Abstract
Background
Palliative chemotherapy is often administered to terminally ill cancer patients to relieve symptoms. Yet, unnecessary use of chemotherapy can worsen patients' quality of life due to treatment-related toxicities. Thus, accurate prediction of survival in terminally ill patients can help clinicians decide on the most appropriate palliative care for these patients. However, studies have shown that clinicians often make imprecise predictions of survival in cancer patients. Hence, the purpose of this study was to create a clinical decision support tool to predict survival in cancer patients beyond 120 days after palliative chemotherapy.
Materials and Methods
Data were obtained from a retrospective study of 400 randomly selected terminally ill cancer patients in the National Cancer Centre Singapore (NCCS) from 2008 to 2009. After removing patients with missing data, there were 325 patients remaining for model development. Three classification algorithms, naive Bayes (NB), neural network (NN), and support vector machine (SVM) were used to create the models. A final model with the best prediction performance was then selected to develop the tool.
Results
The NN model had the best prediction performance. The accuracy, specificity, sensitivity, and area under the curve (AUC) of this model were 78%, 82%, 74%, and 0.857, respectively. Five patient attributes (albumin level, alanine transaminase level (ATL), absolute neutrophil count, Eastern Cooperative Oncology Group (ECOG) status, and number of metastatic sites) were included in the model.
Conclusions
A decision support tool to predict survival in cancer patients beyond 120 days after palliative chemotherapy was created. With further validation, this tool coupled with the professional judgment of clinicians can help improve patient care.
Introduction
Palliative chemotherapy is often administered to terminally ill cancer patients to relieve symptoms and has been shown to be able to extend life in some cases.1,2 However, unnecessary use of chemotherapy can worsen patients' quality of life due to undesirable treatment-related toxicities. Hence, accurate prediction of survival before the initiation of palliative chemotherapy can assist clinicians to make informed decisions on the most appropriate palliative care for their patients and to avoid inappropriate treatments and treatment-related toxicities.
A systematic review of eight studies and a recent prospective cohort study has revealed that clinicians tend to make imprecise and overly optimistic predictions of survival.3,4 Identification of potential prognostic factors and the use of traditional statistical techniques to develop survival prediction models, such as the Palliative Prognostic (PaP) Score, have been studied frequently.5–7 However, the predictive performances of these models are often not optimum due to the complexity of medical data, and the inherent nature of traditional statistical methods in making assumptions on the dataset.8 Furthermore, the prognostic factors of the existing prediction models do not include treatments given to patients. Thus, their survival prediction only reflects the prognosis at the time before treatment and not after treatment has been tried. So, they are not suitable to aid clinicians in weighing the risks and benefits of using palliative chemotherapy.
Data mining (DM) is the process of discovering new patterns and relationships in a large dataset. In comparison to statistical methods, DM methods make minimum or no assumptions on the dataset. This allows complex information to be modeled easily. Moreover, prediction models created using DM methods have been shown to be more accurate compared with those created by statistical methods.8,9 Examples of the use of DM methods in oncology include determination of breast cancer treatment methods10 and development of models for predicting 2-year survival in lung cancer patients treated with radiography.11
In this study, the objective was to develop a decision support tool using DM methods to predict survival in cancer patients beyond 120 days after palliative chemotherapy. One hundred twenty days was chosen because that timeframe was considered as delayed mortality in a benchmarking study (Abstract 101, 2008 ASCO Breast Cancer Symposium). Our institution, National Cancer Centre Singapore (NCCS), also had conducted a benchmark project to establish the 30 and 120 days mortality rates after chemotherapy. Thus, we believed this decision support tool could be useful to assist clinicians in deciding whether it is beneficial to initiate palliative chemotherapy.
Material and Methods
Data collection
The study was approved by the ethics committee of NCCS. Using a random number generator, 400 randomly selected terminally ill cancer patients treated at NCCS between 2008 and 2009 were included in this study. The inclusion criteria were as follows: (1) incurable cancer in the terminal stage (expected survival length to be <3 months) and (2) defined by physician as being on palliative chemotherapy.
Patients' data were collected retrospectively from electronic databases and case notes. Patients' attributes collected are listed in Table 1.
