et al., 2006). It has also been shown that Drosha cleavage is initiated cotranscriptionally (Morlando et al., 2008), and retention of pri-miRNAs at the transcriptional sites enhances their conversion to pre-miRNAs (Pawlicki and Steitz, 2008). Several RNA-binding proteins have been implicated in regulating pri-miRNA maturation (Guil and Caceres, 2007; Trabucchi et al., 2009); however, the detailed mechanisms remain elusive.
Pre-mRNA splicing, including regulated alternative splicing, is another layer of eukaryotic gene regulation. More than 90% of human genes might be alternatively spliced (Wang et al., 2008); in fact, alternative splicing is one major contributor to mammalian proteome diversity (Lander et al., 2001; Maniatis and Tasic, 2002). Intron removal is catalyzed by the spliceosome, a macromolecular complex that consists of 5 different snRNAs and over 300 associated proteins (Wahl et al., 2009). Among them, SR proteins are a family of key splicing factors involved in both constitutive and alternative splicing. In addition to splicing regulation, SR proteins are also involved in diverse events during the life cycle of mRNAs, including transcription elongation, mRNA export, nonsense-mediated mRNA decay, and translational regulation (Long and Caceres, 2009).
We show here that SF2/ASF, a prototypical SR protein splicing factor encoded by the SFRS1 gene (Ge and Manley, 1990; Krainer et al., 1990), is intimately involved in pri-miRNA processing. We found that SF2/ASF and miR-7 can form a negative feedback circuit: SF2/ASF promotes the maturation of miR-7, which negatively regulates SFRS1 expression at the translational level. We further show that SF2/ASF directly binds to pri-miR-7 and has a splicing-independent function to enhance Drosha cleavage. Lastly, SF2/ASF-enhanced miRNA production is not limited to miR-7, suggesting that SF2/ASF might be a key regulator that coordinates splicing regulation and miRNA-mediated gene repression.
RESULTS
Identification of SFRS1-miR-7 Feedback Circuit
SF2/ASF is a well-known splicing factor involved in both constitutive and alternative splicing. Given the functional importance of SF2/ASF in tissue- and cell-type-specific splicing, the expression level of its gene, SFRS1, has to be tightly regulated (Grosso et al., 2008; Karni et al., 2007). It is known that SF2/ASF can negatively regulate its own expression, and several mechanisms are involved at the posttranscriptional and translational steps. First of all, SF2/ASF regulates its own splicing to generate unproductive mRNA isoforms (Lareau et al., 2007; Ni et al., 2007; Sun et al., 2010). In addition, the negative feedback is mainly mediated by the 3′UTR of SFRS1, which is important for polysome association and translation of the mRNA (Sun et al., 2010).
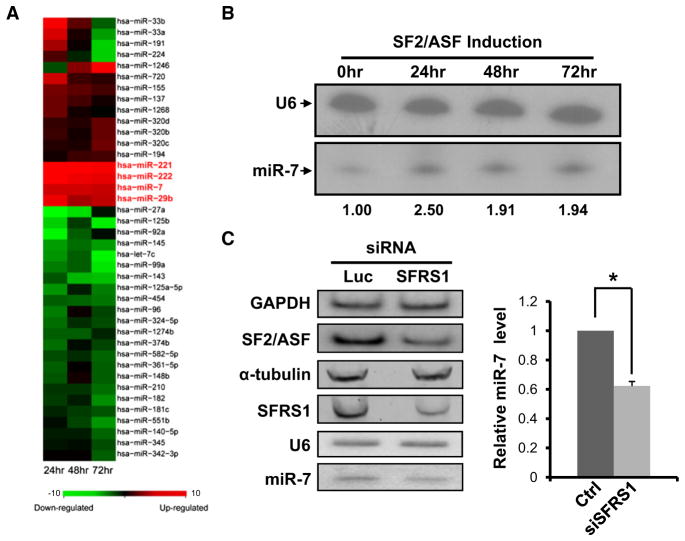
Since miRNAs are known to bind the 3′UTRs of their target genes to repress gene expression via translation inhibition and/or mRNA degradation (Bartel, 2009), we suspected that miRNAs might be involved in the SFRS1 negative feedback. One plausible mechanism is that SF2/ASF may increase the expression of a miRNA or multiple miRNAs, which in turn repress the translation of SFRS1 mRNA. To test this hypothesis, we established a stable HeLa cell line in which the expression of T7-tagged SF2/ASF cDNA is under the control of an inducible Tet-off promoter (Sun et al., 2010). While the overall SF2/ASF protein level was increased upon induction, the endogenous SF2/ASF was significantly reduced (see Figures S1A and S2B available online), which recapitulates the negative feedback regulation of SF2/ASF. To identify differentially expressed miRNAs, Illumina/Solexa sequencing was used to monitor miRNA expression profiles along the time course of SF2/ASF induction (0, 24, 48, and 72 hr). We obtained 1.37 million reads that could be perfectly mapped to 369 known human miRNAs (Figures S1C and S2D). Of these miRNAs, 40 were differentially expressed (>1.5-fold change; Fisher’s exact test, q < 0.01) under one or more SF2/ASF-induced conditions (Figure 1A). Based on TargetScan (Grimson et al., 2007; Lewis et al., 2005), 24 miRNAs could potentially target the 3′UTR of SFRS1. Among them, 22 showed detectable expression in at least one induction time point, and 18 miRNAs could be detected in all four time points. Whereas the majority of them were not differentially expressed, miR-7 clearly stood out in that it was consistently upregulated by ~2-fold across all SF2/ASF-induced conditions (Figure 1A and Figure S1E). Together, these data suggest that SF2/ASF and miR-7 potentially form a negative feedback circuit.
Figure 1. Differentially Expressed miRNAs upon SF2/ASF Induction.
(A) Normalized counts of individual miRNAs at 24, 48, or 72 hr after SF2/ASF induction were compared to that of cells without induction. Differentially expressed miRNAs (>1.5-fold change and q < 0.01) are shown, and z scores are plotted in the heat map.
(B) Northern blotting analysis of mature miR-7 in the stable HeLa cell line with inducible SF2/ASF expression. U6 snRNA was used as an internal control. The changes in relative miR-7 expression levels are shown at the bottom of the panel.
