Abstract
Background
The promise of Alzheimer’s disease (AD) biomarkers has led to their incorporation in new diagnostic criteria and in therapeutic trials; however, significant barriers exist to widespread use. Chief among these is the lack of internationally accepted standards for quantitative metrics. Hippocampal volumetry is the most widely studied quantitative magnetic resonance imaging (MRI) measure in AD and thus represents the most rational target for an initial effort at standardization.
Methods and Results
The authors of this position paper propose a path toward this goal. The steps include: 1) Establish and empower an oversight board to manage and assess the effort, 2) Adopt the standardized definition of anatomic hippocampal boundaries on MRI arising from the EADC-ADNI hippocampal harmonization effort as a Reference Standard, 3) Establish a scientifically appropriate, publicly available Reference Standard Dataset based on manual delineation of the hippocampus in an appropriate sample of subjects (ADNI), and 4) Define minimum technical and prognostic performance metrics for validation of new measurement techniques using the Reference Standard Dataset as a benchmark.
Conclusions
Although manual delineation of the hippocampus is the best available reference standard, practical application of hippocampal volumetry will require automated methods. Our intent is to establish a mechanism for credentialing automated software applications to achieve internationally recognized accuracy and prognostic performance standards that lead to the systematic evaluation and then widespread acceptance and use of hippocampal volumetry. The standardization and assay validation process outlined for hippocampal volumetry is envisioned as a template that could be applied to other imaging biomarkers.
Keywords: Alzheimer’s disease, biomarkers, Magnetic resonance imaging, hippocampus, biomarker standards
1. Introduction
A biomarker is a physiological, biochemical, or anatomic parameter that can be objectively measured as an indicator of normal biologic processes, pathological processes, or responses to a therapeutic intervention (1). Biomarkers used in the Alzheimer’s disease (AD) field include both imaging measures and biofluid analytes. Biofluid analytes in this context can refer to proteins in any biofluid, however cerebrospinal fluid (CSF) biomarkers are presently the most well developed (2). The five most widely studied biomarkers in AD can be divided into two major categories: 1) Biomarkers of cerebral Aβ amyloid accumulation - these are increased radiotracer retention on amyloid-tracer based positron emission tomography (PET) imaging and low CSF Aβ 1-42, and 2) Biomarkers of neuronal degeneration or injury - these are elevated CSF tau (both total and phosphorylated tau); decreased fluorodeoxyglucose (FDG) uptake on PET in the temporo-parietal cortex; and brain atrophy in the medial, basal and lateral temporal lobes and the medial and lateral parietal cortices determined from structural magnetic resonance imaging (MRI) or computed tomography (CT) (3). Three of these five major AD biomarkers are imaging measures and imaging is the primary focus of this position paper. Biomarkers are increasingly important in AD in two contexts: clinical diagnosis/prognosis and therapeutic trials.
Criteria for the clinical diagnosis of AD were established in 1984 (4). These criteria have been widely adopted, validated against neuropathological examination in many studies, and are still used today. A consensus now exists, however, that diagnostic criteria for AD should be updated to reflect the scientific advances of the past quarter of a century. One of most important of these advances is the development of biomarkers for AD. This recognition has inspired recent efforts on several fronts to revise diagnostic criteria for AD. The two most well-known such efforts are those of Dubois et al (5, 6) and the National Institute on Aging (NIA)-Alzheimer’s Association (AA) (7-10). The NIA-AA commissioned three work groups to revise diagnostic criteria. Each was assigned the task of defining or revising criteria for one of three recognized phases of the disease: pre-clinical or asymptomatic AD, symptomatic pre-dementia or mild cognitive impairment (MCI), and the AD dementia phase (7-10). Biomarkers providing evidence of in situ AD pathophysiology are employed in the revised definitions of AD in all three phases of the disease by the NIA-AA and are also included in the criteria of Dubois et al (5, 6).
The second major use for biomarkers of AD is in clinical trials, where biomarkers can be employed for several distinct purposes. As an indicator of AD pathophysiological processes, AD biomarkers may be used for subject inclusion/exclusion – to ensure study subjects are appropriate for targeting of the therapeutic mechanism of action or as an enrichment strategy to improve efficiency of therapeutic trials (2, 11). Biomarkers also provide a biologically-based measure of disease severity. They can be used as a covariate in outcome analyses and as safety measures. Finally, an important application of AD biomarkers in clinical trials is as outcome measures, in which an effect on the biomarker is sought as evidence of modification of the underlying pathological AD process (12-21). However, since AD pathophysiology is increasingly being recognized to be very complex and multifaceted, effects of candidate drugs on some individual pathophysiological aspects of AD may not necessarily be of functional or cognitive relevance. Therefore, increasing efforts are being spent on developing biomarkers which could serve as surrogate endpoints in clinical trials, accurately predicting and reflecting clinically significant outcomes (2, 22) Biomarkers are more objective and reliable quantitative measures of AD pathophysiological processes than traditional cognitive and functional outcomes that are affected by subject motivation and extrinsic factors such as alertness, environmental stresses, and informant mood and distress.
The evaluation of the value of biomarkers is different for therapeutic trials than for clinical diagnosis, but the rationale and methods to standardize and validate the reliability of the measures are very similar. Moreover, if an imaging biomarker is used as an inclusion criterion for subjects participating in a clinical trial of a compound that subsequently achieves regulatory approval, then it is possible, some would say likely, that regulators will require the same biomarker must be approved as a diagnostic to identify patients that are suitable for treatment. This would then require that the biomarker, in our case imaging, be easily implementable in clinical imaging facilities world-wide. Therefore, although requirements in terms of precision and sensitivity to pathology may vary, issues pertaining to standardization of an imaging biomarker for use in clinical trials and for clinical diagnostics are inextricably interwoven.
The potential value of quantitative imaging biomarkers for both clinical diagnosis and clinical trials is clear, but major barriers exist to widespread acceptance and implementation. The most substantive barriers have been the lack of standardized methods for 1) image acquisition, 2) extraction of quantitative information from images, and 3) linking quantitative metrics to internationally recognized performance criteria. These in turn have impeded the establishment of cut points in the continuous range of quantitative values that can be used in diagnosis and evaluating change in clinical trials. Standardization of image acquisition for structural MRI and PET scans has been a major focus of the Alzheimer’s Disease Neuroimaging Initiative (ADNI) project (23, 24) and ADNI acquisition protocols have become the de facto standard for clinical trials and could be applied clinically. On the other hand, little progress has been made in the standardization of techniques for quantitative image analysis, either in ADNI or in the field in general. This is particularly true for MRI where the lack of standardization has led to publication of values that are highly disparate across the literature. For example, greater than two-fold differences in hippocampal volume of cognitively normal elderly subjects have been reported from different centers (25). This is unlikely to have a basis in biology and is almost certainly due to inter-center differences in the measurement tools and the anatomical protocols for delineating the hippocampus. Likewise, a strong methodological dependence is evident in published rates of hippocampal atrophy. Three-fold differences in rates of hippocampal atrophy have been reported in elderly controls as well as wide variations in apparently similar cohorts of AD patients (26). For example, Du et al (27) reported annualized rates of hippocampal atrophy in healthy elderly controls mean age 77 of 0.8%/yr; Jack et al (28) in controls age 78 of 1.4%/yr and Wang et al (29) mean age 73 of 2.3%/yr. This strong dependence upon the method used and its specific implementation undermines the credibility of the results. Both newly proposed diagnostic criteria explicitly point out that extensive work on imaging biomarker standardization is needed prior to widespread adoption for diagnostic purposes.