Table 1.
Patient Data Collected
| Patient attributes | Age | Gender | Race | Height | Weight | Body mass index |
| Tumor attributes | Primary tumor site | Sites of metastases | Recurrent cancer | |||
| Treatment attributes | Chemotherapy drugs given | Cycle of therapy | Number of lines of chemotherapies | Presence of adverse effects | ||
| Prior radiotherapy | Prior surgery | |||||
| Clinical attributes | Co-morbidities | Eastern Cooperative Oncology Group (ECOG) performance status | ||||
| Laboratory attributes | Blood urea nitrogen | Creatinine | Albumin | Alanine transaminase | ||
| Aspartate transaminase | Alkaline phosphatase | White blood cell count | Red blood cell count | |||
| Hemoglobin level | Absolute neutrophil count | Absolute lymphocyte count | Absolute monocyte count | |||
| Platelet count | ||||||
Data preparation
The data were analyzed to identify records that might contain inaccurate or inconsistent data. An example of inaccurate data is that the patient had prostate cancer but gender had been keyed in as female. An example of inconsistent data is that for alanine transaminase (ALT) level, there were some inputs with values such as 0 units and 2.9 units, which were not consistent with the other inputs. These records were checked again with the original source to confirm the accuracy or consistency of the data.
Two approaches were used to handle records with missing values in some patient attributes. The first involved removing patient attributes with numerous missing values. This removed blood urea nitrogen (BUN) as its value was frequently missing. The second approach was to exclude patients with missing values. This removed 75 patients, leaving 325 patients for model development.
From the initial 28 attributes, 48 new attributes were derived. For example, age was further categorized into 2 different bands (age <65 and age ≥65) represented by binary codes, hence giving rise to 2 new attributes. The attribute, recurrent cancer, was removed due to all patients having the same data value. Thus, the final list of attributes used in model building was compiled (see supplementary Table S1 at www.liebertpub.com).
After cleaning the dataset, the study population was separated into a training set and a validation set. The training set was used in model development and internal validation. The purpose of the validation set was to validate the performance of the final model, and therefore it was not used during model building and selection. A 60/40 strategy was used to divide the study population. The training set consisted of 225 patients who received chemotherapy in 2008 and early 2009, whereas the validation set consisted of 100 patients who received chemotherapy at a later part of 2009. This strategy of dividing the dataset according to time ensures that the validation set will be able to reliably assess the historical generalizability of the final model.12
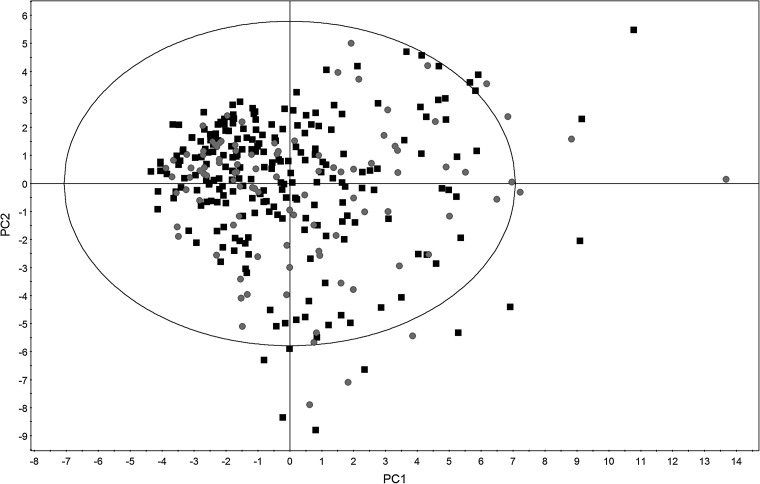
After finalizing the attributes, principal component analysis (PCA) was performed on the processed dataset. PCA provides a means for understanding the variability of the patients in the dataset by extracting a smaller series of principal components from the patients' data. The result is presented in the form of a score scatter plot constructed using the first two principal components to allow a visual analysis of the variability in the dataset. In addition, any differences between the training and validation set can also be determined.