(C) HeLa cells were transfected with small interfering RNAs (siRNAs) against either luciferase or SFRS1. Two days after transfection, radiolabeled RT-PCR and western blotting were used to monitor the SFRS1 mRNA level (top two panels; GAPDH as an internal control) and protein level (middle two panels; α-tubulin as an internal control), respectively. Northern blotting analysis of endogenous miR-7 level with U6 as an internal control (bottom two panels). Results from triplicate experiments are plotted in the right panel. The * indicates p < 0.05 (t test, n = 3). Error bar represents SEM.
SF2/ASF Is Required for Efficient Production of miR-7
As a logical step to validate the SF2/ASF-miR-7 circuit, we first examined whether miR-7 expression can indeed be enhanced by elevated SF2/ASF levels. Northern blotting was performed with the inducible HeLa cell line (Figure 1B and data not shown). The results agreed well with the miRNA profiling data (~2-fold change). Three additional upregulated miRNAs (miR-29b, miR-221, and miR-222) were also validated (Figure S1F). Together, these data suggest that deep sequencing is a reliable method to detect differentially expressed miRNAs.
In addition, loss-of-function analysis was carried out by knocking down the expression of endogenous SFRS1 in HeLa cells. Western blotting and quantitative RT-PCR assays con-firmed that endogenous SFRS1 expression was reduced to approximately 30% of its normal level, resulting in a 40% decrease of the mature miR-7 level (p < 0.05, Figure 1C). Taken together, these results demonstrate that SF2/ASF is required for efficient production of mature miR-7 in HeLa cells.
SFRS1 Is a Physiological Target of miR-7
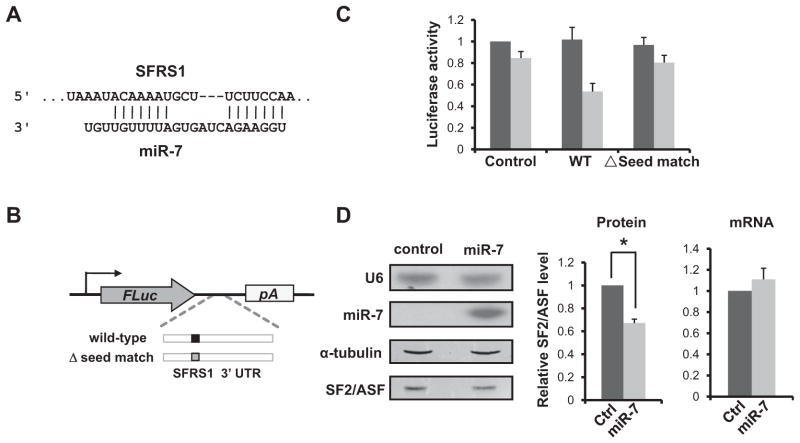
We next examined whether SFRS1 mRNA is a bona fide target of miR-7. Dual luciferase assay was initially used to confirm the target relationship. TargetScan predicts that the 3′UTR of SFRS1 has a highly conserved seed match (m6+u1A) for miR-7 (Figure 2A). We cloned the putative target site and its surrounding region into a Firefly luciferase reporter (Figure 2B). The resulting construct or the empty vector control was cotransfected into HeLa cells together with synthetic miR-7 precursors or scrambled RNAs. A Renilla luciferase construct was also included to normalize transfection efficiency. With the control plasmid, miR-7 had little effect on the relative luciferase activity, compared to the scrambled RNAs. In contrast, the normalized luciferase activity of the SFRS1 reporter was reduced to half by miR-7 (Figure 2C). The downregulation is dependent on the predicted miR-7 seed match, because the luciferase activity was restored when the site was deleted (Figure 2C).
Figure 2. SFRS1 Is a Physiological Target of miR-7.
(A) Base-pairing between mature miR-7 and its putative target site in the 3′UTR of SFRS1.
(B) Schematic diagram of the luciferase reporter constructs. The putative miR-7 target site is shown.
(C) Dual luciferase assays were performed in triplicate. For each construct, the relative luciferase activity was plotted by normalizing between cells transfected with control RNAs (dark gray) and miR-7 precursors (light gray).
(D) HeLa cells were transfected with either control RNAs or synthetic miR-7 precursors. Two days after transfection, the levels of mature miR-7 and endogenous SF2/ASF protein were quantified by northern (U6 as internal control; top two panels) and western blotting (α-tubulin as an internal control; bottom two panels), respectively. The relative levels of SF2/ASF protein from three separate transfections are plotted (t test, p < 0.05; middle panel). Quantitative RT-PCR results of endogenous SFRS1 mRNA in the transfected cells are shown in the right panel; GAPDH mRNA was used as an internal control. Error bars represent SEM.
We next transiently transfected HeLa cells with synthetic miR-7 precursors. Compared to control RNAs, ectopic miR-7 expression significantly reduced the level of endogenous SF2/ASF protein (p < 0.05) while the mRNA levels of SFRS1 remained constant (Figure 2D). We did not detect significant upregulation of SF2/ASF protein when endogenous miR-7 was blocked in HeLa cells (data not shown), possibly due to the relatively low level of miR-7 in these cells. As an alternative, we used HEK293 cells, in which the mature miR-7 level is considerably higher (Reddy et al., 2008). Knocking down endogenous miR-7 led to a reproducible increase of SF2/ASF protein level by over 30% (Figure S2). Taken together, these results argue that SFRS1 is a physiological target of miR-7, and the repression occurs at the translational level.
The reciprocal targeting between SF2/ASF and miR-7 confirmed that they can potentially form a negative feedback loop. SF2/ASF promotes the production of miR-7, which in turn targets SFRS1 mRNA to repress its translation. The circuit structure implies that miR-7 might in part contribute to the negative feedback regulation observed for SF2/ASF. Conversely, SF2/ASF may play a role in maintaining the steady-state level of mature miR-7, an important regulatory molecule involved in diverse cellular processes (see the Discussion).
