Abstract
Prostate cancer (PrCa) is the most frequently diagnosed male cancer in developed countries. To identify common PrCa susceptibility alleles, we conducted a multi-stage genome-wide association study and previously reported the results of the first two stages, which identified 16 novel susceptibility loci for PrCa. Here we report the results of stage 3 in which we evaluated 1,536 SNPs in 4,574 cases and 4,164 controls. Ten novel association signals were followed up through genotyping in 51,311 samples in 30 studies through the international PRACTICAL consortium. In addition to previously reported loci, we identified a further seven new prostate cancer susceptibility loci on chromosomes 2p, 3q, 5p, 6p, 12q and Xq (P=4.0 ×10−8 to P=2.7 ×10−24). We also identified a SNP in TERT more strongly associated with PrCa than that previously reported. More than 40 PrCa susceptibility loci, explaining ~25% of the familial risk in this disease, have now been identified.
Genome-wide association studies (GWAS) provide a powerful approach to identify common disease alleles. We previously conducted a GWAS based on genotyping of 541,129 SNPs in 1,854 clinically detected PrCa cases and 1,894 controls (stage 1)1. In a second stage, 43,671 SNPs showing evidence for association in stage 1 were genotyped in 3,650 PrCa cases and 3,940 controls (stage 2). These studies, together with further follow-up through the PRACTICAL consortium identified 16 PrCa susceptibility loci1–3. Taken together with loci identified through other GWAS, more than 30 PrCa susceptibility loci have been reported4, and these account for approximately 23% of the familial risk of the disease.
Since the risks associated with common susceptibility alleles are modest (per-allele odds ratios, OR, ranging from 1.10–1.25), it is likely that other PrCa predisposition loci will have been missed by previous studies, and that such loci should be detectable by studies with larger sample sizes5. We therefore conducted a more extensive follow-up of SNPs showing evidence of association in our GWAS. We first used imputation, utilising the HapMap phase II CEU data as a reference, to estimate genotypes in our stage 1 and 2 data at ~2.6M SNPs. To improve power, we then combined these data with imputed data from the Cancer Genetic Markers of Susceptibility (CGEMS;) PrCa study, a GWAS of 1,117 PrCa cases and 1,105 controls, using a stratified 1df P-trend (see statistical methods).
We used these combined results to select SNPs for a stage 3 analysis. We selected 1,263 SNPs based on their ranked P-values, together with 178 additional SNPs selected for fine-mapping of three regions, of known susceptibility loci or from candidate gene studies (see methods). These SNPs were genotyped using an Illumina Golden Gate assay in 4,999 PrCa cases and 4,939 controls from the United Kingdom (UK) and Australia (see Methods and Supplementary Notes. After quality control (QC) exclusions (see Methods), data were utilised from 1,347 SNPs in 4,574 PrCa cases and 4,164 controls (Supplementary Figure 1). Genotype frequencies in cases and controls were compared using a 1 degree of freedom (df) Cochran-Armitage trend test, stratified by study.
After exclusion of SNPs in previously known susceptibility regions, there was a clear excess of nominally significant associations in stage 3, with 11 SNPs significant at P<0.001 (in 7 regions on chromosomes 2, 5, 6 and 12) compared with 1.1 that would be expected by chance. We then combined data from the stage 3 with those from the previous stages and with the CGEMS data. In the combined dataset, we identified 16 SNPs from 10 regions in which the highest ranking SNP reached a significance of P<10−7 in the combined analysis or P<10−3 in stage 3. Multiple logistic regression analysis indicated that a single SNP was sufficient to explain the association signal at each region, in that no further SNPs were significantly associated with risk after adjustment for the most strongly associated SNP.