Model development
During model development, a subset of the available attributes was randomly selected and the forward selection (FS) method13 was used to identify prognostic factors for the prediction model. A classification DM algorithm was then used to create the prediction models from the selected prognostic factors and the performance of the model was assessed using a fivefold cross validation method. This random selection of attributes followed by FS was repeated 30 times to produce 30 different prediction models for the classification DM algorithm. Three classification DM algorithms, naive Bayes (NB), neural network (NN) and support vector machine (SVM), were used in this study. These algorithms have been extensively described elsewhere, and thus only a brief description is provided for NN, which was found to have the best performance.
Neural network (NN)
The NN is a network structure consisting of a number of nodes connected through directional links. Typically, a NN will consist of a set of inputs that represent the input layer of the network, one or more hidden layers of computational nodes, and, finally, an output layer of computational nodes. The type of NN used in this study is a single-layer feed-forward NN, which has only a single, hidden layer.
NN has a high pattern recognition-like ability and is a robust classifier with the ability to generalize and make decisions from a large and fuzzy dataset. Another feature of NN is its ability to handle a large number of variables despite their underlying nonlinearity. Nevertheless, the “black box” nature of NN may limit its use in the medical domain.
Selection of a model from each classification algorithm
The selection of the model with the best prediction performance from the 30 models for each classification algorithm was based on the average of the performance training (PT) and the performance cross validation (PCV) of the models. The main difference between PT and PCV is that the former refers to the area under the curve (AUC) of the model evaluated using the entire training set, whereas the latter refers to the AUC of the model evaluated during the fivefold cross validation. By calculating the average performance, equal weighting is assigned to both PT and PCV.
The receiver operating characteristic (ROC) curve plots the true positive rate or sensitivity on the vertical axis, and the false positive rate or 1 minus specificity on the horizontal axis. The AUC is equivalent to the probability that the model will rank a randomly chosen positive instance higher than a randomly chosen negative instance. Usually, the larger the AUC, the greater will be the performance of the model. Henceforth, AUC serves as a metric of overall model quality.
Final model selection and external validation
A final model was selected from the three models based on the average performance. The performance of this model was then evaluated using the validation set (100 patients). The respective accuracy, specificity, and sensitivity of the model were calculated using the following equations and the AUC of this final model was also determined.
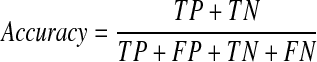 |
(2) |
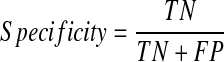 |
(3) |
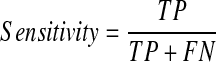 |
(4) |
Software tools
SIMCA-P+ version 12.0.1 (Umetrics AB, Umea, Sweden) was used to perform PCA. Statistical Package for the Social Sciences (SPSS) version 19.0 (IBM, New York) was used to perform statistical analysis on the dataset. RapidMiner version 5.0.010 (Rapid-I, Dortmund, Germany), an open-source system for DM, was used to create the models.
Results
Patients characteristics and data collection
Demographics, clinical information, and laboratory parameters were collected from the 325 patients (see supplementary Tables S2 to S4 at www.liebertpub.com). A score scatter plot was obtained after performing PCA on the dataset (Fig. 1). From the score scatter plot, it was observed that both the training and validation sets were randomly scattered throughout the four quadrants. In addition, the dense and sparse regions for both sets were similar. These results suggested that the patient characteristics between the training and validation sets were similar.
FIG. 1.
Score scatter plot constructed using PCA. The black squares represent patients in the training set, whereas the gray circles represent patients in the validation set.
Model selection from each classification algorithm
A total of 90 models were built, and 3 models were selected based on the criterion mentioned earlier. Details on the chosen models are summarized in Table 2.
Table 2.