The Domain Requirements of SF2/ASF for Promoting miR-7 Expression
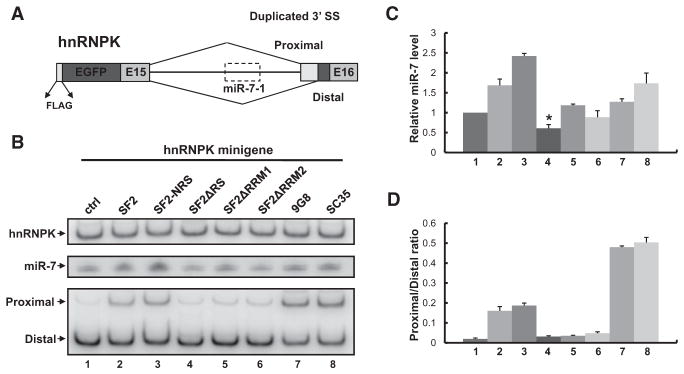
We next sought to characterize the molecular mechanism underlying SF2/ASF-enhanced miR-7 expression. Mature miR-7 is encoded by three distinct loci in the human genome. One of them, hsa-miR-7-1, is embedded in the last intron of the hnRNPK gene, which is alternatively spliced via two duplicated 3′ splice sites (3′ss). The resulting hnRNPK variants are identical in protein sequence, except for the last six amino acids (ADVEGF versus SGKFF). Interestingly, SF2/ASF is known to promote proximal splice-site usage for substrates with duplicated 5′ or 3′ss (Fu et al., 1992). In the case of hnRNPK, ectopic expression of SF2/ASF significantly increased the ratio between the proximal and distal variants, without affecting the overall mRNA level (data not shown). Because splicing efficiency could affect the processing of intronic miRNAs (Pawlicki and Steitz, 2008), one attractive model is that SF2/ASF might enhance miR-7 production through alternative splicing regulation.
To test this hypothesis, we constructed a minigene in which the miR-7-containing intron and its flanking exons were inserted downstream of the EGFP open reading frame (Figure 3A). The use of a minigene avoids potential complications due to multiple endogenous miR-7 loci and unforeseen transcriptional or post-transcriptional regulations. When transfected into HeLa cells, the hnRNPK minigene produced both mature miR-7 and the two expected splicing variants, of which the distal 3′ss was preferentially used (Figure S3; Figure 3B, lane 1). Cotransfection of SF2/ASF cDNA increased the level of mature miR-7 (1.7-fold) as well as the ratio between proximal and distal splicing variants (Figure 3B, lanes 1 and 2), indicating that the minigene can recapitulate the effects of SF2/ASF at the endogenous hnRNPK locus. Similar results were obtained for an SF2/ASF variant with a nuclear retention signal (SF2-NRS) (Cazalla et al., 2002), except that it was even more active in promoting miR-7 production (2.4-fold; Figures 3B and 3C, lanes 1–3). Moreover, the level of miR-7 precursors was also increased (Figure S3D), suggesting that enhanced miR-7 expression is likely to take place at the nuclear processing step (Drosha cleavage).
Figure 3. The Domain Requirement of SF2/ASF for Promoting miRNA Production and Alternative Splicing.
(A) Diagram of the hnRNPK minigene reporter. Proximal/distal splice sites and miR-7-1 precursor (dashed box) are shown. (B) The hnRNPK mini-gene was cotransfected with control vector (ctrl), wild-type/mutant SF2/ASF, SC35, or 9G8 cDNAs into HeLa cells. Cells were harvested 2 days after transfection. The overall level of hnRNPK (top panel) and its alternative splice-site usage (bottom panel) were monitored by radiolabeled RT-PCR. The mature miR-7 level was determined by northern blotting (middle panel). The relative miR-7 levels (normalized to total hnRNPK levels) and the alternative splicing ratios (proximal/distal) are plotted in (C) and (D), respectively (t test, p < 0.05; n = 3). Error bars represent SEM.
To further dissect the domain requirement of SF2/ASF for promoting miR-7 expression, three mutants (SF2ΔRS, SF2ΔRRM1, and SF2ΔRRM2) were examined (Caceres and Krainer, 1993). All mutants failed to promote proximal splice-site usage, indicating that both the RS domain and the RNA-recognition motifs are required for the splicing regulation. In contrast, SF2ΔRS reproducibly repressed miR-7 expression to two-thirds of its normal level (p < 0.05), whereas deletion of RRM1 or RRM2 rendered SF2/ASF inactive in promoting miR-7 production (Figure 3B, lane 1 and lanes 4–6; Figure 3C). The dominant-negative effect of SF2ΔRS suggests a previously uncharacterized function of SF2/ASF in miRNA biogenesis, which is separable from its activities in splicing regulation.
Two additional SR proteins, SFRS2 (SC35) and SFRS7 (9G8), were also tested. Despite the fact that both SR proteins showed much stronger activation of proximal splicing than SF2/ASF, their activities in miR-7 promotion were not correspondingly increased (Figure 3, lanes 7 and 8). Notably, 9G8 had little effect on miR-7 expression, suggesting that different SR proteins may have distinct substrate specificities in regulating miRNA expression. In agreement with the observations above, these results demonstrate that enhanced miR-7 expression is unlikely due to increased proximal splice-site usage, indicating that there are two separate functions of SF2/ASF, in alternative splicing and in miRNA processing.
A Splicing-Independent Function of SF2/ASF in Pri-miR-7 Processing
To uncouple SF2/ASF’s functions in alternative splicing and miRNA processing, we mutated the proximal or distal splice site of the hnRNPK minigene (PM or DM). When transfected into HeLa cells, the two mutants gave rise to opposite splicing patterns (Figure 4A, lanes 1, 3, and 5). Notably, we could detect both proximal and distal variants, due to background expression of the endogenous hnRNPK gene, which contributes to less than 5% of the total hnRNPK transcripts and can be ignored in miR-7 expression analysis (Figure 4A and data not shown). As expected, cotransfection of SF2/ASF no longer affected alternative splicing choice. In contrast, both precursor and mature miR-7 levels were significantly increased by SF2/ASF for the PM and DM constructs (Figure 4B and Figure S4A). The levels of miR-7 enhancement were slightly different between the DM and PM constructs (p = 0.076, Figure 4B), indicating possible context dependence (see the Discussion). We also made a construct for which both the proximal and distal splice sites were mutated; however, it led to several cryptic splicing variants (Figure S4B). As an alternative, we cloned the miR-7-1 precursor and its flanking regions into a heterologous context to eliminate all possible splicing (Figure 4C). Once again, the levels of both precursor and mature miR-7 were increased by SF2/ASF over-expression, and the enhancement level was comparable to that in the endogenous context (Figures 4C and 4D; Figure S4C). Together, these data clearly demonstrate that enhanced miR-7 expression is mediated by a splicing-independent function of SF2/ASF.