2. Why hippocampal volume?
Qualification or general acceptance of the validity of a biomarker in clinical trials must rest on a well-established body of evidence beginning with widespread agreement that there is clinical significance to the result of the biomarker and that it can be measured with appropriate accuracy and reproducibility. Quantitative measurement of hippocampal volume fulfills these basic criteria. The advantages of hippocampal volume as a target for an initial standardization and assay validation exercise are: 1) The hippocampus is an anatomically defined structure with boundaries that are visually definable in a properly acquired MRI scan. 2) The hippocampus is involved early and progressively with neuronal loss and neurofibrillary tangles, which is one of the primary hallmarks of AD pathology (30). 3) A large imaging and pathology literature provides evidence that loss of hippocampal volume is significant in AD. Numerous studies have shown the association of hippocampal atrophy with neurodegenerative pathology at autopsy (31-36), with clinical diagnoses of AD or MCI (37-43), and with the severity of cognitive disorders and episodic memory deficits due to AD pathophysiology (44, 45). In addition, longitudinal measures of change in hippocampal volume both predict the future cognitive decline and correlate with contemporary indices of clinical decline (46, 47), and quantitative measures of the hippocampus predict progression from MCI to AD (48-63).. 4) Fully automated software tools are now available that can measure hippocampal volume efficiently and reproducibly (21, 37, 58, 64-71). Visual rating (72-74), while convenient and currently used in some diagnostic settings, does not lend itself to detecting subtle size differences, lacks precision relative to quantitative methods, and does not take advantage of the power of current technology. Formal computer-aided manual tracing of the entire hippocampus was introduced over two decades ago to aid in seizure lateralization (75). Although manual hippocampal tracing has been effective for research studies in different diseases, and still serves as the best available Reference Standard measure of the hippocampus on MRI (76), it is time consuming and requires highly trained operators. Thus it is not feasible in routine clinical practice and due to its expense it is impractical in clinical trials. Fully-automated hippocampal volumetry using standardized methods would be a practical alternative to manual methods. Automated hippocampal volumetry has successfully enabled the discovery of novel genes associated with hippocampal volume in over 7000 subjects scanned at multiple internationally distributed sites. This result supports the assertion that such methods can be efficiently and reproducibly applied on a worldwide scale (77). Furthermore, software methods that employ within-subject registration permit sensitive measures of volume change over time (51, 78). 5) While more complex MRI measures of disease-related atrophy consisting of combinations of multiple regions of interest (ROI) might have superior diagnostic properties compared to hippocampal volume (79-84), the analysis of hippocampal volume is less complex than multi-ROI approaches so a reference standard is easier to generate. Specifically, the hippocampus can be delineated by hand, but the disease signatures of more complex analytic methods are a result of training and machine learning methods that would present a further challenge to validate, and are likely to evolve over time.
Further supporting hippocampal volumetry as a target for initial AD imaging biomarker standardization and assay validation is the fact that clinical guidelines in many countries (85, 86) dictate that all patients investigated for cognitive impairment should undergo structural brain imaging to exclude treatable causes such as tumors and hematoma. An MRI acquisition sequence that would permit quantitative analysis of hippocampal volume is easy to include in a routine clinical MRI examination, only lengthens the exam by a few minutes, and is currently considered to be an essential part of a clinically diagnostic imaging protocol at some centers. Moreover, a significant effort has already been expended to standardize acquisition parameters for the high resolution 3D anatomical MR imaging sequence needed for quantitative volume measures across MRI vendors in the ADNI study (23). The ADNI 3D T1 anatomical sequence used for volumetric measurements can be performed in a standardized manner in an overwhelming majority of imaging centers worldwide. Finally, there is an ongoing international initiative led by one of the co-authors (GBF) to establish a Reference Standard in hand-drawn hippocampal volumes, which is the European Alzheimer’s Disease Centers (EADC) – ADNI Hippocampal Harmonization Effort (87, 88).
The issue of validating imaging biomarkers for AD has recently drawn the attention of non-profit organizations, including the Radiological Society of North America (RSNA) and the Coalition Against Major Disease (CAMD). CAMD is part of Critical Path Institute a nonprofit public private partnership dedicated to more efficient drug development. Qualification of hippocampal atrophy for use in clinical trial enrichment is being pursued by CAMD with the US Food and Drug Administration (FDA) and European Medicines Agency (EMA). At a meeting of The Radiological Society of North America Quantitative Imaging Biomarkers consortium in September, 2010 a work group was convened to address the issue of standardizing quantitative imaging of AD. Among the candidate imaging modalities discussed, measures of hippocampal volume on structural MRI were identified as the most widely used in the context of multicenter clinical trials, and therefore were the most obvious candidates for an initial (exemplar) effort to standardize quantitative imaging biomarkers. This position paper follows from the recommendations of this RSNA work group.
3. Biomarker development
In general terms, three separate steps are required for biomarker development: 1) Assay validation (also called technical or analytical performance validity) to show that, when following defined standardized procedures, the biomarker can be measured precisely and accurately compared to a reference standard (89), 2) Clinical Validation to establish that the biomarker has value for a specific intended task and context of use, and 3) Qualification of the biomarker with the appropriate regulatory agencies based upon wide-spread consensus that the biomarker is “fit for purpose” for a particular use. Each proposed task (e.g., diagnostic, prognostic, outcome) needs to be considered separately. Qualification of a biomarker for clinical trials may be a stepping stone to a qualification for its use as a clinical diagnostic. However, the use of a biomarker in clinical diagnosis is distinct from its use in therapeutic trials, and development may focus on one or the other first. The use of a biomarker in clinical trials is at the discretion of the trial sponsor, but mechanisms have been introduced by which regulatory bodies (e.g., the US Food and Drug Administration Center for Drug Evaluation and Research, FDA CDER; or the European Medicines Agency EMA) qualify biomarkers for use in clinical trials. The use of a biomarker for clinical diagnosis requires regulatory approval in the relevant jurisdiction (e.g., approval by FDA Center for Devices and Radiological Health, CDRH, in the USA; or CE marking in Europe), and may separately also require approval from healthcare funders for reimbursement.
4. Steps to standardization and validation of hippocampal volumetry as a biomarker of AD
Below we outline the steps of a proposed work plan that would lead to standardization of quantitative (automated or manual) hippocampal volumetry as a biomarker for AD in evaluative studies in the context of clinical trials and for diagnosis.
Establish an Oversight Board to manage the effort and empower this body with authority to make decisions necessary to assess the results as outlined below. The Oversight Board should have the following attributes: a) include all necessary areas of expertise, b) be unbiased, c) represent both academia as well as industry, and d) be international. All potential conflicts of interest must be fully disclosed. Our recommendation is that this oversight board be linked to the Alzheimer’s Association.
Identify a standardized definition of anatomic hippocampal boundaries on MRI with the assistance of expert neuroanatomists for use as a Reference Standard. Anatomic boundary criteria should be acceptable to the international scientific community and consistent with use in all neuroscience disciplines. We recognize that for hippocampal volume measures to be widely used diagnostically in clinical practice and in clinical trials, automated techniques are essential. However, manual tracing of the hippocampus using a consensus-from-experts approach in accordance with a standardized definition provides the most effective Reference Standard to evaluate automated methods. Expert opinion is an accepted method to create a reference standard. This is preferable to the alternative, arbitrarily picking one automated method and anointing it as the Reference Standard, which would be problematic. Because most, if not all, automated techniques rely on some a priori anatomical notion of hippocampal boundaries, such an arbitrary approach would not reflect a consensus from the scientific community as a whole and would not result in a Reference Standard with broad-based support from all stakeholders. Since an international effort is currently in place with precisely this aim, leveraging the work of the EADC-ADNI Hippocampal Harmonization effort (87, 88) is the most logical and practical approach. The Reference Standard recommended by the authors of this position paper is therefore the manual hippocampal tracing of ADNI subjects who will be developed by the EADC-ADNI effort.
- Establish a Reference Standard Dataset based on manual delineation of the hippocampus in accordance with the standardized definition. The Reference Standard Dataset should have the following attributes:
- All subjects in the reference database must have given informed consent for public access under an ethics board-approved protocol. Compliance with relevant privacy legislation to the jurisdiction where the data were collected, and permission of a research ethics committee for use of the data should be obtained. In the US, the relevant guidelines are those of the Health Insurance Portability and Accountability Act (HIPAA); however, other jurisdictions will have different regulations.