Performance of the Three Models and Attributes Used for Each Model
| Model 1 | Model 2 | Model 3 | |
|---|---|---|---|
| Classification algorithm | NB | NN | SVM |
| Performance training | 0.895 | 0.995 | 0.924 |
| Performance cross validation | 0.897 | 0.869 | 0.916 |
| Average of the two performances | 0.896 | 0.932 | 0.920 |
| Types of attributes used | Albumin, ALP, ECOG status 0-1, platelets <140×109 L, number of metastatic sites | Albumin, ALT, absolute neutrophil count, ECOG status 0-1, number of metastatic sites | Albumin, ECOG status 0-1, liver metastatic site, lung metastatic site, platelets >440×109 L, RBC, WBC |
ALP, alkaline phosphatase; ALT, alanine aminotransferase; ECOG, Eastern Cooperative Oncology Group; RBC, red blood cell count; WBC, white blood cell count.
Final model selection and external validation
According to Table 2, model 2 had the highest average performance of 0.932 among the three models; hence it was selected as the final model to be used as our decision support tool. The validation set was used to validate the performance of this final model. The final model achieved an accuracy of 78% with specificity of 82% and sensitivity of 74%. The AUC of this model was 0.857.
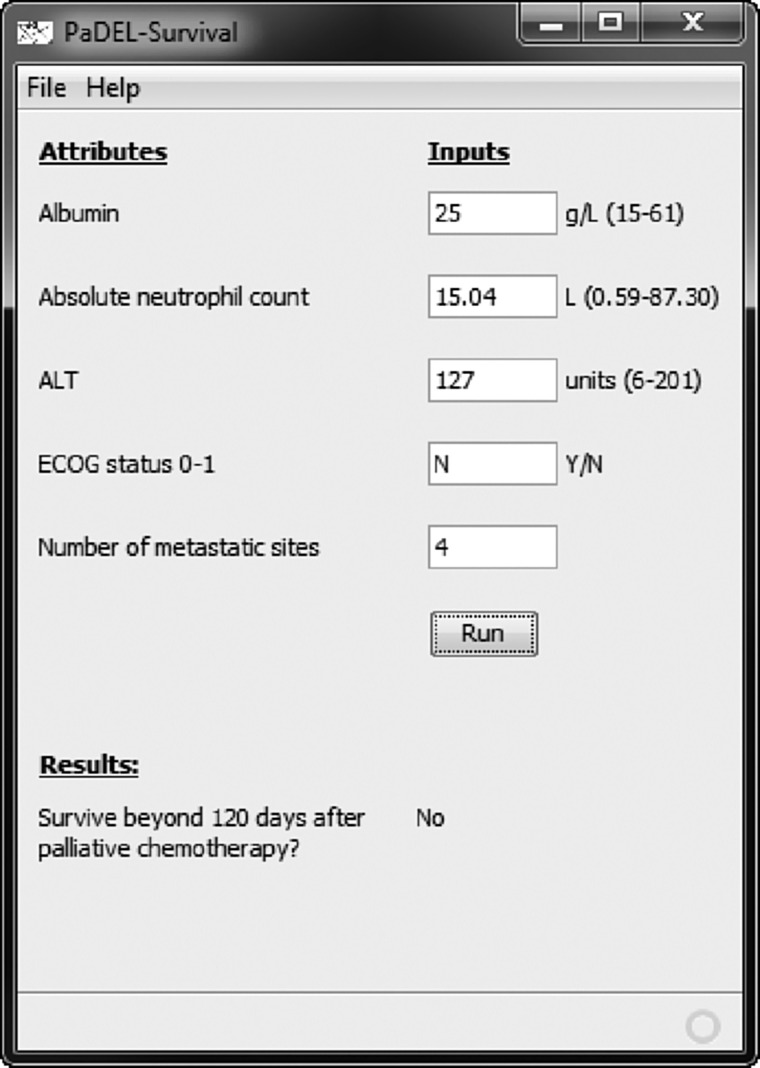
Development of a simple decision support tool
A simple clinical decision support tool was developed to ease the use of the final model (Fig. 2). This tool is available for download at http://padel.nus.edu.sg/software/padelsurvival.
FIG. 2.
Simple clinical decision support tool for determining whether a patient will survive beyond 120 days after chemotherapy.