Figure 4. A Splicing-Independent Function of SF2/ASF in Promoting miR-7 Expression.
(A and B) (A) Wild-type hnRNPK minigene, the proximal 3′ splice-site mutant (PM) or the distal 3′ splice-site mutant (DM), was transfected into HeLa cells together with a control vector or a cDNA expressing SF2/ASF. Alternative splicing of hnRNPK and miR-7 expression were monitored by radiolabeled RT-PCR and northern blotting, respectively. The levels of mature miR-7 were normalized to total hnRNPK levels. Results from triplicate experiments were plotted in (B) (t test, p < 0.05; n = 3).
(C) Intronless minigene expressing miR-7 (pCG-miR-7) was transfected into HeLa cells together with a control vector or SF2/ASF cDNA. The levels of EGFP and miR-7 were monitored by radiolabeled RT-PCR and northern blotting, respectively. (D) The miR-7 levels were normalized to EGFP, and the results from triplicate experiments are plotted (t test, p < 0.05; n = 3). Error bars represent SEM.
SF2/ASF Directly Binds to Pri-miR-7 In Vivo
Two RNA-binding protein splicing factors (hnRNPA1 and KSRP) were recently shown to directly interact with primary miRNA transcripts and serve as auxiliary factors for more efficient Drosha cleavage (Michlewski et al., 2008; Trabucchi et al., 2009). A genome-wide survey found that SF2/ASF may bind in close proximity to the stem loop of three miRNAs in HEK293T cells, although the functional significance remains unclear (Sanford et al., 2009). We therefore examined whether SF2/ASF can directly bind to the primary miR-7 transcript to promote its cropping. To this end, a UV crosslinking and immunoprecipitation (CLIP) assay was employed, which can capture in vivo interactions between an RNA-binding protein and its cognate RNAs (Ule et al., 2003).
Since the activities of SF2/ASF in splicing and miRNA processing converge at the endogenous hnRNPK locus, the intronless miR-7-expressing minigene (Figure 4C) was used for CLIP analysis to avoid potential binding of SF2/ASF to neighboring exons. We transfected HeLa cells with a moderate level of the pCG-miR-7 construct. UV crosslinking was carried out 48 hr after transfection, followed by partial RNase digestion and immunoprecipitation with a monoclonal antibody against SF2/ASF (Hanamura et al., 1998). The antibody is highly specific for SF2/ASF with an excellent IP efficiency (Figures S5A and S5B). RNA fragments associated with SF2/ASF were then analyzed by radiolabeled RT-PCR against the EGFP and miR-7 regions. The primer pair specific for miR-7 is located in the flanking regions of its stem loop, such that the primary transcripts rather than the miR-7 precursors are probed. The EGFP region, on the other hand, serves as an internal control to estimate the enrichment of SF2/ASF binding.
As excepted, correct PCR products were detected only in the SF2/ASF CLIP, but not the mock immunoprecipitation (Figure S5C, lanes 3–6, and data not shown). We observed a 2-fold enrichment of the pri-miR-7-1 fragment by CLIP compared to the EGFP amplicon (Figure S5D). Although we cannot rule out that the EGFP region might also be bound by SF2/ASF, our data demonstrate that SF2/ASF preferentially binds to pri-miR-7-1 RNA. Using ESEfinder (Cartegni et al., 2003), we identified a putative SF2/ASF-binding site within the stem-loop region of miR-7-1 (Figure 5A). The motif was then mutated and the CLIP experiment was repeated to examine its effect on SF2/ASF binding. In contrast to the wild-type minigene, the mutant construct showed similar ratios between the EGFP and miR-7 amplicons before and after CLIP, suggesting that the binding of SF2/ASF to pri-miR-7 transcripts is mediated by the predicted binding site (Figure 5B, lanes 4 and 8; Figure 5C).
Figure 5. Direct Interaction between SF2/ASF and the Stem-Loop Region of miR-7-1.
(A) The stem-loop region of hsa-miR-7-1: the high-score SF2/ASF motif is shown in red (dashed box); mature miR-7 and miR-7* are shown in pink. The SF2/ASF motif was disrupted by mutations in four nucleotides (upper case, blue) with compensatory mutations in the opposite arm (upper case, green).
(B) Wild-type or mutant pCG-miR-7 was transiently transfected into HeLa cells. RNA samples obtained from input and SF2/ASF-specific CLIP were analyzed by radiolabeled RT-PCR. Two primer pairs to specifically amplify the EGFP and the miR-7 stem-loop regions are shown.
(C) The relative enrichments of the miR-7 stem-loop region before and after SF2/ASF CLIP are plotted for wild-type and mutant pCG-miR-7 transcripts (t test, p < 0.05; n = 3). Error bars represent SEM.
(D) In vitro-transcribed pri-miR-7-1 RNA or its mutant was coupled to agarose beads and incubated with HeLa extract. The bound proteins were analyzed by western blotting with α-tubulin as a negative control. Approximately 1/200 of the input extract was loaded as positive control (lanes 1 and 3). The relative SF2/ASF protein levels are shown at the bottom of the panel.
The CLIP results were further corroborated by an RNA affinity purification assay. The stem-loop regions of the corresponding miR-7-1 transcripts were in vitro transcribed and covalently coupled to agarose beads, followed by incubation with HeLa cell extract. Whereas SF2/ASF protein could be pulled down by both RNAs, the pull-down efficiency was markedly reduced when the mutant RNA was used (Figure 5D). Extending the CLIP data, these results confirmed that the interaction between SF2/ASF and miR-7 is mediated at least in part by the SF2/ASF-binding site within the miR-7 stem loop.