- Access to the database must be straightforward, open, and readily available.
- Appropriate subjects, in clinical characteristics and number, must be included in the reference database – in this case, elderly cognitively normal control, MCI and AD subjects diagnosed according to internationally recognized diagnostic criteria.
- MRI scans must have been acquired with a standardized protocol that is amenable to widespread use.
- Appropriate clinical meta-data must be linked to the MRI scans and readily available to users – i.e., demographics, clinical diagnosis, basic neuropsychology, and longitudinal clinical course. The subjects, 3D volume T1-weighted images, and clinical data of ADNI represent a data set that meets these criteria. The authors recommend that the EADC-ADNI harmonization traces or masks of the 1.5T ADNI MPRAGE data serve as the hippocampal volume Reference Standard Dataset.
Extend the Reference Standard Dataset to enable a thorough evaluation of technical aspects of MR acquisition on measurement performance. This includes the effects of MR vendor, receiver coil type, accelerated acquisition methods, and field strength. Although the EADC-ADNI harmonization plan focuses are on 1.5T data, a significant portion of neuroimaging in the future will be performed at 3T, with acquisition acceleration, and with increasingly complex coil arrays. The potential effects of these technical advances on measurement standardization should be investigated (90).
Split the complete sample of traced hippocampi into balanced training and test data sets for assessing the technical performance characteristics of new analysis methods. This would enable automated methods to be trained on a portion of the reference data and then test performance against an independent subset of the reference data. Careful attention to the composition of these subsets is important so that age, gender or clinical variables are not inadvertently unbalanced.
Develop standards for reporting measurement units including a standardized approach for normalization of raw hippocampal volume measures. This will include defining correct measures of head size through standardization of intracranial volume measures. In addition to disease severity, hippocampal volume is affected by other variables that are easily ascertained such as age, sex, and head size (taller people tend to have larger brains and thus larger intracranial volume) (91). Experience indicates that normalization of raw hippocampal volumes for these descriptive or confounds variables improves the performance of hippocampal volumetry in evaluation studies, and thus recommendations for standardized normalization procedures for adjusting raw hippocampal volumes (e.g., by head size, age, sex) in the reference data set will be necessary.
- Define minimum technical performance metrics as benchmarks to judge new analysis methods (89). At a minimum these metrics should include:
- Accuracy with respect to the manually traced Reference Standard Dataset. We note that automated techniques will likely not precisely match a manually traced Reference Standard. However, a straightforward mathematical transformation of the output an accurate automated algorithm to match the reference standard should be possible. Criteria would need to be set as to how close the automated method would have to match the manual tracing in order for it to be credentialed by the oversight board.
- Test/re-test precision. This would include not just numeric precision at the volume level, but also more exacting indices of area/pixel overlap such as Dice coefficients.
- Compliance with regulatory requirements (Good Clinical Practice (GCP), FDA 21 CFR part 11, EU GMP Annex 11 on Computerized Systems) for any computer systems running these algorithms.
Define minimum prognostic performance metrics for new analysis methods based upon benchmarks established from Reference Standard Dataset: We recommend metrics that predict conversion from MCI to AD within 24 months, progression of dementia severity at 24 months in patients with AD, and maintenance of normal cognition at 24 months in cognitively normal subjects (sensitivity, specificity, positive and negative predictive value, ROC analysis). This will serve as further assay validation for new analysis methods.
Empower the oversight board to oversee credentialing of applications for analysis methods. While the Reference Standard Dataset can be used to credential new manual tracers, its primary use is envisioned as a means of validating and credentialing automated hippocampal quantification methods for use in therapeutic trials and for new clinical diagnostic criteria. The board could also make context of use recommendations based on limitations identified during the evaluation of a particular method. In order for a potential hippocampal volume measurement application to be credentialed by the oversight board it would have to meet established technical and prognostic performance benchmarks using the reference data set described above.
Ideally, the work plan would follow the timeline above where initial steps would focus on establishing the reference standard of manual hippocampus traces, generating a standardized approach to volume normalization and benchmark performance metrics. Once the reference standard is established, then the focus likely would be on evaluation studies and qualifying the reference standard with the FDA and EMA for diagnostic, prognostic and outcome use in clinical trials. Standardized acquisition of MRI scans suitable for hippocampal volumetry are already widely performed and support from the pharmaceutical industry is likely. Subsequently, we expect evaluation studies will be conducted to show the diagnostic value of hippocampal volumetry use outside the context of clinical trials. We wish to emphasize that the intent of this position paper is not to stifle existing alternative methods or innovative development of new methods, but rather to facilitate the development of widely available implementations of automated hippocampal volumetry methods, and to serve as a template for an initial effort which can then be used for other imaging biomarkers.
5. Illustration
As an example illustrating the approach discussed above we identified 373 ADNI subjects diagnosed as MCI at baseline who qualified for an analysis of time to progression to AD. Of the 397 ADNI subjects diagnosed as MCI at baseline, 16 had no follow-up visits, and 8 failed quality control, leaving 373 for this analysis (Table 1). A list of the ADNI subject ID numbers used in the example MCI analyses is included as a Supplement. All subjects had hippocampal volume measured in three ways, labeled Methods A, B and C here. In this exercise, we considered Method A to represent the Reference Standard Dataset, and assessed Methods B and C in two ways: technical performance accuracy relative to the Reference Standard Dataset and prognostic performance in predicting conversion from MCI to AD at 2 years post baseline. While the data presented below are real, and not hypothetical, the specific methods are left undefined because we do not wish to have this position paper misconstrued as evidence that the authors endorse a particular method for credentialing.
Table 1.
Descriptive Characteristics
| Characteristic | All | Stable MCI | AD Converter |
|---|---|---|---|
| N | 373 | 178 | 135 |
| Age, years | 75 (70, 80) | 75 (71, 81) | 75 (70, 80) |
| Female gender, no. (%) | 136 (36) | 63 (35) | 51 (38) |
| Education, years | 16 (14, 18) | 16 (14, 18) | 16 (14, 18) |
| MMSE | 27 (26, 28) | 28 (26, 29) | 27 (25, 28) |
| Hippocampal Volume, cm3 | |||
| Method A | 6.3 (5.6, 7.1) | 6.7 (6.0, 7.4) | 6.0 (5.2, 6.6) |
| Method B | 6.3 (5.6, 7.1) | 6.6 (5.9, 7.2) | 5.9 (5.1, 6.6) |
| Method C | 6.9 (6.2, 7.5) | 7.1 (6.5, 7.6) | 6.6 (6.0, 7.2) |
All values are reported as median (inter quartile range - IQR) unless otherwise noted Stable/Converter is defined as progression to AD by 24 months
MMSE, Mini Mental State Exam
Of the 373 patients, 166 progressed from MCI to AD during follow-up and 8 progressed to non-AD dementia based upon clinical criteria. We also examined a subset of 313 subjects that either progressed to AD at or prior to the 24 month visit (n=135) or had available follow-up through the 24 month visit without progressing to AD (n=178) to evaluate differences in hippocampal volume for those that progressed at 24 months vs. those that remain stable. Subjects who progressed to non-AD dementia at or before 24 months were excluded from this analysis.