Discussion
Model performance and application
A NN predictive model for prediction of survival beyond 120 days in terminally ill cancer patients after chemotherapy was created. To compare the predictive performance of our NN model with other existing DM models, several studies were selected for comparison. The first was a study conducted by Jayasurya and colleagues to create predictive models for 2-year survival prediction in lung cancer patients treated with radiotherapy.11 The AUCs for their Bayesian network and SVM models using the Ghent validation set were 0.77 and 0.71, respectively, which were lower than for our NN model. Nonetheless, the higher performance of our NN model must be interpreted with caution due to differences in context between the studies. To further evaluate our NN model, several prediction studies that also used the NN algorithm in different cancer types were selected for comparison. The AUCs of these previously created NN models fall in the range of 0.76 to 0.89.14–16 Therefore, our final NN model with an AUC of 0.857 can be considered to have a satisfactory predictive performance.
As mentioned earlier, clinicians tend to make imprecise and overly optimistic prediction of survival. A prospective cohort study conducted to describe doctors' prognostic accuracy in terminally ill patients found that only 20% of the predictions were accurate; 63% were overoptimistic, and 17% were overpessimistic.17 The poor accuracy of clinicians' predictions could be attributed to their tendency to overestimate survival due to the fear of diminishing hope in their patients.18 In comparison, our NN model had an accuracy of 78% with 9% overoptimistic and 13% overpessimistic. This shows the potential of our model in helping clinicians estimate survival more accurately, as our model greatly reduces the number of overoptimistic predictions without increasing the number of overpessimistic predictions.
For each patient, the NN model computes a confidence score that ranges from 0 to 1. A threshold value is then applied to the confidence score to determine whether the patient will be predicted to be able to survive beyond 120 days or not survive beyond 120 days after chemotherapy. The threshold value used in our model is set at 0.5, where values above 0.5 will be predicted as positive (survive beyond 120 days) and values below 0.5 will be predicted as negative (not survive beyond 120 days). Hence, the model can be modified to predict either true positive or true negative better by adjusting the threshold of the model. A higher threshold will improve the accuracy of the model to identify patients who will not survive beyond 120 days (fewer false positives), whereas a lower threshold will enhance the accuracy of the model to identify patients who will survive beyond 120 days (fewer false negatives).
Attribute selection
The FS method selected five attributes (albumin level, ALT level, Eastern Cooperative Oncology Group (ECOG) status of 0 or 1, absolute neutrophil count, and number of metastatic sites) to construct the NN model.
The selection of albumin level in our NN model is in agreement with several studies that have also identified albumin level as a prognostic factor in patients with advanced cancer.19,20 Likewise, albumin level was also included in the NB and SVM models (Table 2), suggesting the significance of this attribute in the prediction of survival in cancer patients. The level of albumin can reflect the nutritional status of patients, and correlates well with a poor survival rate in patients with advanced cancer.21 Also, cancer patients are in an inflammation state, and this can result in low albumin level. Systemic inflammatory response had been found to be associated with poor survival outcome in cancer patients.22
Both ALT level and absolute neutrophil count were included in our model. ALT is derived from the heart, liver, muscle, kidney, and pancreas. However, ALT is more liver specific than aspartate aminotransferase (AST). Elevated level of ALT may indicate liver disease, but the diagnosis of liver disease will normally encompass other indices such as AST, bilirubin, and albumin levels. Nonetheless, a study conducted by Proctor and coworkers identified ALT level to be associated with a reduced 5-year overall survival rate.23 Both neutropenia and leukocytosis (primarily due to neutrophils) has been associated with a poor outcome in cancer patients,24 and in our model, absolute neutrophil count was included. Number of metastatic sites has also been identified in several studies as a prognostic factor in terminally ill cancer patients, where a high number of metastatic sites is correlated to a poor outcome.25,26
Performance status has been the most extensively studied variable to be associated with survival in terminally ill cancer patients. The Karnofsky Performance Status Scale and ECOG status are the two most widely used tools to measure performance status, and in our study, ECOG status was preferred due to its simplicity. According to a systematic review of 44 studies, a poor ECOG status is generally associated with poor survival, whereas the converse is true.24 In our final model, ECOG status of 0 or 1 was included. Likewise, ECOG status was included in both the NB and SVM models (Table 2), and this suggests the importance of performance status in the prediction of cancer survival. Even though performance status is recognized as a significant prognostic factor for survival in terminally ill cancer patients, one must be made aware that it is a subjective rating that may be influenced by acute but self-limited events.27 An ECOG status of 0 or 1 in an ambulatory and relatively asymptomatic patient may temporarily drop to an ECOG status of 3 or 4 due to the occurrence of acute infectious illnesses or a bone pathologic fracture.27 Such subjectivity may potentially affect the predictive outcome when using the model.