SF2/ASF Is Involved in the Drosha Cleavage Step of miR-7 Maturation
We next examined the effect of SF2/ASF binding on enhanced miR-7 production. When the wild-type or mutant pCG-miR-7 minigene was transfected into HeLa cells, the basal levels of miR-7 expression were comparable. This is possibly due to the mutations (G-C to A-T base-pairing) which might relax the local secondary structure and partially compensate for the loss of the SF2/ASF-binding site. However, the two constructs differed substantially under SF2/ASF overexpression conditions: SF2/ASF-enhanced miR-7 expression was significantly reduced with the mutant construct (p = 0.017; Figure 6A), suggesting that the binding site is required for SF2/ASF-enhanced miR-7 production.
Figure 6. Sequence-Dependent Promotion of miR-7 Maturation by SF2/ASF.
(A) Wild-type or mutant miR-7 construct was cotransfected into HeLa cells with a control vector or SF2/ASF-expressing plasmid. Transfection efficiencies were normalized to the EGFP levels (radiolabeled RT-PCR). Precursor and mature miR-7 levels were monitored by northern blotting. The relative activities of SF2/ASF in promoting miR-7 expression are shown in the right panel (t test, p < 0.05; n = 3). Error bars represent SEM. (B) HeLa cells were transfected with either luciferase or SFRS1-specific siRNA. Two days after transfection, cells were harvested to prepare whole-cell extracts, for which the levels of SF2/ASF protein were analyzed by western blotting. In vitro pri-miRNA processing assay was performed with wild-type or mutant pri-miR-7-1 substrates (Michlewski et al., 2008); the results are shown in (C) and (D), respectively.
Notably, the levels of both precursor and mature miR-7 were upregulated by SF2/ASF in vivo (Figure 6A), indicating that the Drosha cleavage step is likely to be involved. This is further supported by the observation that SF2-NRS is more potent than wild-type SF2/ASF for promoting miR-7 expression (Figure 3B). As a more direct approach, an in vitro pri-miRNA processing assay was performed, which monitors the conversion of pri-miRNAs to pre-miRNAs (Guil and Caceres, 2007). Whole-cell extracts were prepared from HeLa cells with or without SF2/ASF knockdown (Figure 6B). Consistent with the in vivo data, the processing of wild-type pri-miR-7 was significantly reduced when SF2/ASF-knockdown cell extract was used (Figure 6C, lanes 2 and 3). Adding back purified SF2/ASF protein restored the processing efficiency in a dose-dependent manner (Figure 6C, lanes 4–6). These results strongly argue that SF2/ASF indeed functions at the Drosha cleavage step. Furthermore, depletion and add-back of SF2/ASF had little effect on the miR-7 substrate with a mutated SF2/ASF-binding site (Figure 6D). These data agreed with the in vivo data showing that the level of pre-miR-7 was not significantly changed by cotransfection of the mutant pri-miR-7 construct with an SF2/ASF expression vector. Overall, our data clearly demonstrate that SF2/ASF promotes the Drosha cleavage step of pri-miR-7 processing, although additional effects on postcropping steps cannot be ruled out.
SF2/ASF Is Broadly Involved in miRNA Processing
In addition to miR-7, expression profiling identified three additional miRNAs (miR-29b, miR-221, and miR-222) whose expressions were consistently upregulated upon SF2/ASF induction (Figure 1A). The results were further validated by northern blotting using the samples obtained from the same stable cell line (Figure S1F). For each miRNA, a minigene reporter was constructed using the intronless vector (analogous to pCG-miR-7) and cotransfected into HeLa cells with SF2/ASF cDNAs or a control vector (Figure 7A). Similar to the miR-7 case, both the precursor and the mature miRNAs were increased upon SF2/ASF overexpression for miR-221 and miR-222, suggesting that the Drosha cleavage step is involved. Interestingly, SF2/ASF overexpression increased the level of mature miR-29b but not the miRNA precursor, implying that SF2/ASF might act at a post-Drosha-cleavage step (e.g., Dicer cleavage). The enhancement could only be observed for miR-29b-1, but not miR-29b-2. The latter construct serves as a negative control for substrate specificity. Taken together, these results suggest that SF2/ASF is broadly involved in miRNA biogenesis and that its function may not be limited to the Drosha cleavage step.
Figure 7. SF2/ASF Is Broadly Involved in miRNA Biogenesis.
(A) HeLa cells were transfected with intronless miR-29b-1/2, miR-221, or miR-222 constructs, together with an empty or SF2/ASF expression vector. The levels of EGFP and miR-7 were monitored by radiolabeled RT-PCR and northern blotting, respectively.
(B) Schematic diagram of the SFRS1/miR-7 negative feedback loop. SF2/ASF directly binds to pri-miR-7 to promote its nuclear cropping, although effects on later steps (pre-miRNA export and/or Dicer cleavage; dashed lines) cannot be ruled out. Additional factors (circle with a question mark) may also be involved in efficient pri-miRNA maturation, possibly through the interaction with the RS domain of SF2/ASF. In the cytoplasm, mature miR-7 (yellow line) binds to the 3′UTR of the SFRS1 mRNA (rectangle box) and represses the production of SF2/ASF protein via translational inhibition.
DISCUSSION
We show here that SF2/ASF and miR-7 can form a negative feedback circuit. SF2/ASF directly binds to the primary miR-7 transcript to promote its maturation; mature miR-7 in turn represses the translation of SFRS1 mRNA by targeting its 3′UTR (Figure 7B). Notably, negative feedback does not always lead to a stable steady state, as overcorrection and/or time delay could result in oscillation (Elowitz and Leibler, 2000). Because gene repression by miRNA is relatively modest, with little time delay, miRNA-mediated negative feedback loops are advantageous for noise dampening and have been shown as a recurrent circuit motif in mammalian gene regulatory networks (Tsang et al., 2007).
Negative feedback is also a common mechanism to maintain the steady-state levels of SR proteins (Jumaa and Nielsen, 1997; Sureau et al., 2001). In the case of SF2/ASF, autofeedback has been proposed to occur at multiple levels, including unproductive alternative splicing (Lareau et al., 2007; Ni et al., 2007) and inhibition of translation initiation (Sun et al., 2010). Thus, the miR-7-mediated negative feedback loop is expected to synergize with other feedback mechanisms to precisely control the protein level of SF2/ASF in cells. The relative contribution of each mechanism may vary under different conditions, such as in different cell types or physiological states. This may explain the different phenotypes we observed when knocking down endogenous miR-7 in HEK293T and HeLa cells. Because the regulation observed in the SFRS1/miR-7 circuit is relatively modest (2-fold effects in both directions), miR-7-mediated negative feedback does not appear to be a major contributor to the robust feedback regulation observed for SF2/ASF. Interestingly, Dicer null embryonic stem cells (ESCs) showed compromised but detectable feedback of SF2/ASF on its own expression (Sun et al., 2010), consistent with the notion that both miRNA-dependent and -independent mechanisms are involved.