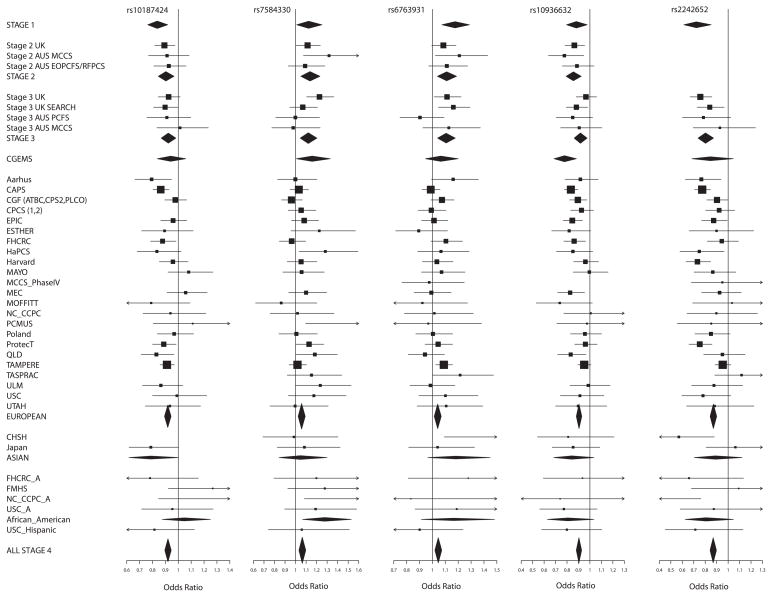
These 10 SNPs were then subjected to further replication analysis (stage 4) in an international study, involving 26,055 cases and 25,256 controls from 30 studies participating in the PRACTICAL Consortium (Supplementary Figure 1 and Supplementary Notes). Nine SNPs showed evidence of replication in stage 4 (P≤0.007) each in the same direction as in stages 1–3, with a significance of P=4.0 ×10−8 to P= 2.7 ×10−24 in a combined analysis across all stages (Table 1., Fig 1. and Supplementary Figure 1). rs37181 on 5q22 showed no evidence of association in stage 4 and did not reach “genome-wide” significance in an overall combined analysis (P=.17 in stage 4, P=5.3×10−5 overall). Of the nine loci reaching genome-wide significance, SNP rs7584330 is in the same region as a locus recently reported in a parallel GWAS of PrCa (Schumacher et al, Hum Mol Genet. 2011, in press). The second stage of that study also included data from stages 1 and 2 of our GWAS. The associated SNP in that study, rs2292884, is in LD with rs7584330 (r2=0.59) reported here and is likely to reflect the same signal, although rs7584330 exhibited a stronger association in our GWAS (Supplementary Figure 2. a).
Table 1.
Summary results for the 10 SNPs selected for genotyping in stage 4.
| Marker (Comm./Rare) | Chr | Position | Stage | AF1 | R2* | Per allele OR (95%CI) | P-value | |
|---|---|---|---|---|---|---|---|---|
| Stage | Combined | |||||||
| rs10187424 (A/G) | 2p11 | 85647807 | Stage 1 | .40 | 1.00 | .84 (.76–.92) | 1.7×10−4 | 3.1×10−15 |
| Stage 2 | .39 | .99 | .90 (.85–.97) | .003 | ||||
| Stage 3 | .41 | NA | .92 (.87–.98) | .01 | ||||
| CGEMS | .42 | .99 | .94 (.84–1.06) | .31 | ||||
| Stage 4 | .41 | NA | .92 (.89–.94) | 2.1×10−9 | ||||
| rs7584330 (T/C) | 2q37 | 238051966 | Stage 1 | .25 | .98 | 1.12 (1.01–1.25) | .03 | 3.2×10−9 |
| Stage 2 | .27 | .90 | 1.14 (1.05–1.23) | .002 | ||||
| Stage 3 | .26 | NA | 1.12 (1.04–1.20) | .002 | ||||
| CGEMS | .22 | .98 | 1.16 (1.01–1.33) | .04 | ||||
| Stage 4 | .22 | NA | 1.06 (1.02–1.09) | .001 | ||||
| rs6763931 (C/T) | 3q23 | 142585522 | Stage 1 | .495 | 1.00 | 1.18 (1.07–1.29) | .001 | 2..0×10−8 |
| Stage 2 | .46 | 1.00 | 1.11 (1.04–1.19) | .002 | ||||
| Stage 3 | .45 | NA | 1.11 (1.04–1.18) | .001 | ||||
| CGEMS | .44 | 1.00 | 1.07 (.95–1.20) | .30 | ||||
| Stage 4 | .45 | NA | 1.04 (1.01–1.07) | .003 | ||||
| rs10936632 (A/C) | 3q26 | 171612795 | Stage 1 | .48 | .77 | .88 (.80–.98) | .015 | 6.6×10−22 |
| Stage 2 | .47 | .72 | .86 (.79–.92) | 6.9×10−5 | ||||
| Stage 3 | .47 | NA | .92 (.86–.98) | .006 | ||||