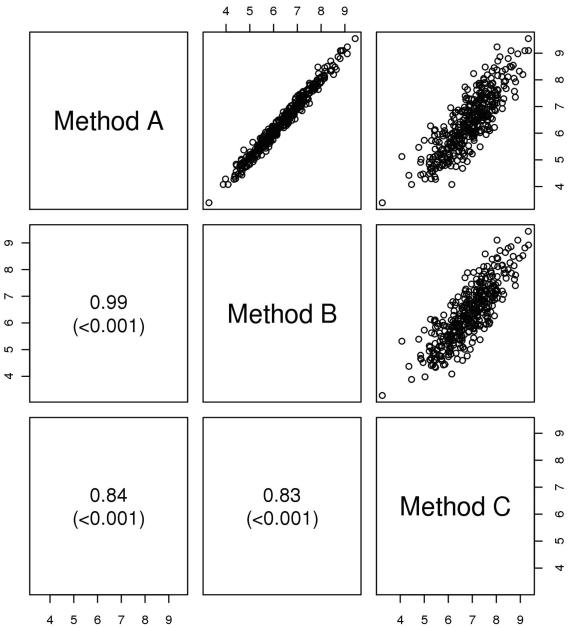
Method B potentially meets two major criteria for credentialing – it is highly accurate in the group-wise and individual measurement of hippocampal volume relative to Method A as shown in the table and scatter plots, and it also has essentially identical performance in predicting conversion from MCI to AD (Fig. 1, Table 2). Method C has a similar prognostic performance in predicting conversion to AD as Method A as shown in the ROC analysis, but in its current form might not meet technical accuracy criteria relative to the reference standard dataset. This is how we would envision the credentialing process would proceed for most automated applications, with the EADC-ADNI harmonization data set of manually traced hippocampi serving as the Reference Standard Dataset and the oversight committee setting predetermined minimal benchmark criteria to judge the performance of individual methods.
Figure 1.
Scatterplots of hippocampal volume (cm3) by method. Spearman correlations and p-values are shown for each pair.
Table 2.
ROC Curves Comparing Prognostic Performance of Methods A, B, and C for Progression from MCI to AD within two years
| Area under the ROC curve for each hippocampal volume method predicting stable vs. converter at 24 months. |
AUROC |
|---|---|
| Method A | 0.675 |
| Method B | 0.678 |
| Method C | 0.625 |
One important feature of the process for critically evaluating automated hippocampal segmentation algorithms is the failure rate. For a variety of reasons, usually related to poor scan quality, automated algorithms will fail to produce a plausible result in some proportion of cases in a study. Taken to the extreme, imagine, for example, a method that produced perfect predictive results in cases that underwent successful hippocampal segmentation, but the method failed in 99% of the time. The method would score quite well on prognostic metrics, but would not be practical. A fair and objective approach therefore is needed to penalize automated segmentation algorithms that fail in an unacceptably high proportion of cases.
6. Future efforts
Although a position paper is a first step, the objective of standardizing hippocampal volumetry as an AD biomarker will require active participation by stakeholders in academia and industry. The authors’ objective is to see hippocampal volumetry evolve from its current state, a measure that is valid only in specific studies or a single institution, to a universally accepted biomarker with standardized units of measure. In some cases, this could simply involve having developers of automated measurement tools directly import the EADC-ADNI anatomic definition of the hippocampal boundaries into the atlas of the automated application.
Standardizing single time point hippocampal volume as an AD biomarker is the most logical and readily achievable initial goal; however, the authors recognize that other more complex topographic structural MRI measures might be more specific, or ultimately more powerful. The major difficulty here is identifying an appropriate reference standard if an anatomically based classifier does not conform to the boundaries of a classically defined anatomic structure as the hippocampus does.
Longitudinal change measures on structural MRI should be standardized using the approach outlined above as a template. This could include an extension of the EADC-ADNI effort to include expert manual tracing of serial hippocampi to create a longitudinal reference standard dataset using the same model as the single time point dataset proposed in this position paper.
FDG PET, amyloid PET imaging, and possibly other MRI modalities (e.g., resting state functional connectivity, diffusion tensor imaging, and arterial spin labeled perfusion imaging) are also important imaging biomarkers for AD. Pursuing standardized quantitative metrics for these imaging modalities is a high priority. The efforts to standardize, validate and evaluate quantitative measures in these modalities could roughly follow the same approach outlined above for hippocampal volume.
For all imaging biomarkers, future efforts will need to focus on developing a quantitative score to allow the assessment of individual imaging biomarker measures against well-developed norms that incorporate other appropriate covariates, such as age, sex and head size are for the hippocampus (91, 92).
Ideally, diagnostic biomarkers should be evaluated against post-mortem histopathological findings. It is well established that hippocampal atrophy while a feature of AD, is not specific for AD as it occurs in other conditions (32, 93).
To optimize the use of biomarkers in new AD diagnostic criteria - future efforts will need to focus on establishing diagnostic cut points in the continuous range of quantitative values to identify normal, abnormal, and indeterminate levels in individual subjects. For use in clinical practice, quantitative metrics will need to be developed and then tested in clinically typical and representative populations. Diagnostic biomarkers in AD should function analogously to those in other diseases where, for example, cut points in the continuous range of blood pressure and fasting serum glucose are universally recognized as useful in aiding the diagnosis of hypertension and diabetes and standardized treatment protocols are based on these biomarker cut points. For the purposes of diagnosis in typical clinical settings, cut points should be derived from carefully characterized groups of subjects chosen in such a way that the results can be generalized to the overall population. For example, ADNI subjects were selected to represent a typical AD clinical trial, with specific inclusion/exclusion criteria. Thus the results from ADNI are not generalizable to the overall population and are not optimal to generate normative data for general diagnostic purposes. Selecting meaningful diagnostic cut points is complicated by the fact the many cognitively normal elderly subjects harbor significant AD pathology. Thus the definition of normal is not straightforward. Consensus guidelines have been established for evaluating and reporting the clinical utility of diagnostic biomarkers and should be followed in studies using the results of the assay validation steps described here. In clinical settings, the sensitivity of detecting AD should exceed 80% and specificity for distinguishing AD from other similar dementias also should exceed 80% (94). Standardized reporting of results should follow STARD criteria (95) and for clinical settings additional reporting criteria to demonstrate pragmatic utility are needed (96).
Supplementary Material
Acknowledgements
Heather J Wiste, Mayo Clinic, data analysis
Footnotes
Disclosures
Clifford R. Jack Jr. serves as a consultant for Janssen, Lilly, GE, Johnson and Johnson, Eisai, and Élan. He is an investigator in clinical trials sponsored by Pfizer, Allon and Baxter, Inc. He receives research funding from the NIA [R01-AG11378 (PI), P50-AG16574 (Co-I), R21-AG38736 (Co-I), and U01 AG024904-01 (Co-I)], and the Alexander Family Alzheimer’s Disease Research Professorship of the Mayo Foundation. He owns stock in Johnson and Johnson. Frederik Barkhof has received compensation for consulting from Roche, Lundbeck, Janssen Alz Immunotherapy, and GE Medical Systems. Matthew A. Berstein receives research contract funding from Pfizer Inc., and Baxter Allon. Marc Cantillon is an employee of Coalition Against Major Diseases.
Patricia E. Cole reports no conflicts. Charles DeCarli reports no conflicts. Bruno Dubois has served on the advisory boards of GE healthcare and Eisai/Pfizer. Simon Duchesne reports no conflicts. Nick C. Fox served on the scientific advisory boards of Alzheimer’s Research Forum, Alzheimer’s Society and Alzheimer’s Research Trust and editorial boards of Alzheimer’s Disease and Associated Disorders; Neurodegenerative Diseases and BioMed Central - Alzheimer’s Research and Therapy. He holds a patent for QA Box that may accrue revenue. In the last five years his research group has received payment for consultancy or for conducting studies from Abbott Laboratories, Elan Pharmaceuticals, Eisai, Eli Lilly, GE Healthcare, IXICO, Lundbeck, Pfizer Inc, Sanofi-Aventis and Wyeth Pharmaceuticals. He receives research support from MRC [G0801306 (PI), G0601846 (PI)] NIH [U01 AG024904 (Co-investigator(sub contract)], Alzheimer Research Trust [ART/RF/2007/1 (PI)], NIHR (Senior Investigator) and EPSRC [GR/S48844/01 (PI)]. Giovanni B. Frisoni serves/has served on the advisory boards for Lilly, BMS, Bayer, Lundbeck, Elan, Astra Zeneca, Pfizer, Taurx, and Wyeth. He is a member of the editorial boards of Lancet Neurology, Aging Clinical & Experimental Research, Alzheimer’s Disease & Associated Disorders, and Neurodegenerative Diseases. He serves as the Imaging Selection Editor of Neurobiology of Aging. He has received grants from Wyeth Int’l, Lilly Int’l, Lundbeck Italia, and the Alzheimer’s Association. Harald Hampel disclosed no conflicts. Derek L.G. Hill is an employee and stock holder in IXICO Ltd. Keith Johnson reports no conflicts. Jean-Francois Mangin reports no conflicts. Philip Scheltens serves/has served on the advisory boards of Genentech, Novartis, Roche, Danone, Nutricia, Baxter and Lundbeck. He has been a speaker at symposia organized by Lundbeck, Merz, Danone, Novartis, Roche and Genentech. For all his activities he receives no personal compensation.