Comparison of the models (with and without ECOG status)
Due to the subjective nature of performance status, a NN model without using ECOG status was created. The rationale was to determine whether a NN model using objective attributes (albumin level, ALT level, absolute neutrophil count, and number of metastatic sites) can perform as well as a NN model with ECOG status.
Using the validation set to evaluate the performance of this new model, the classification accuracy, specificity, sensitivity, and AUC were 71%, 68%, 74%, and 0.765, respectively. The new model (without ECOG status) did not perform as well as our final selected model (with ECOG status). Thus, even though determination of ECOG status is subjective, it is an important attribute in predicting the survival of terminally ill cancer patients.
Limitations and future work
A major limitation of this study was its retrospective nature. It was difficult to validate the accuracy and integrity of data recordings, and not all data were captured during data collection. In addition, some patient attributes that can potentially affect survival were not included during the development of the model. An example would be psychological distress. A prospective cohort study had found that psychological distress, namely depression and anxiety, had a strong impact on survival.4 However, due to the retrospective nature of our study, we were unable to incorporate potentially useful attributes such as psychological distress in our model development stage, and this could have reduced the predictive performance of our model. Other valuable attributes include cancer anorexia-cachexia syndrome and C-reactive protein level.19
Conclusions
In this study, we demonstrated the feasibility of using DM techniques in creating a clinical decision support tool to predict survival in cancer patients beyond 120 days after palliative chemotherapy. With the user-friendly interface, clinicians can conveniently use the model to make better prediction of survival, especially when deciding on the types of palliative care for their patients.
Supplementary Material
Acknowledgments
This work was supported by a National University of Singapore (NUS) start-up grant (R-148-000-105-133) to Chun Wei Yap, and an NUS Department of Pharmacy Final Year Project grant (R-148-000-003-001) to Terence Ng.
Author Disclosure Statement
No conflicting financial interests exist.
References
- 1.Browner I. Carducci MA. Palliative chemotherapy: Historical perspective, applications, and controversies. Semin Oncol. 2005;32(2):145–155. doi: 10.1053/j.seminoncol.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 2.McCall K. Johnston B. Treatment options in end-of-life care: The role of palliative chemotherapy. Int J Palliat Nurs. 2007;13:486–488. doi: 10.12968/ijpn.2007.13.10.27491. [DOI] [PubMed] [Google Scholar]
- 3.Glare P. Virik K. Jones M. Hudson M. Eychmuller S. Simes J. Christakis N. A systematic review of physicians' survival predictions in terminally ill cancer patients. BMJ. 2003;327:195–198. doi: 10.1136/bmj.327.7408.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gripp S. Moeller S. Bolke E. Schmitt G. Matuschek C. Asgari S. Asgharzadeh F. Roth S. Budach W. Franz M. Willers R. Survival prediction in terminally ill cancer patients by clinical estimates, laboratory tests, and self-rated anxiety and depression. J Clin Oncol. 2007;25:3313–3320. doi: 10.1200/JCO.2006.10.5411. [DOI] [PubMed] [Google Scholar]
- 5.Chow E. Harth T. Hruby G. Finkelstein J. Wu J. Danjoux C. How accurate are physicians' clinical predictions of survival and the available prognostic tools in estimating survival times in terminally ill cancer patients? A systematic review. Clin Oncol (R Coll Radiol) 2001;13:209–218. doi: 10.1053/clon.2001.9256. [DOI] [PubMed] [Google Scholar]