Given the reciprocal nature of the SFRS1/miR-7 circuit, an alternative possibility is that SF2/ASF may be critical for the homeostasis of miR-7, an important regulatory molecule orchestrating diverse cellular functions (Li et al., 2009). In this scenario, SF2/ASF serves as a “rheostat” to sense the cellular level of mature miR-7. Fluctuations in miR-7 level are expected to drive the expression of endogenous SF2/ASF in an opposite direction. Because SF2/ASF is required for efficient pri-miR-7 maturation, the negative feedback loop in effect buffers the noise in miR-7 expression to better maintain its steady state.
Besides the SFRS1/miR-7 feedback loop, a circuit with the same architecture has been reported between the transcriptional factor E2F and the miR-17-92 cluster (Woods et al., 2007). Mathematical modeling showed that miR-17-92 is essential for the E2F/Myc cancer network to balance between cell proliferation and apoptosis (Aguda et al., 2008). Therefore, it will be of interest to further investigate the SFRS1/miR-7 feedback loop from a network perspective in order to fully understand its functional significance.
One key finding of this study is a splicing-independent function of SF2/ASF in pri-miRNA processing. We provided multiple lines of evidence that SF2/ASF promotes miR-7 maturation at the Drosha cleavage step. HnRNPA1, another well-known alternative splicing factor, serves as an auxiliary factor in pri-miR-18a processing (Guil and Caceres, 2007). In fact, binding of hnRNPA1 to pri-miR-18a introduces a conformational change in its terminal loop to allow more efficient Drosha cleavage (Michlewski et al., 2008). It is plausible that SF2/ASF may function in a similar manner in promoting pri-miR-7 maturation. In addition, we observed a dominant-negative effect of SF2ΔRS on miR-7 production. Since the RS domain of SR proteins often mediates protein-protein interactions, this result suggests that an additional factor(s) might also be involved to enhance pri-miRNA maturation.
Both alternative splicing and miRNA processing coincide in the last intron of the hnRNPK gene, which provides a unique opportunity to examine the potential cooperation and/or competition between the spliceosome and microprocessor (Drosha/DGCR8 complex). Our results showed that SF2/ASF promotes proximal 3′ splice-site usage of the miR-7-containing intron. However, the ability of SF2/ASF to promote miR-7 expression is slightly reduced when the proximal 3′ss is used (Figures 4A and 4B), suggesting a context dependence. It has been shown that processing of intronic miRNAs takes place before intron removal (Kim and Kim, 2007). Therefore, our data imply that spliceosome assembly at the nearby splice sites may affect the processing of intronic miRNAs. One attractive model is that the spliceosome might compete with the miRNA processing machinery for common auxiliary factors (e.g., SF2/ASF). Alternatively, intronic miRNAs might adopt different local conformations depending on alternative splice-site usage. We therefore propose that the functions of SF2/ASF in mRNA and miRNA processing might not be mutually exclusive; instead they might modulate each other in a context-dependent manner.
The functional involvement of SF2/ASF in pri-miRNA processing is not limited to miR-7. We acquired initial evidence that SF2/ASF is also involved in the maturation of miR-221, miR-222, and miR-29b-1 (Figure 7A). While enhanced Drosha cleavage is likely to be involved in the case of miR-221 and miR-222, SF2/ASF might promote the expression of miR-29b-1 (but not miR-29b-2) at a postcropping step (e.g., pre-miRNA export and/or Dicer cleavage). Conversely, our profiling results showed that the expression of a subset of miRNAs can also be repressed by SF2/ASF. It will be interesting to find out whether SF2/ASF is directly involved and can play a negative role in pri-miRNA processing in a substrate-specific manner. Supporting these notions, SF2/ASF is a shuttling protein that plays diverse roles in both the nucleus and cytoplasm (Caceres et al., 1998; Sanford et al., 2004). Furthermore, there is a precedent that KSRP, a well-known factor involved in alternative splicing and mRNA degradation, regulates the biogenesis of a subset of miRNAs at multiple distinct steps (Trabucchi et al., 2009). Lastly, RNA-binding proteins other than SF2/ASF (e.g., hnRNP A1 or KSRP) also participate in miRNA processing. It will be important to determine the substrate specificities of, as well as the potential cooperation/competition between, different RNA-binding proteins in controlling tissue- and cell type-specific miRNA expression.
Both splicing factors and miRNAs regulate the expression of a large number of protein-coding genes. Therefore, they may share common downstream targets and/or signaling pathways. Such a “wiring” structure has been reported between transcriptional factors and their regulated miRNAs and is a recurring motif in transcriptional gene networks (Lee et al., 2007; Shalgi et al., 2007). One well-known example is the miR-34 family of miRNAs, which are direct transcriptional targets of p53 and act in concert with other p53 downstream effectors to inhibit inappropriate cell proliferation (Chang et al., 2007; He et al., 2007). Notably, SF2/ASF can also act as an oncoprotein by activating the mTOR pathway (Karni et al., 2007). In addition, several SF2/ASF-upregulated miRNAs (e.g., miR-221 and miR-222) have been implicated in tumorigenesis (Sun et al., 2009; Terasawa et al., 2009). It is possible that these miRNAs may contribute to SF2/ASF-driven tumorigenesis. One attractive scenario is that splicing regulation and miRNA-mediated gene repression may be broadly coordinated in posttranscriptional gene regulatory networks, a possibility that deserves systematic characterization.
EXPERIMENTAL PROCEDURES
MicroRNA Deep Sequencing
Cells were harvested at the indicated time points after SF2/ASF induction. Total RNAs were isolated using mirVana miRNA isolation kit (Ambion). miRNA sequencing libraries were constructed as described (Lau et al., 2001) with several minor modifications. Detailed sections covering the library construction and data analysis are presented in the Supplemental Data.