He serves on the editorial board of Alzheimer’s Research & Therapy and Alzheimers Disease and Associated Disorders, is a member of the scientific advisory board of the EU Joint Programming Initiative and the French National Plan Alzheimer. The Alzheimer Center receives unrestricted funding from various sources through the VUmc Fonds. Adam J. Schwartz is an employee and shareholder of Eli Lilly and Company. Reisa Sperling has been a consultant for Pfizer, Janssen, Elan, Bristol-Myers-Squibb, Bayer and Avid (unpaid). She is a site investigator for clinical trials for Bristol-Myers-Squibb, Pfizer, Janssen, and Avid. Joyce Suhy is an employee of Synarc. Paul M. Thompson reports no conflicts. Michael Weiner serves/has served on the advisory boards of Elan/Wyeth, Novartis, Banner, VACO, Lilly, Araclon and Insitut Catala de Neurociencies Aplicades, Biogen Idec, and Pfizer. He serves/has served as a consultant for Elan/Wyeth, Novartis, Forest, Ipsen, Daiichi Sankyo, Inc., Astra Zeneca, Araclon, Medivation/Pfizer, TauRx Therapeutics LTD, Bayer Healthcare, Biogen Idec, Exonhit Therapeutics, SA, Servier, Synarc, and Pfizer. Funding for his travel has been provided by Elan/Wyeth, Alzheimer’s Association, Forest, University of California, Davis, Tel-Aviv University Medical School, Colloquium Paris, Ipsen, Wenner-Gren Foundations, Social Security Administration, Korean Neurological Association, National Institutes of Health, Washington University at St. Louis, Banner Alzheimer’s Institute, CTAD, Veterans Affairs Central Office, Beijing Institute of Geriatrics, Innogenetics, New York University, NeuroVigil, Inc., CHRU-Hospital Roger Salengro, Siemens, AstraZeneca, Geneva University Hospitals, Lilly, University of California, San Diego – ADNI, Paris University, Institut Catala de Neurociencies Aplicades, University of New Mexico School of Medicine, Pfizer, Paul Sabatier University, and Novartis. He serves on the editorial advisory boards of Alzheimer’s and Dementia and MRI. He has received honoraria from American Academy of Neurology, Ipsen, NeuroVigil, Inc., and Insitut Catala de Neurociencies Aplicades. He receives research support from Merck and Avid. He owns stock in Synarc and Elan. Norman L. Foster disclosed no conflicts.
References
- 1.Biomarkers Definitions Working Group Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001;69(3):89–95. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]
- 2.Hampel H, Frank R, Broich K, Teipel SJ, Katz RG, Hardy J, et al. Biomarkers for Alzheimer’s disease: academic, industry and regulatory perspectives. Nat Rev Drug Discov. 2010;9(7):560–74. doi: 10.1038/nrd3115. [DOI] [PubMed] [Google Scholar]
- 3.Jack CR, Jr., Knopman DS, Jagust WJ, Shaw LM, Aisen PS, Weiner MW, et al. Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade. Lancet Neurol. 2010;9(1):119–28. doi: 10.1016/S1474-4422(09)70299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology. 1984;34(7):939–44. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 5.Dubois B, Feldman HH, Jacova C, Dekosky ST, Barberger-Gateau P, Cummings J, et al. Research criteria for the diagnosis of Alzheimer’s disease: revising the NINCDS-ADRDA criteria. Lancet Neurol. 2007;6(8):734–46. doi: 10.1016/S1474-4422(07)70178-3. [DOI] [PubMed] [Google Scholar]
- 6.Dubois B, Feldman HH, Jacova C, Cummings JL, Dekosky ST, Barberger-Gateau P, et al. Revising the definition of Alzheimer’s disease: a new lexicon. Lancet Neurol. 2010 doi: 10.1016/S1474-4422(10)70223-4. [DOI] [PubMed] [Google Scholar]
- 7.Albert MS, DeKosky ST, Dickson D, Dubois B, Feldman HH, Fox NC, et al. The diagnosis of mild cognitive impairment due to Alzheimer’s disease: Recommendations from the National Institute on Aging and Alzheimer’s Association Workgroup. Alzheimers Dement. 2011 doi: 10.1016/j.jalz.2011.03.008. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jack CR, Jr., Albert MS, Knopman DS, McKhann GM, Sperling RA, Carillo M, et al. Introduction to the recommendations from the National Institute on Aging and the Alzheimer Association workgroup on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011 doi: 10.1016/j.jalz.2011.03.004. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR, Jr., Kawas CH, et al. The diagnosis of dementia due to Alzheimer’s disease: Recommendations from the National Institute on Aging and the Alzheimer’s Assocation Workgroup. Alzheimers Dement. 2011 doi: 10.1016/j.jalz.2011.03.005. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sperling RA, Aisen PS, Beckett LA, Bennett DA, Craft S, Fagan AM, et al. Toward defining the preclinical stages of Alzheimer’s disease: Recommendations from the National Institute on Aging and the Alzheimer Assocation Workgroup. Alzheimers Dement. 2011 doi: 10.1016/j.jalz.2011.03.003. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McEvoy L, Hagler D, Holland D, Roddey J, Fennema-Notestine C, Salmon D, et al. Neuroimaging Enrichment Strategy for Secondary Prevention Trials in Alzheimer’s Disease. Alzheimer’s Disease and Related Disorders. 2010;24(3):269–77. doi: 10.1097/WAD.0b013e3181d1b814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fox NC, Black RS, Gilman S, Rossor MN, Griffith SG, Jenkins L, et al. Effects of Abeta immunization (AN1792) on MRI measures of cerebral volume in Alzheimer disease. Neurology. 2005;64(9):1563–72. doi: 10.1212/01.WNL.0000159743.08996.99. [DOI] [PubMed] [Google Scholar]
- 13.Holland D, Brewer JB, Hagler DJ, Fenema-Notestine C, Dale AM, Weiner M, et al. Subregional neuroanatomical change as a biomarker for Alzheimer’s disease. Proc Natl Acad Sci U S A. 2009;106(49):20954–20959. doi: 10.1073/pnas.0906053106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jack CR, Jr., Slomkowski M, Gracon S, Hoover TM, Felmlee JP, Stewart K, et al. MRI as a biomarker of disease progression in a therapeutic trial of milameline for AD. Neurology. 2003;60(2):253–60. doi: 10.1212/01.wnl.0000042480.86872.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schott JM, Frost C, Whitwell JL, Macmanus DG, Boyes RG, Rossor MN, et al. Combining short interval MRI in Alzheimer’s disease: Implications for therapeutic trials. J Neurol. 2006;253(9):1147–53. doi: 10.1007/s00415-006-0173-4. [DOI] [PubMed] [Google Scholar]
- 16.Fox NC, Cousens S, Scahill R, Harvey RJ, Rossor MN. Using serial registered brain magnetic resonance imaging to measure disease progression in Alzheimer disease: power calculations and estimates of sample size to detect treatment effects. Arch Neurol. 2000;57(3):339–44. doi: 10.1001/archneur.57.3.339. [DOI] [PubMed] [Google Scholar]