| CGEMS | .47 | .85 | .78 (.69–.88) | 8.9×10−5 | ||||
| Stage 4 | .48 | NA | .90 (.88–.93) | 1.0×10−13 | ||||
| rs2242652 (G/A) | 5p15 | 1333027 | Stage 1 | .13 | .60 | .72 (.61–.85) | 1.1×10−4 | 2.7×10−24 |
| Stage 2 | .15 | .27 | ||||||
| Stage 3 | .17 | NA | .80 (.74–.87) | 3.2×10−7 | ||||
| CGEMS | .14 | .60 | .85 (.68–1.05) | .12 | ||||
| Stage 4 | .19 | NA | .87 (.84–.90) | 2.2×10−16 | ||||
| rs2121875 (T/G) | 5p12 | 44401301 | Stage 1 | .34 | 1.00 | 1.09 (.99–1.21) | .07 | 4.0×10−8 |
| Stage 2 | .35 | .37 | 1.13 (1.01–1.27) | .04 | ||||
| Stage 3 | .34 | NA | 1.11 (1.04–1.18) | .001 | ||||
| CGEMS | .34 | 1.00 | 1.15 (1.02–1.30) | .03 | ||||
| Stage 4 | .34 | NA | 1.05 (1.02–1.08) | 4.0×10−4 | ||||
| rs37181 (A/C) | 5q22 | 115657902 | Stage 1 | .24 | 1.00 | 1.16 (1.04–1.30) | .007 | 5.3×10−5 |
| Stage 2 | .24 | 1.00 | 1.11 (1.03–1.20) | .008 | ||||
| Stage 3 | .23 | NA | 1.14 (1.06–1.23) | 3.3×10−4 | ||||
| CGEMS | .25 | 1.00 | 1.06 (.92–1.21) | .45 | ||||
| Stage 4 | .23 | NA | 1.02 (.99–1.05) | .17 | ||||
| rs130067 (T/G) | 6p21 | 31226489 | Stage 1 | .23 | 1.00 | 1.20 (1.07–1.34) | .002 | 3.2×10−8 |
| Stage 2 | .21 | 1.00 | 1.07 (.98–1.16) | .12 | ||||
| Stage 3 | .21 | NA | 1.16 (1.08–1.25) | 1.4×10−4 | ||||
| CGEMS | .21 | 1.00 | 1.13 (.97–1.30) | .11 | ||||
| Stage 4 | .21 | NA | 1.05 (1.02–1.09) | .001 | ||||
| rs10875943 (T/C) | 12q13 | 47962276 | Stage 1 | .27 | 1.00 | 1.18 (1.06–1.31) | .002 | 6.9×10−12 |
| Stage 2 | .28 | .99 | 1.10 (1.03–1.19) | .008 | ||||
| Stage 3 | .28 | NA | 1.12 (1.05–1.20) | .001 | ||||
| CGEMS | .30 | 1.00 | 1.02 (.89–1.15) | .81 | ||||
| Stage 4 | .31 | NA | 1.07 (1.04–1.10) | 6.7×10−7 | ||||
| rs5919432 (A/G) | Xq12 | 66938274 | Stage 1 | .18 | 1.00 | .92 (.83–.999) | .038 | 1.2×10−8 |
| Stage 2 | .19 | 1.00 | .87 (.81–.93) | 5.9×10−6 | ||||
| Stage 3 | .18 | NA | .92 (.82–1.03) | .13 | ||||
| CGEMS | .19 | 1.00 | .93 (.83–1.04) | .20 | ||||
| Stage 4 | .19 | NA | .94 (.89–.98) | .007 | ||||
Allele frequency of the second allele.
R2 is the estimate of the precision of the imputation provided by MACH program and for chromosome X, the precision of imputation provided by IMPUTE program. SNPs with R2 <0.3 were omitted from analyses. “NA” means that we used the actual genotype data for analysis.
Figure 1.
Forest plots for the 10 SNPs genotyped in stages 1–4. Squares represent the estimated per-allele odds ratio (OR) for individual studies. The area of square is inversely proportional to the precise of the estimate. Diamonds represent the summary OR estimates for the subgroups indicated. Horizontal lines represent 95% confidence limits.