Plasmids
T7-tagged SF2/ASF, SC35, and 9G8 were cloned into the pCGT7 vector (Caceres et al., 1997). To construct the hnRNPK minigene reporter, an EGFP cDNA fragment was first cloned into the pTag2A vector (Stratagene). A genomic hnRNPK fragment, which corresponds to the last intron and its neighboring exons, was amplified and cloned in-frame with the EGFP gene with the primers 5′-CGTCATGAGTCGGGAGCTTC-3′ and 5′-GCAGGACTCCT TCAGTTCTTCA-3′. For the pCG-miR-7 construct, we first cloned EGFP into the pcDNA3.1+ vector (Invitrogen). MiR-7-1 precursor sequence was then cloned downstream of the EGFP gene. All clones were verified by sequencing.
Cell Culture and Transient Transfection
HeLa and HEK293T cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS). To generate the stable cell line, HeLa Tet-off cells were transfected with STP retroviral vectors containing a human SF2/ASF cDNA; stable transductants were selected with puromycin (2 μg/ml). Transient transfection was performed with Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions.
Luciferase Assay
HEK293T cells were grown to ~50% confluence in 24-well plates. For each transfection, two luciferase reporters, pRL-SV40 (Renilla luciferase, Promega) and pcDNA-Luc with or without the 3′UTR region of SFRS1 (Firefly luciferase, see above), were mixed at a 1:2.5 molar ratio. Synthetic miR-7 precursors or control RNAs (Applied Biosystems) were cotransfected at a final concentration of 25 nM. Firefly and Renilla luciferase activities were analyzed by a Dual-Glo luciferase assay system (Promega).
Crosslinking and Immunoprecipitation
CLIP analysis of SF2/ASF was performed as described (Ule et al., 2003) with a few minor modifications. Briefly, HeLa cells were cultured in 10 cm dishes; UV crosslinking was carried out at 50 mJ/cm2. Crosslinked cells were collected and lysed in RIPA buffer. After DNase treatment, the cell lysate was treated with RNase A (Promega) for 10 min at a final dilution of 1:1,000,000. The reaction was stopped by adding 200 U RNase inhibitor (Invitrogen). Immunoprecipitation was carried out at 4°C for 2 hr with protein A/G PLUS-agarose beads (Santa Cruz) coupled with SF2/ASF monoclonal antibody AK96. After extensive washing, SF2/ASF-bound RNAs were released by Proteinase K treatment, followed by phenol extraction and ethanol precipitation. The resulting RNAs were treated with DNase I and reverse transcribed with Superscript II and random hexamers. Quantitative PCR was then performed with radiolabeled primer pairs specific for EGFP and the miR-7 stem loop.
Northern Blotting
Total RNAs were resolved on a 15% polyacrylamide TBE-Urea gel and blotted onto a Hybond-N+ membrane (Amersham). An LNA probe (5′-A+CAA+ CAA+A AT+CACTA+GTCTT+CCA-3′; +N stands for LNA base), which is anti-sense to mature miR-7, was labeled with 32P using T4 polynucleotide kinase (Invitrogen). After hybridization, the resulting membrane was exposed to a phosphorimaging screen (Amersham) for 1 hr to detect ectopically expressed miR-7, or overnight to visualize the endogenous miR-7. The signal was analyzed by ImageQuant (Amersham). For the hnRNPK/EGFP minigene experiments, the levels of minigene mRNA and mature miR-7 were first normalized to that of GAPDH mRNA and U6 snRNA, respectively. To normalize the transfection efficiency, gels were reloaded by keeping the minigene expression relatively constant, such that the differences in miR-7 levels can be better visualized.
Semiquantitative PCR
Fifteen to twenty cycles of radiolabeled PCR were carried out to ensure that the amplifications were in the linear range. The resulting PCR products were resolved by 8% TBE-PAGE gel and detected with a Storm 840 PhosphorImager. All primers used are listed in the Supplemental Data.
Western Blotting
Proteins were separated on 8% SDS-PAGE gels and blotted with monoclonal antibody against β-catenin (Sigma), α-tubulin (DM1A; Upstate), T7 epitope tag (Novagen), or SF2/ASF (AK96; Hanamura et al., 1998). The signal was detected with Alexa Fluor 488 goat anti-mouse IgG (H+L) antibody (Invitrogen) and quantified with a Storm 840 PhosphorImager.
RNA Affinity Purification
DNA templates for in vitro transcription were prepared by PCR from the pCG-miR-7 and its SF2/ASF-binding site mutant with the primers 5′-TAATACGACT CACTATAGGGTAGAAGATTCATTGGATGTTGG-3′ and 5′-TTGTCCTGTAGA GGCATG-3′. In vitro transcription was carried out with T7 RNA polymerase (NEB) following the manufacturer’s protocol. After gel purification, the resulting RNAs were coupled to agarose beads (Sigma), and affinity purification of miRNA binding factors from HeLa extract was performed as described (Caputi et al., 1999). Protein factors associated with the immobilized RNAs were analyzed by western blotting.
RNA Interference
Dicer substrate small interfering RNA (DsiRNA) against SFRS1 (forward 5′-CCAAGGACAUUGAGGACGUGUUCUA-3′; reverse 5′-UAGAACACGUCC UCAAUGUCCUUGGUU-3′) was custom designed and synthesized (IDT). 106 HeLa cells were transfected with 100 pmol DsiRNA duplex using TriFECTin transfection reagent (IDT). Cells were harvested 48 hr after transfection. The knockdown efficiency was determined by RT-PCR and western blotting.
In Vitro Pri-miRNA Processing Assay
In vitro processing was performed as described (Michlewski et al., 2008). Briefly, each reaction (30 μl) contained 20% (v/v) wild-type or depleted HeLa cell extract, 0.5 mM ATP, 20 mM creatine phosphate (Sigma), 3.2 mM MgCl2, and 200,000 cpm (~100 fmol) of in vitro-transcribed pri-miRNAs. The reactions were assembled on ice followed by incubation at 30°C for 30 min. After phenol/chloroform extraction and ethanol precipitation, the RNA samples were resolved by 10% (w/v) TBE-Urea gel electrophoresis and exposed overnight on X-ray film at −80°C.