- 17.Leung KK, Clarkson MJ, Bartlett JW, Clegg S, Jack CR, Jr., Weiner MW, et al. Robust atrophy rate measurement in Alzheimer’s disease using multi-site serial MRI: Tissue-specific intensity normalization and parameter selection. Neuroimage. 2010;50(2):516–523. doi: 10.1016/j.neuroimage.2009.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hua X, Leow AD, Lee S, Klunder AD, Toga AW, Lepore N, et al. 3D characterization of brain atrophy in Alzheimer’s disease and mild cognitive impairment using tensor-based morphometry. Neuroimage. 2008;41(1):19–34. doi: 10.1016/j.neuroimage.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hua X, Gutman B, Boyle C, Rajagopalan P, Leow AD, Yanovsky I, et al. Accurate measurement of brain changes in longitudinal MRI scans using tensor-based morphometry. Neuroimage. 2011 doi: 10.1016/j.neuroimage.2011.01.079. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vemuri P, Wiste HJ, Weigand SD, Knopman DS, Trojanowski JQ, Shaw LM, et al. Serial MRI and CSF Biomarkers in Normal Aging, MCI and AD. Neurology. 2010;75(2):143–151. doi: 10.1212/WNL.0b013e3181e7ca82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolz R, Heckemann RA, Aljabar P, Hajnal JV, Hammers A, Lotjonen J, et al. Measurement of hippocampal atrophy using 4D graph-cut segmentation: application to ADNI. Neuroimage. 2010;52(1):109–18. doi: 10.1016/j.neuroimage.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 22.Hampel H, Wilcock G, Andrieu S, Aisen P, Blennow K, Broich K, et al. Biomarkers for Alzheimer’s disease therapeutic trials. Prog Neurobiol. 2010 doi: 10.1016/j.pneurobio.2010.11.005. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 23.Jack CR, Jr., Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, et al. The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008;27(4):685–91. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jagust WJ, Bandy D, Chen K, Foster NL, Landau SM, Mathis CA, et al. The Alzheimer’s Disease Neuroimaging Initiative positron emission tomography core. Alzheimers Dement. 2010;6(3):221–9. doi: 10.1016/j.jalz.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geuze E, Vermetten E, Bremner JD. MR-based in vivo hippocampal volumetrics: 1. Review of methodologies currently employed. Mol Psychiatry. 2005;10(2):147–59. doi: 10.1038/sj.mp.4001580. [DOI] [PubMed] [Google Scholar]
- 26.Barnes J, Bartlett JW, van de Pol LA, Loy CT, Scahill RI, Frost C, et al. A meta-analysis of hippocampal atrophy rates in Alzheimer’s disease. Neurobiol Aging. 2009;30(11):1711–23. doi: 10.1016/j.neurobiolaging.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Du AT, Schuff N, Kramer JH, Ganzer S, Zhu XP, Jagust WJ, et al. Higher atrophy rate of entorhinal cortex than hippocampus in AD. Neurology. 2004;62(3):422–7. doi: 10.1212/01.wnl.0000106462.72282.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jack CR, Jr., Shiung MM, Gunter JL, O’Brien PC, Weigand SD, Knopman DS, et al. Comparison of different MRI brain atrophy rate measures with clinical disease progression in AD. Neurology. 2004;62(4):591–600. doi: 10.1212/01.wnl.0000110315.26026.ef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Swank JS, Glick IE, Gado MH, Miller MI, Morris JC, et al. Changes in hippocampal volume and shape across time distinguish dementia of the Alzheimer type from healthy aging. Neuroimage. 2003;20(2):667–82. doi: 10.1016/S1053-8119(03)00361-6. [DOI] [PubMed] [Google Scholar]
- 30.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82(4):239–59. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 31.Bobinski M, de Leon MJ, Wegiel J, Desanti S, Convit A, Saint Louis LA, et al. The histological validation of post mortem magnetic resonance imaging-determined hippocampal volume in Alzheimer’s disease. Neuroscience. 2000;95(3):721–5. doi: 10.1016/s0306-4522(99)00476-5. [DOI] [PubMed] [Google Scholar]
- 32.Jack CR, Jr., Dickson DW, Parisi JE, Xu YC, Cha RH, O’Brien PC, et al. Antemortem MRI findings correlate with hippocampal neuropathology in typical aging and dementia. Neurology. 2002;58(5):750–7. doi: 10.1212/wnl.58.5.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gosche KM, Mortimer JA, Smith CD, Markesbery WR, Snowdon DA. Hippocampal volume as an index of Alzheimer neuropathology: findings from the Nun Study. Neurology. 2002;58(10):1476–82. doi: 10.1212/wnl.58.10.1476. [DOI] [PubMed] [Google Scholar]
- 34.Csernansky JG, Hamstra J, Wang L, McKeel D, Price JL, Gado M, et al. Correlations between antemortem hippocampal volume and postmortem neuropathology in AD subjects. Alzheimer Dis Assoc Disord. 2004;18(4):190–5. [PubMed] [Google Scholar]
- 35.Silbert LC, Quinn JF, Moore MM, Corbridge E, Ball MJ, Murdoch G, et al. Changes in premorbid brain volume predict Alzheimer’s disease pathology. Neurology. 2003;61(4):487–92. doi: 10.1212/01.wnl.0000079053.77227.14. [DOI] [PubMed] [Google Scholar]
- 36.Zarow C, Vinters HV, Ellis WG, Weiner MW, Mungas D, White L, et al. Correlates of hippocampal neuron number in Alzheimer’s disease and ischemic vascular dementia. Ann Neurol. 2005;57(6):896–903. doi: 10.1002/ana.20503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Colliot O, Chetelat G, Chupin M, Desgranges B, Magnin B, Benali H, et al. Discrimination between Alzheimer disease, mild cognitive impairment, and normal aging by using automated segmentation of the hippocampus. Radiology. 2008;248(1):194–201. doi: 10.1148/radiol.2481070876. [DOI] [PubMed] [Google Scholar]
- 38.Jack CR, Jr., Petersen RC, O’Brien PC, Tangalos EG. MR-based hippocampal volumetry in the diagnosis of Alzheimer’s disease. Neurology. 1992;42(1):183–8. doi: 10.1212/wnl.42.1.183. [DOI] [PubMed] [Google Scholar]
- 39.Killiany RJ, Gomez-Isla T, Moss M, Kikinis R, Sandor T, Jolesz F, et al. Use of structural magnetic resonance imaging to predict who will get Alzheimer’s disease. Ann Neurol. 2000;47(4):430–9. [PubMed] [Google Scholar]
- 40.McEvoy LK, Fennema-Notestine C, Roddey JC, Hagler DJ, Jr., Holland D, Karow DS, et al. Alzheimer disease: quantitative structural neuroimaging for detection and prediction of clinical and structural changes in mild cognitive impairment. Radiology. 2009;251(1):195–205. doi: 10.1148/radiol.2511080924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Desikan RS, Cabral HJ, Hess CP, Dillon WP, Glastonbury CM, Weiner MW, et al. Automated MRI measures identify individuals with mild cognitive impairment and Alzheimer’s disease. Brain. 2009;132(Pt 8):2048–57. doi: 10.1093/brain/awp123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gerardin E, Chetelat G, Chupin M, Cuingnet R, Desgranges B, Kim HS, et al. Multidimensional classification of hippocampal shape features discriminates Alzheimer’s disease and mild cognitive impairment from normal aging. Neuroimage. 2009;47(4):1476–86. doi: 10.1016/j.neuroimage.2009.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dickerson BC, Goncharova I, Sullivan MP, Forchetti C, Wilson RS, Bennett DA, et al. MRI-derived entorhinal and hippocampal atrophy in incipient and very mild Alzheimer’s disease. Neurobiol Aging. 2001;22(5):747–54. doi: 10.1016/s0197-4580(01)00271-8. [DOI] [PubMed] [Google Scholar]
- 44.McDonald CR, Gharapetian L, McEvoy LK, Fennema-Notestine C, Hagler DJ, Jr., Holland D, et al. Relationship between regional atrophy rates and cognitive decline in mild cognitive impairment. Neurobiol Aging. 2010 doi: 10.1016/j.neurobiolaging.2010.03.015. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarazin M, Chauvire V, Gerardin E, Colliot O, Kinkingnehun S, de Souza LC, et al. The amnestic syndrome of hippocampal type in Alzheimer’s disease: an MRI study. J Alzheimers Dis. 2010;22(1):285–94. doi: 10.3233/JAD-2010-091150. [DOI] [PubMed] [Google Scholar]