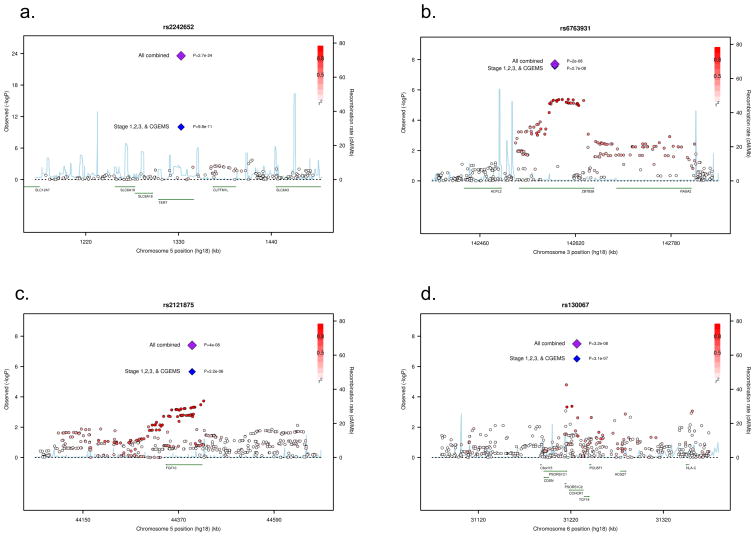
SNP rs2242652 on 5p15 lies in intron 3 of TERT (telomerase reverse transcriptase) (Figure 2. a.). SNPs in this region have been associated with multiple cancers, including basal cell carcinoma, lung cancer, bladder cancer, glioma and testicular cancer6. Some evidence for an association with PrCa risk has also previously been reported for SNPs rs401681 and rs2736098 in this region (P=3.6×10−4 and P=1.3×10−4)6. In contrast, we found much stronger evidence of an association with SNP rs2242652 in the same region. The novel SNP identified in this GWAS, rs2242652, is weakly correlated with the previously reported rs401681 (r2= 0.19) and showed a much stronger association with PrCa risk in our original GWAS (P =9.8 ×10−11 in stages 1, 3 and CGEMS vs. P=0.001 for rs401681). Multiple logistic regression analysis indicated that rs2242652 was associated with PrCa risk after adjustment for rs401681 (P= 4.6×10−9 and P= 0.07 respectively). In a stepwise logistic regression including both SNPs, rs2242652 alone remained in the model (P= 4.4×10−11). rs2242652 is also modestly correlated with rs2736098 (r2=0.10), although the latter SNP was not genotyped in any stage of our GWAS. Thus, rs2242652 may be more strongly correlated to the variant(s) causally related to PrCa risk than either rs401681 or rs2736098. This may also indicate that the variant(s) functionally related to PrCa risk in the TERT region differs from those for other cancers. The major role of telomerase is to catalyze the de novo addition of telomeric repeat sequences onto chromosome ends and thereby counterbalance telomere-dependent replicative ageing. Several studies have reported an association between short telomeres and increased risk of cancer at several sites although the cancer-associated SNPs in TERT have thus far not been associated with telomere length.
Figure 2.
Regional plots of four of the associated SNPs at 5p15 (a.), 3q25 (b.), 5p12 (c.), and 6p21 (d.). Plots show the genomic regions associated with PrCa and −log10 association P values of SNPs. Also shown are the SNP build 36/hg18 coordinates in kilobases, recombination rates and genes in the region. The intensity of red shading indicates the strength of LD (r2) with the index SNP. Plots drawn with a modified Rscript from http://www.broadinstitute.org/mpg/snap/ldplot.php
All but one of the autosomal SNPs associated with PrCa risk exhibit a pattern of association consistent with a log-additive model, as observed for most common cancer susceptibility alleles (Supplementary Table 1). For rs2121875 at 5p12, the estimated OR in stage 4 for rare allele homozygotes is 1.14 (95%CI 1.07–1.21) which was greater than expected under a log-additive model (P=0.02).
There was some evidence for a difference in the per-allele ORs among European, Asian and African-American populations for rs2121875 (P=0.03 for heterogeneity in the OR by population, Supplementary Table 2). However, the sample sizes for the Asian and African-American populations were too small to evaluate the associations in these populations reliably.
We were able to examine the associations between genotypes of the 10 SNPs selected for replication in PRACTICAL and serum PSA levels in 1089 control samples from stage 3 of our scan and 1540 controls from stage 4 (Supplementary Table 3). One SNP, rs2242652 (chromosome 5p15, TERT) showed an association with PSA level (P=0.006), in the direction consistent with its association with PrCa risk.
Data on Gleason score were available for 19,959 PrCa European cases. There was some evidence of a higher per-allele OR for Gleason score ≥ 8 disease for one SNP, rs5919432 (Xq12; Supplementary Table 4). This general lack of association with aggressiveness is broadly consistent with the other susceptibility loci identified through GWAS. Four SNPs, however, showed evidence of a differential association with age, in each case with bigger effect (smaller or bigger OR) at younger ages: rs10187424 (2p11.2; P=0 .02), rs2242652 (5p15.33; P=0.01), rs130067 (6p21.33; P=0.03), and rs5919432 (Xq12; p=0.04) (Supplementary Table 5). This age effect has not been seen consistently for previously identified susceptibility SNPs, but would be consistent with the higher familial relative risks at younger age. In addition, four SNPs exhibited stronger associations when analyses were restricted to cases with a family history of PrCa: rs10187424 (2p11.2; P=.006), rs7584330 (2q37.2; P=0.01), rs6763931 (3q23; P =0.03) and rs130067 (6p21.33; P=0.002). This is consistent with the expected enrichment of effect in familial cases under a multiplicative polygenic model (Supplementary Table 6). A PrCa locus at 2q37.2 has recently been reported in a genome-wide linkage scan in Finnish families7.