Supplementary Material
Acknowledgments
We thank Drs. Sandeep Dave and Mariano Garcia-Blanco (Duke University Medical Center) for comments and critical reading of the manuscript. We are grateful to Drs. Uwe Ohler and Zhong Wang (Duke Institute for Genome Sciences and Policy) for valuable advice on computational analysis, and Mr. Matt Gemberling for constructing the mutant minigene reporters. This work was supported by the National Science Foundation (MCB 0822033 to J.Z.).
Footnotes
Supplemental Information includes five figures and Supplemental Experimental Procedures and can be found with this article at doi:10.1016/j.molcel.2010.02.021.
References
- Aguda BD, Kim Y, Piper-Hunter MG, Friedman A, Marsh CB. MicroRNA regulation of a cancer network: consequences of the feedback loops involving miR-17-92, E2F, and Myc. Proc Natl Acad Sci USA. 2008;105:19678–19683. doi: 10.1073/pnas.0811166106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres JF, Krainer AR. Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO J. 1993;12:4715–4726. doi: 10.1002/j.1460-2075.1993.tb06160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres JF, Misteli T, Screaton GR, Spector DL, Krainer AR. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J Cell Biol. 1997;138:225–238. doi: 10.1083/jcb.138.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres JF, Screaton GR, Krainer AR. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 1998;12:55–66. doi: 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputi M, Mayeda A, Krainer AR, Zahler AM. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 1999;18:4060–4067. doi: 10.1093/emboj/18.14.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESE-finder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalla D, Zhu J, Manche L, Huber E, Krainer AR, Caceres JF. Nuclear export and retention signals in the RS domain of SR proteins. Mol Cell Biol. 2002;22:6871–6882. doi: 10.1128/MCB.22.19.6871-6882.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Fu XD, Mayeda A, Maniatis T, Krainer AR. General splicing factors SF2 and SC35 have equivalent activities in vitro, and both affect alternative 5′ and 3′ splice site selection. Proc Natl Acad Sci USA. 1992;89:11224–11228. doi: 10.1073/pnas.89.23.11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H, Manley JL. A protein factor, ASF, controls cell-specific alternative splicing of SV40 early pre-mRNA in vitro. Cell. 1990;62:25–34. doi: 10.1016/0092-8674(90)90236-8. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosso AR, Gomes AQ, Barbosa-Morais NL, Caldeira S, Thorne NP, Grech G, von Lindern M, Carmo-Fonseca M. Tissue-specific splicing factor gene expression signatures. Nucleic Acids Res. 2008;36:4823–4832. doi: 10.1093/nar/gkn463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guil S, Caceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- Hanamura A, Caceres JF, Mayeda A, Franza BR, Jr, Krainer AR. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA. 1998;4:430–444. [PMC free article] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumaa H, Nielsen PJ. The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. EMBO J. 1997;16:5077–5085. doi: 10.1093/emboj/16.16.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol. 2007;14:185–193. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krainer AR, Conway GC, Kozak D. Purification and characterization of pre-mRNA splicing factor SF2 from HeLa cells. Genes Dev. 1990;4:1158–1171. doi: 10.1101/gad.4.7.1158. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultra-conserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Li Z, Brower-Sinning R, John B. Regulatory circuit of human microRNA biogenesis. PLoS Comput Biol. 2007;3:e67. doi: 10.1371/journal.pcbi.0030067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW. A microRNA imparts robustness against environmental fluctuation during development. Cell. 2009;137:273–282. doi: 10.1016/j.cell.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- Michlewski G, Guil S, Semple CA, Caceres JF. Posttranscriptional regulation of miRNAs harboring conserved terminal loops. Mol Cell. 2008;32:383–393. doi: 10.1016/j.molcel.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlando M, Ballarino M, Gromak N, Pagano F, Bozzoni I, Proudfoot NJ. Primary microRNA transcripts are processed co-transcriptionally. Nat Struct Mol Biol. 2008;15:902–909. doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O’Brien G, Shiue L, Clark TA, Blume JE, Ares M., Jr Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlicki JM, Steitz JA. Primary microRNA transcript retention at sites of transcription leads to enhanced microRNA production. J Cell Biol. 2008;182:61–76. doi: 10.1083/jcb.200803111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy SD, Ohshiro K, Rayala SK, Kumar R. MicroRNA-7, a homeobox D10 target, inhibits p21-activated kinase 1 and regulates its functions. Cancer Res. 2008;68:8195–8200. doi: 10.1158/0008-5472.CAN-08-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JR, Gray NK, Beckmann K, Caceres JF. A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 2004;18:755–768. doi: 10.1101/gad.286404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JR, Wang X, Mort M, Vanduyn N, Cooper DN, Mooney SD, Edenberg HJ, Liu Y. Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome Res. 2009;19:381–394. doi: 10.1101/gr.082503.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalgi R, Lieber D, Oren M, Pilpel Y. Global and local architecture of the mammalian microRNA-transcription factor regulatory network. PLoS Comput Biol. 2007;3:e131. doi: 10.1371/journal.pcbi.0030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Wang Q, Balk S, Brown M, Lee GS, Kantoff P. The role of microRNA-221 and microRNA-222 in androgen-independent prostate cancer cell lines. Cancer Res. 2009;69:3356–3363. doi: 10.1158/0008-5472.CAN-08-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Zhang Z, Sinha R, Karni R, Krainer AR. SF2/ASF autoregulation involves multiple layers of post-transcriptional and translational control. Nat Struct Mol Biol. 2010;17:306–312. doi: 10.1038/nsmb.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureau A, Gattoni R, Dooghe Y, Stevenin J, Soret J. SC35 its expression by promoting splicing events that destabilize its mRNAs. EMBO J. 2001;20:1785–1796. doi: 10.1093/emboj/20.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasawa K, Ichimura A, Sato F, Shimizu K, Tsujimoto G. Sustained activation of ERK1/2 by NGF induces microRNA-221 and 222 in PC12 cells. FEBS J. 2009;276:3269–3276. doi: 10.1111/j.1742-4658.2009.07041.x. [DOI] [PubMed] [Google Scholar]
- Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem. 2007;282:2130–2134. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.