- 46.Fox NC, Warrington EK, Freeborough PA, Hartikainen P, Kennedy AM, Stevens JM, et al. Presymptomatic hippocampal atrophy in Alzheimer’s disease. A longitudinal MRI study. Brain. 1996;119(Pt 6):2001–7. doi: 10.1093/brain/119.6.2001. [DOI] [PubMed] [Google Scholar]
- 47.Jack CR, Jr., Petersen RC, Xu Y, O’Brien PC, Smith GE, Ivnik RJ, et al. Rates of hippocampal atrophy correlate with change in clinical status in aging and AD. Neurology. 2000;55(4):484–89. doi: 10.1212/wnl.55.4.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jack CR, Jr., Petersen RC, Xu YC, O’Brien PC, Smith GE, Ivnik RJ, et al. Prediction of AD with MRI-based hippocampal volume in mild cognitive impairment. Neurology. 1999;52(7):1397–403. doi: 10.1212/wnl.52.7.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Visser PJ, Scheltens P, Verhey FR, Schmand B, Launer LJ, Jolles J, et al. Medial temporal lobe atrophy and memory dysfunction as predictors for dementia in subjects with mild cognitive impairment. J Neurol. 1999;246(6):477–85. doi: 10.1007/s004150050387. [DOI] [PubMed] [Google Scholar]
- 50.Risacher SL, Saykin AJ, West JD, Shen L, Firpi HA, McDonald BC. Baseline MRI predictors of conversion from MCI to probable AD in the ADNI cohort. Curr Alzheimer Res. 2009;6(4):347–61. doi: 10.2174/156720509788929273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Henneman WJ, Sluimer JD, Barnes J, van der Flier WM, Sluimer IC, Fox NC, et al. Hippocampal atrophy rates in Alzheimer disease: added value over whole brain volume measures. Neurology. 2009;72(11):999–1007. doi: 10.1212/01.wnl.0000344568.09360.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brys M, Glodzik L, Mosconi L, Switalski R, De Santi S, Pirraglia E, et al. Magnetic resonance imaging improves cerebrospinal fluid biomarkers in the early detection of Alzheimer’s disease. J Alzheimers Dis. 2009;16(2):351–62. doi: 10.3233/JAD-2009-0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Landau SM, Harvey D, Madison CM, Reiman EM, Foster NL, Aisen PS, et al. Comparing predictors of conversion and decline in mild cognitive impairment. Neurology. 2010;75(3):230–8. doi: 10.1212/WNL.0b013e3181e8e8b8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kantarci K, Weigand SD, Przybelski SA, Shiung MM, Whitwell JL, Negash S, et al. Risk of dementia in MCI: combined effect of cerebrovascular disease, volumetric MRI, and 1H MRS. Neurology. 2009;72(17):1519–25. doi: 10.1212/WNL.0b013e3181a2e864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karas G, Sluimer J, Goekoop R, van der Flier W, Rombouts SA, Vrenken H, et al. Amnestic mild cognitive impairment: structural MR imaging findings predictive of conversion to Alzheimer disease. AJNR Am J Neuroradiol. 2008;29(5):944–9. doi: 10.3174/ajnr.A0949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jack CR, Jr., Wiste HJ, Vemuri P, Weigand SD, Senjem ML, Zeng G, et al. Brain beta-amyloid measure and magnetic resonance imaging atophy both predict time-to-progression from mild cognitive impairment to Alzheimer’s disease. Brain. 2010;133(11):3336–3348. doi: 10.1093/brain/awq277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fleisher AS, Sun S, Taylor C, Ward CP, Gamst AC, Petersen RC, et al. Volumetric MRI vs clinical predictors of Alzheimer disease in mild cognitive impairment. Neurology. 2008;70(3):191–9. doi: 10.1212/01.wnl.0000287091.57376.65. [DOI] [PubMed] [Google Scholar]
- 58.Leung KK, Barnes J, Ridgway GR, Bartlett JW, Clarkson MJ, Macdonald K, et al. Automated cross-sectional and longitudinal hippocampal volume measurement in mild cognitive impairment and Alzheimer’s disease. Neuroimage. 2010;51(4):1345–59. doi: 10.1016/j.neuroimage.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Devanand DP, Pradhaban G, Liu X, Khandji A, De Santi S, Segal S, et al. Hippocampal and entorhinal atrophy in mild cognitive impairment: prediction of Alzheimer disease. Neurology. 2007;68(11):828–36. doi: 10.1212/01.wnl.0000256697.20968.d7. [DOI] [PubMed] [Google Scholar]
- 60.Apostolova LG, Dutton RA, Dinov ID, Hayashi KM, Toga AW, Cummings JL, et al. Conversion of mild cognitive impairment to Alzheimer disease predicted by hippocampal atrophy maps. Arch Neurol. 2006;63(5):693–9. doi: 10.1001/archneur.63.5.693. [DOI] [PubMed] [Google Scholar]
- 61.Apostolova LG, Mosconi L, Thompson PM, Green AE, Hwang KS, Ramirez A, et al. Subregional hippocampal atrophy predicts Alzheimer’s dementia in the cognitively normal. Neurobiol Aging. 2010;31(7):1077–88. doi: 10.1016/j.neurobiolaging.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chetelat G, Fouquet M, Kalpouzos G, Denghien I, De la Sayette V, Viader F, et al. Three-dimensional surface mapping of hippocampal atrophy progression from MCI to AD and over normal aging as assessed using voxel-based morphometry. Neuropsychologia. 2008;46(6):1721–31. doi: 10.1016/j.neuropsychologia.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 63.Beckett LA, Harvey DJ, Gamst A, Donohue M, Kornak J, Zhang H, et al. The Alzheimer’s Disease Neuroimaging Initiative: Annual change in biomarkers and clinical outcomes. Alzheimers Dement. 2010;6(3):257–64. doi: 10.1016/j.jalz.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33(3):341–55. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- 65.Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage. 1999;9(2):179–94. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- 66.Barnes J, Foster J, Boyes RG, Pepple T, Moore EK, Schott JM, et al. A comparison of methods for the automated calculation of volumes and atrophy rates in the hippocampus. Neuroimage. 2008;40(4):1655–71. doi: 10.1016/j.neuroimage.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 67.Morra JH, Tu Z, Apostolova LG, Green AE, Avedissian C, Madsen SK, et al. Automated 3D mapping of hippocampal atrophy and its clinical correlates in 400 subjects with Alzheimer’s disease, mild cognitive impairment, and elderly controls. Hum Brain Mapp. 2009;30(9):2766–88. doi: 10.1002/hbm.20708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim H, Besson P, Colliot O, Bernasconi A, Bernasconi N. Surface-based vector analysis using heat equation interpolation: a new approach to quantify local hippocampal volume changes. Med Image Comput Comput Assist Interv. 2008;11(Pt 1):1008–15. doi: 10.1007/978-3-540-85988-8_120. [DOI] [PubMed] [Google Scholar]