All of the newly associated loci lie in LD blocks that include plausible causative genes Besides TERT, particularly notable is rs6763931 at 3q23, which lies in intron 4 of ZBTB38, a zinc finger transcriptional repressor that binds methylated DNA8 (Figure 2. b). The murine homologue of ZBTB38, cibz, is a repressor of apoptosis9. An association between rs6763931 and human adult height has previously been reported, with the PrCa risk allele (A) being associated with increased height10, 11. Previous studies have suggested that tallness may be associated with an elevated risk of aggressive PrCa12; Gudbjartsson et al. found that rs6763931 was the SNP most significantly correlated with ZBTB38 expression in blood and adipose tissue.
rs2121875 at 5p12 is intronic in FGF10, a fibroblast growth factor essential for a range of developmental processes (Figure 2. c). FGF10 is often over-expressed in breast carcinomas13 and there is some evidence to indicate a role in the growth of normal prostatic epithelial cells14, 15. rs130067 at 6p21 is a coding SNP (Glu>Asp) in CCHCR1 (coding for coiled-coil alpha-helical rod protein1) a gene linked with psoriasis, an inflammatory condition (Figure 2. d). CCHCR1 is up-regulated in skin cancer and is associated with EGFR expression16. CCHCR1 promotes steroidogenesis by interacting with the steroidogenic acute regulator protein (StAR)17 and has a regulatory role in transcription factor binding18.
The other four associated loci are on chromosome 2p, 3q, 12q and X. (Supplementary Fig. 2. b–e.). rs10187424 on 2p11 is located in a gene-rich region that includes GGCX, VAMP8, VAMP5, and RNF181, a gene for a DNA damage-regulated RING finger protein. rs10936632 on 3q26 resides between the claudin gene CLDN11 and SKIL, a SKI-like oncogene. rs10875943 on 12q13 is between an α-tubulin gene cluster and the peripherin gene PRPH. The X-chromosomal SNP rs5919432 is located 77 kb from AR (androgen receptor).
With the identification of these new loci, more than 40 susceptibility loci for PrCa have now been identified. Most of the per-allele ORs estimated for these variants in this study population were modest, the strongest being an OR of 0.87 associated with the minor allele of rs2242652. These results were based on imputation of common variants using the Hapmap phase II reference panel and results from genotyped SNPs at stages 3 and 4. Taken together with previous GWAS analyses, these results strongly suggest that no further common variants that are imputable from the GWAS completed from current genotyping platforms confer substantially higher ORs (e.g. greater than 1.2, as estimated for rs10993994 at MSMB, or susceptibility SNPs on 8q24).
Based on an overall two-fold familial relative risk to first-degree relatives of PrCa cases, and on the assumption that the SNPs combine multiplicatively, the new loci reported here together explain approximately 1.5% of the familial risk of PrCa. When previously reported loci are included, approximately 25% of familial risk in PrCa may now be explained. Under this model, the top 10% of the population at highest risk has a relative risk approximately 2.4 fold greater than the average risk in the general population, while the top 1% has an estimated 4.1 fold increased relative risk. Such risk prediction may become important for facilitating targeted screening and prevention programs.
Methods
Samples
PrCa cases and controls used in stage 1 and 2 of the GWAS have been described previously1, 3. PrCa cases and controls for stage 3 were selected from studies in the UK and Australia. UK cases were drawn from the UK Genetic Prostate Cancer Study (UKGPCS) and Studies of Epidemiology and Risk Factors in Cancer Heredity (SEARCH). UKGPCS includes cases PrCa recruited from urologists throughout the UK, and a series of cases recruited from PrCa clinics in the Urology Unit at The Royal Marsden NHS Foundation Trust over a 17 year period. The controls (n=1947) were selected from men in the ProtecT (Prostate testing for cancer and Treatment) study19. ProtecT is a national study of community-based PSA testing and a randomised trial of subsequent PrCa treatment. Approximately 110,000 men between the ages of 50 and 69 years, (with a small set of men aged 45–49 years from one centre), were ascertained through general practices in nine regions in the UK. For this study we selected as controls men who had a PSA of <10ng/ml and negative prostate biopsies. Men with PSA ≥3ng/ml were excluded if they had a positive prostatic biopsy. We excluded, from both cases and controls, men who self-reported to be non-white. The majority of men in the UK are diagnosed via a clinical presentation; amongst the cases in this study 100% of those from the ProtecT study were diagnosed through asymptomatic PSA screening.
SEARCH cases (n=1468) were recruited through the Eastern Cancer Registration and Information Centre (ECRIC), a regional cancer registry covering the counties of Cambridgeshire, Norfolk, Suffolk, Bedfordshire, Hertfordshire and Essex. Cases diagnosed below the age of 70 since August 2003 who were within three years of diagnosis at notification are eligible to participate. The first 1468 patients recruited were included in this study. Controls were identified from the registers of general practices in the Eastern region that had recruited cases and were broadly age-group matched to the cases.