- 69.Mouiha A, Duchesne S. HIPPOCAMPAL ATROPHY RATES IN ALZHEIMER’S DISEASE: AUTOMATED SEGMENTATION VARIABILITY ANALYSIS. Neurosci Lett. 2011 doi: 10.1016/j.neulet.2011.02.065. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 70.Chupin M, Hammers A, Liu RS, Colliot O, Burdett J, Bardinet E, et al. Automatic segmentation of the hippocampus and the amygdala driven by hybrid constraints: method and validation. Neuroimage. 2009;46(3):749–61. doi: 10.1016/j.neuroimage.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brewer JB, Magda S, Airriess C, Smith ME. Fully-automated quantification of regional brain volumes for improved detection of focal atrophy in Alzheimer disease. AJNR Am J Neuroradiol. 2009;30(3):578–80. doi: 10.3174/ajnr.A1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Scheltens P, Leys D, Barkhof F, Huglo D, Weinstein HC, Vermersch P, et al. Atrophy of medial temporal lobes on MRI in “probable” Alzheimer’s disease and normal ageing: diagnostic value and neuropsychological correlates. J Neurol Neurosurg Psychiatry. 1992;55(10):967–72. doi: 10.1136/jnnp.55.10.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.de Leon MJ, Golomb J, George AE, Convit A, Tarshish CY, McRae T, et al. The radiologic prediction of Alzheimer disease: the atrophic hippocampal formation. AJNR Am J Neuroradiol. 1993;14(4):897–906. [PMC free article] [PubMed] [Google Scholar]
- 74.DeCarli C, Frisoni GB, Clark CM, Harvey D, Grundman M, Petersen RC, et al. Qualitative estimates of medial temporal atrophy as a predictor of progression from mild cognitive impairment to dementia. Arch Neurol. 2007;64(1):108–15. doi: 10.1001/archneur.64.1.108. [DOI] [PubMed] [Google Scholar]
- 75.Jack CR, Jr., Sharbrough FW, Twomey CK, Cascino GD, Hirschorn KA, Marsh WR, et al. Temporal lobe seizures: lateralization with MR volume measurements of the hippocampal formation. Radiology. 1990;175(2):423–9. doi: 10.1148/radiology.175.2.2183282. [DOI] [PubMed] [Google Scholar]
- 76.Teipel SJ, Ewers M, Wolf S, Jessen F, Kolsch H, Arlt S, et al. Multicentre variability of MRI-based medial temporal lobe volumetry in Alzheimer’s disease. Psychiatry Res. 2010;182(3):244–50. doi: 10.1016/j.pscychresns.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 77.The ENIGMA Consortium Genome-Wide Association Meta-Analysis of Hippocampal Volume via the ENIGMA Consortium; Organization for Human Brain Mapping Meeting; Quebec City, Canada. 2011. [Google Scholar]
- 78.van de Pol LA, Barnes J, Scahill RI, Frost C, Lewis EB, Boyes RG, et al. Improved reliability of hippocampal atrophy rate measurement in mild cognitive impairment using fluid registration. Neuroimage. 2007;34(3):1036–41. doi: 10.1016/j.neuroimage.2006.10.033. [DOI] [PubMed] [Google Scholar]
- 79.Vemuri P, Gunter JL, Senjem ML, Whitwell JL, Kantarci K, Knopman DS, et al. Alzheimer’s disease diagnosis in individual subjects using structural MR images: validation studies. Neuroimage. 2008;39(3):1186–97. doi: 10.1016/j.neuroimage.2007.09.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Davatzikos C, Fan Y, Wu X, Shen D, Resnick SM. Detection of prodromal Alzheimer’s disease via pattern classification of magnetic resonance imaging. Neurobiol Aging. 2008;29(4):514–23. doi: 10.1016/j.neurobiolaging.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fan Y, Shen D, Davatzikos C. Classification of structural images via high-dimensional image warping, robust feature extraction, and SVM. Med Image Comput Comput Assist Interv Int Conf Med Image Comput Comput Assist Interv. 2005;8(Pt 1):1–8. doi: 10.1007/11566465_1. [DOI] [PubMed] [Google Scholar]
- 82.Stonnington CM, Tan G, Kloppel S, Chu C, Draganski B, Jack CR, Jr., et al. Interpreting scan data acquired from multiple scanners: a study with Alzheimer’s disease. Neuroimage. 2008;39(3):1180–5. doi: 10.1016/j.neuroimage.2007.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kloppel S, Stonnington CM, Chu C, Draganski B, Scahill RI, Rohrer JD, et al. Automatic classification of MR scans in Alzheimer’s disease. Brain. 2008;131(Pt 3):681–9. doi: 10.1093/brain/awm319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Duchesne S, Caroli A, Geroldi C, Barillot C, Frisoni GB, Collins DL. MRI-based automated computer classification of probable AD versus normal controls. IEEE Trans Med Imaging. 2008;27(4):509–20. doi: 10.1109/TMI.2007.908685. [DOI] [PubMed] [Google Scholar]
- 85.Knopman DS, DeKosky ST, Cummings JL, Chui H, Corey-Bloom J, Relkin N, et al. Practice parameter: diagnosis of dementia (an evidence-based review). Report of the Quality Standards Subcommittee of the American Academy of Neurology. Neurology. 2001;56(9):1143–53. doi: 10.1212/wnl.56.9.1143. [DOI] [PubMed] [Google Scholar]
- 86.Hort J, O’Brien JT, Gainotti G, Pirttila T, Popescu BO, Rektorova I, et al. EFNS guidelines for the diagnosis and management of Alzheimer’s disease. Eur J Neurol. 2010;17(10):1236–48. doi: 10.1111/j.1468-1331.2010.03040.x. [DOI] [PubMed] [Google Scholar]
- 87.Boccardi M, Ganzola R, Duchesne S, Redolfi A, Bartzokis G, Csernansky J, et al. ICAD - Alzheimer’s Imaging Consortium . Survey of segmentation protocols for manual hippocampal volumetry: Prepatory phase for an EADC-ADNI harmonization protocol. Honolulu, HI: 2010. [Google Scholar]
- 88.Frisoni GB, Jack CR. Harmonization of magnetic resonance-based manual hippocampal segmentation: A mandatory step for wide clinical use. Alzheimers Dement. 2011;7(2):171–4. doi: 10.1016/j.jalz.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 89.Fox NC, Ridgway GR, Schott JM. Algorithms, atrophy and Alzheimer’s disease: Cautionary tales for clinical trials. Neuroimage. 2011 doi: 10.1016/j.neuroimage.2011.01.077. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 90.Cover KS, van Schijndel RA, van Dijk BW, Redolfi A, Knol DL, Frisoni GB, et al. Assessing the reproducibility of the SienaX and Siena brain atrophy measures using the ADNI back-to-back MP-RAGE MRI scans. Psychiatry Research. doi: 10.1016/j.pscychresns.2011.02.012. In Press. [DOI] [PubMed] [Google Scholar]
- 91.Barnes J, Ridgway GR, Bartlett J, Henley SM, Lehmann M, Hobbs N, et al. Head size, age and gender adjustment in MRI studies: a necessary nuisance? Neuroimage. 2010;53(4):1244–55. doi: 10.1016/j.neuroimage.2010.06.025. [DOI] [PubMed] [Google Scholar]
- 92.Jack CR, Jr., Petersen RC, Xu YC, Waring SC, O’Brien PC, Tangalos EG, et al. Medial temporal atrophy on MRI in normal aging and very mild Alzheimer’s disease. Neurology. 1997;49(3):786–94. doi: 10.1212/wnl.49.3.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jagust WJ, Zheng L, Harvey DJ, Mack WJ, Vinters HV, Weiner MW, et al. Neuropathological basis of magnetic resonance images in aging and dementia. Ann Neurol. 2008;63(1):72–80. doi: 10.1002/ana.21296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Reagan Working Group. The Ronald and Nancy Reagan Research Institute of the Alzheimer’s Association and the National Institute on Aging Working Group Consensus report of the Working Group on: “Molecular and Biochemical Markers of Alzheimer’s Disease. Neurobiol Aging. 1998;19(2):109–16. [PubMed] [Google Scholar]
- 95.Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig LM, et al. Towards complete and accurate reporting of studies of diagnostic accuracy: the STARD initiative. Fam Pract. 2004;21(1):4–10. doi: 10.1093/fampra/cmh103. [DOI] [PubMed] [Google Scholar]
- 96.Glasgow RE, Magid DJ, Beck A, Ritzwoller D, Estabrooks PA. Practical clinical trials for translating research to practice: design and measurement recommendations. Med Care. 2005;43(6):551–7. doi: 10.1097/01.mlr.0000163645.41407.09. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.