The Australian samples (1379 cases and 855 controls) in stage 3 were ascertained from two studies; MCCS and EOPCFS1, 3. Stage 4 included samples from 30 PrCa case-control studies participating in the PRACTICAL consortium (PRACTICAL Phase IV) (Supplementary Table 7 and Supplementary Notes). All studies were approved by the appropriate ethics committees.
Genotyping
Stage 3 genotypes were generated using an Illumina Golden Gate Assay. We filtered out all SNPs with a call rate <95%, a minor allele frequency in controls of <1%, or whose genotype frequency in controls departed from Hardy-Weinberg equilibrium at p<0.00001. After these exclusions, we analyzed 1347 out of 1439 genotyped SNPs. Duplicate concordance was 99.99%.
In stage 4, genotyping of samples from 14 studies was performed by KASPar assay (www.kbioscience.co.uk), while 16 study sites performed the 5′exonuclease assay (Taqman™) using the ABI Prism 7900HT sequence detection system according to the manufacturer’s instructions. Primers and probes were supplied directly by Applied Biosystems as Assays-By-Design™. Assays at all sites included at least four negative controls and 2–5% duplicates on each 384-well plate. Quality control guidelines were followed by all the participating groups as previously described3. In addition, all sites also genotyped 16 CEPH samples. We excluded individuals that were not typed for at least 80% of the SNPs attempted. Data on a given SNP for a given site were also excluded if they failed any of the following QC criteria: SNP call rate >95%, no deviation from Hardy-Weinberg equilibrium in controls at P<.00001; <2% discordance between genotypes in duplicate samples and in the CEPH samples. Cluster plots for SNPs that were close to failing any of the QC criteria were re-examined centrally.
Statistical methods
Imputation
We first combined data from stage 1 and 2 of our PrCa GWAS and stage 1 of CGEMS. Stage 1 included data on 1894 controls and 1854 cases genotyped for 541,129 SNPs using an Illumina Infinium 550k array. Stage 2 included 3940 controls and 3650 cases genotyped for 43,671 SNPs using an iSELECT. CGEMS included 2277 samples (1101 controls and 1176 cases) genotyped for 546,613 SNPs using the Illumina 317K and 240K arrays. We used MACH 1.0 (http://www.sph.umich.edu/csg/abecasis/MACH/) to impute genotypes of autosomal markers, with HapMap phase 2 of European population (CEU) as a reference panel. We included imputed data from a SNP if the estimated correlation between the genotype scores and the true genotypes (r2) was >0.3. We used IMPUTE v120 in order to perform the imputation for chromosome X. The imputed genotype probabilities were used to derive a 1df association score statistic for each SNP, and its corresponding variance. The scores and variances were then added to obtain a combined X2 trend statistic for each SNP, (equivalent to the Mantel extension test, or as in a fixed effects meta-analysis), in R. The imputed results were combined with the results of genotyped SNPs from the second stage of CGEMS.
SNP selection for stage 3
SNPs for stage 3 were primarily selected from the ranked list of 1df P-values from the combined analysis of the previous stages. Only SNPs with design scores >0.8 were considered. This list was “thinned” to exclude correlated SNPs. Where SNPs were correlated at r2>0.8, we preferentially included the SNP with the smallest P-value from among all SNPs previously genotyped, or, if no such SNP was available the SNP with the smallest P-value from among the imputed SNPs. We included 1263 SNPs from this list. In addition, we included 132 SNPs based on dense genotyping for fine-mapping purposes of three regions (covering the NKX3.1, ITGA6 and LMTK2 loci), together with 46 known PrCa susceptibility SNPs or proposed candidate SNPs. These latter candidate SNPs were not considered further here.
Stage 3 and stage 4 analysis
We assessed associations between each SNP and PrCa at stages 3 and 4 using a 1df Cochran-Armitage trend test stratified by study. The combined P-values over all stages were generated in similarly (using a 1df trend test based on summing the scores and variances from each stage). SNPs were selected for validation in stage 4 on the basis of either a significance level of P<10−3 in a 1df trend test at stage 3 or a significance level of P<10−7 in a combined analyses of stage 1, 2, 3 and CGEMS. Where there was more than one SNP in a region, multiple logistic regression was used to define the minimal set of SNPs that showed evidence of association after adjustment for other SNPs.
In stage 4 we stratified analyses by study and ethnic group (European, African-American, Asian). Where <100 individuals were recorded in a minority ethnic group, these individuals were excluded. After exclusions, analyses were performed based on 26,055 PrCa cases and 25,256 controls. ORs and 95% confidence limits were estimated using unconditional logistic regression, stratified by study and ethnic group. In the text we have reported the combined tests of association over all stages in European populations, but have emphasized the OR estimates from stage 4 to minimize the effect of “winner’s curse”. Tests of homogeneity of the ORs across strata and populations were assessed using likelihood ratio tests. Modification of the ORs by disease aggressiveness and family history was assessed by using the binary endpoint of family history (Yes vs. No); and Gleason score (<8 vs. ≥ 8). A test for association between genotype and Gleason score as an ordinal variable was also performed, using polytomous regression. Modification of the ORs by age was assessed using a case-only analysis, assessing the association between age and SNP genotype in the cases using polytomous regression. The associations between SNP genotypes and PSA level were assessed using linear regression, after log-transformation of PSA level to correct for skewness. Analyses were performed in R (principally using GenABEL21, SNPTEST, ProbABEL22 and Stata.
Supplementary Material
Acknowledgments
Competing Financial Interests
None
Footnotes
URLs
http://www.srl.cam.ac.uk/consortia/practical/index.html
http://www.stats.ox.ac.uk/~marchini/software/gwas/snptest.html
http://www.sph.umich.edu/csg/abecasis/MACH/
These are detailed in the Supplementary Note
Author contribution
RAE and DFE designed the study, and are joint PIs on the GWAS. RAE is PI of the UKGPCS and project managed the overall study.
ZKJ, RAE, DFE and AA wrote the paper, ZKJ coordinated and managed the Stage 3 and the PRACTICAL Stage 4 genotyping.
ZKJ, DAL, MT, EJS, NM coordinated sample collation for Stage 3 and the PRACTICAL
Stage 4 set genotyped in the UK.
AA and DFE performed the statistical analyses; SB collated the dataset.
GGG, JH, DRE, and GS are PIs of the Australian studies; and MS manages the molecular work.
JS is PI of the Tampere study; TW collected clinical data, performed sample selection, and collated data. TLT coordinated sample collection.
MW is the PI of the CPCS1 and 2 studies, PK, BGE, MAR, ATH and SEB have collected samples data and contributed to genotyping of this study.
FCH, DN and JD are joint PIs of ProtecT; AL is study coordinator and MD the data base manager. AC assisted with sample selection, retrieval and processing.
DA and JV are PIs of the ATBC Study, and were responsible for the original collection of the ATBC DNA samples. SC was responsible for assembly and genotyping.
SIB is the PI of the PLCO study, AG is the PI for St. Louis screening centre for PLCO, and MY oversaw the genotyping for PLCO. HB is PI of the ESTHER study; DR, CS contributed to design and data collection; HM is study coordinator.
CC and JL of the Poland study coordinated sample collection. CC and DWgenotyped the samples.
CM and WV are PIs of the Ulm study. AER identified and collected clinical material/processed samples/undertook genotyping/collated data for Ulm.
TD is PI of the Hannover Prostate Cancer Study; AM and JS coordinated sample collation, provided molecular advice and conducted molecular work.
JLD is PI of the Tasprac study: JRM led the Tasmanian genotyping and collated data; BP provided molecular advice assistance with collating data.
PK coordinated data collection and management for the HPFS.
TØ and KDS are PIs of the Aarhus study. MB coordinated sample collection and registration of clinical data. KDS led the sample genotyping.
TK is PI of the EPIC-Oxford cohort and collected clinical material. RT collated data.
SMG and MJT are PIs of the ACS CPS-II study, WRD is the data manager for this study.
BEH and LL are PIs of the MEC; CH and FS are CI.
YJL and HWZ are joint PIs of CHSH; YJL is study coordinator and HWZ participates and closely supervised the CHSH study.
JLS is PI of the Fred Hutchinson study and EAO is PI of the NHGRI genotyping for PROGRESS; LMF and JSK coordinated data collation.
SAI is PI of the USC study, EMJ is PI of the NCCC study; MCS and RC led the genotyping of both studies.
SNT and DS are PIs of the Mayo clinic study; SKD coordinated data collation.
JYP is PI of the Moffitt study, TAS and HYL are contributors to this study.
JAC, ABS are PIs of the molecular genetics arm of the ProsCan study, and with JB and FL co-ordinated all risk factor data and genetic data collection for prostate cancer cases from Proscan, the Brisbane Retrospective Study, the Australian Prostate Cancer BioResource Brisbane node, and controls from two Queensland control sets. SC, JA and RAG are PIs of the Proscan study and were responsible for the original platform study initiation, conceptualisation and collection of the Proscan study cases.
KAC is PI of the FMHS study, LCA is PI of the Utah study,
RK is PI of the PCMUS study, ATAJ, ALH, LTO’B, RAW, ECP, EJS, DPD, AH, RAH, VSK, CCP, NVA, CJW, AT, TC, CO, LNK, LLM, AA, AC, DMK, EMK, ADJ, AS, TAS, JPS, SC, JA, RAG, JB, MAK, FL, AP, BP, JS, AM, AER, KL, AMR, EML, JF, HK, CS, identified and collected clinical material and VM coordinated data collation. Other members of The UK Genetic Prostate Cancer Study Collaborators/British Association of Urological Surgeons’ Section of Oncology, The UK ProtecT Study Collaborators, and The PRACTICAL Consortium members (membership lists provided in the Supplementary Note) collected clinical samples, assisted in genotyping and provided data management.
Reference List
- 1.Eeles RA, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 2.Al Olama AA, et al. Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat Genet. 2009;41:1058–1060. doi: 10.1038/ng.452. [DOI] [PubMed] [Google Scholar]
- 3.Eeles RA, et al. Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat Genet. 2009;41:1116–1121. doi: 10.1038/ng.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varghese JS, Easton DF. Genome-wide association studies in common cancers--what have we learnt? Curr Opin Genet Dev. 2010;20:201–209. doi: 10.1016/j.gde.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 5.Park JH, et al. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat Genet. 2010;42:570–575. doi: 10.1038/ng.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rafnar T, et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat Genet. 2009;41:221–227. doi: 10.1038/ng.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cropp CD, et al. Genome-wide linkage scan for prostate cancer susceptibility in finland: Evidence for a novel locus on 2q37.2 and confirmation of signal on 17q21–q22. Int J Cancer. 2011 doi: 10.1002/ijc.25906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Filion GJ, et al. A family of human zinc finger proteins that bind methylated DNA and repress transcription. Mol Cell Biol. 2006;26:169–181. doi: 10.1128/MCB.26.1.169-181.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oikawa Y, Matsuda E, Nishii T, Ishida Y, Kawaichi M. Down-regulation of CIBZ, a novel substrate of caspase-3, induces apoptosis. J Biol Chem. 2008;283:14242–14247. doi: 10.1074/jbc.M802257200. [DOI] [PubMed] [Google Scholar]
- 10.Gudbjartsson DF, et al. Many sequence variants affecting diversity of adult human height. Nat Genet. 2008;40:609–615. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- 11.Soranzo N, et al. Meta-analysis of genome-wide scans for human adult stature identifies novel Loci and associations with measures of skeletal frame size. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zuccolo L, et al. Height and prostate cancer risk: a large nested case-control study (ProtecT) and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2008;17:2325–2336. doi: 10.1158/1055-9965.EPI-08-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Theodorou V, et al. Fgf10 is an oncogene activated by MMTV insertional mutagenesis in mouse mammary tumors and overexpressed in a subset of human breast carcinomas. Oncogene. 2004;23:6047–6055. doi: 10.1038/sj.onc.1207816. [DOI] [PubMed] [Google Scholar]
- 14.Ropiquet F, Giri D, Kwabi-Addo B, Schmidt K, Ittmann M. FGF-10 is expressed at low levels in the human prostate. Prostate. 2000;44:334–338. doi: 10.1002/1097-0045(20000901)44:4<334::aid-pros11>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 15.Thomson AA, Cunha GR. Prostatic growth and development are regulated by FGF10. Development. 1999;126:3693–3701. doi: 10.1242/dev.126.16.3693. [DOI] [PubMed] [Google Scholar]
- 16.Suomela S, et al. CCHCR1 is up-regulated in skin cancer and associated with EGFR expression. PLoS One. 2009;4 doi: 10.1371/journal.pone.0006030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sugawara T, Shimizu H, Hoshi N, Nakajima A, Fujimoto S. Steroidogenic acute regulatory protein-binding protein cloned by a yeast two-hybrid system. J Biol Chem. 2003;278:42487–42494. doi: 10.1074/jbc.M302291200. [DOI] [PubMed] [Google Scholar]
- 18.Corbi N, et al. RNA polymerase II subunit 3 is retained in the cytoplasm by its interaction with HCR, the psoriasis vulgaris candidate gene product. J Cell Sci. 2005;118:4253–4260. doi: 10.1242/jcs.02545. [DOI] [PubMed] [Google Scholar]
- 19.Donovan J, et al. Prostate Testing for Cancer and Treatment (ProtecT) feasibility study. Health Technol Assess. 2003;7:1–88. doi: 10.3310/hta7140. [DOI] [PubMed] [Google Scholar]
- 20.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 21.Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 22.Aulchenko YS, Struchalin MV, van Duijn CM. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics. 2010;11:134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.