- 6.Glare PA. Eychmueller S. McMahon P. Diagnostic accuracy of the palliative prognostic score in hospitalized patients with advanced cancer. J Clin Oncol. 2004;22:4823–4828. doi: 10.1200/JCO.2004.12.056. [DOI] [PubMed] [Google Scholar]
- 7.Pirovano M. Maltoni M. Nanni O. Marinari M. Indelli M. Zaninetta G. Petrella V. Barni S. Zecca E. Scarpi E. Labianca R. Amadori D. Luporini G. A new palliative prognostic score: A first step for the staging of terminally ill cancer patients. Italian Multicenter and Study Group on Palliative Care. J Pain Symptom Manage. 1999;17:231–239. doi: 10.1016/s0885-3924(98)00145-6. [DOI] [PubMed] [Google Scholar]
- 8.Breiman L. Statistical modeling: The two cultures. Stat Sci. 2001;16:199–231. [Google Scholar]
- 9.Bostwick DG. Burke HB. Prediction of individual patient outcome in cancer: Comparison of artificial neural networks and Kaplan-Meier methods. Cancer. 2001;91(8 Suppl):1643–1646. doi: 10.1002/1097-0142(20010415)91:8+<1643::aid-cncr1177>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 10.Cakir A. Demirel B. A software tool for determination of breast cancer treatment methods using data mining approach. J Med Syst. 2010;35:1503–1511. doi: 10.1007/s10916-009-9427-x. [DOI] [PubMed] [Google Scholar]
- 11.Jayasurya K. Fung G. Yu S. Dehing-Oberije C. De Ruysscher D. Hope A. De Neve W. Lievens Y. Lambin P. Dekker AL. Comparison of Bayesian network and support vector machine models for two-year survival prediction in lung cancer patients treated with radiotherapy. Med Phys. 2010;37:1401–1407. doi: 10.1118/1.3352709. [DOI] [PubMed] [Google Scholar]
- 12.Justice AC. Covinsky KE. Berlin JA. Assessing the generalizability of prognostic information. Ann Intern Med. 1999;130:515–524. doi: 10.7326/0003-4819-130-6-199903160-00016. [DOI] [PubMed] [Google Scholar]
- 13.Xu L. Zhang WJ. Comparison of different methods for variable selection. Anal Chim Acta. 2001;446:475–481. [Google Scholar]
- 14.Snow PB. Kerr DJ. Brandt JM. Rodvold DM. Neural network and regression predictions of 5-year survival after colon carcinoma treatment. Cancer. 2001;91(8 Suppl):1673–1678. doi: 10.1002/1097-0142(20010415)91:8+<1673::aid-cncr1182>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 15.Sato F. Shimada Y. Selaru FM. Shibata D. Maeda M. Watanabe G. Mori Y. Stass SA. Imamura M. Meltzer SJ. Prediction of survival in patients with esophageal carcinoma using artificial neural networks. Cancer. 2005;103:1596–1605. doi: 10.1002/cncr.20938. [DOI] [PubMed] [Google Scholar]
- 16.Dolgobrodov SG. Moore P. Marshall R. Bittern R. Steele RJ. Cuschieri A. Artificial neural network: Predicted vs observed survival in patients with colonic cancer. Dis Colon Rectum. 2007;50:184–191. doi: 10.1007/s10350-006-0779-8. [DOI] [PubMed] [Google Scholar]
- 17.Christakis NA. Lamont EB. Extent and determinants of error in doctors' prognoses in terminally ill patients: Prospective cohort study. BMJ. 2000;320(7233):469–472. doi: 10.1136/bmj.320.7233.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loprinzi CL. Johnson ME. Steer G. Doc, how much time do I have? J Clin Oncol. 2003;21(9 Suppl):5s–7s. doi: 10.1200/JCO.2003.01.155. [DOI] [PubMed] [Google Scholar]
- 19.Maltoni M. Caraceni A. Brunelli C. Broeckaert B. Christakis N. Eychmueller S. Glare P. Nabal M. Viganò A. Larkin P. De Conno F. Hanks G. Kaasa S. Steering Committee of the European Association for Palliative Care: Prognostic factors in advanced cancer patients: Evidence-based clinical recommendations: A study by the Steering Committee of the European Association for Palliative Care. J Clin Oncol. 2005;23:6240–6248. doi: 10.1200/JCO.2005.06.866. [DOI] [PubMed] [Google Scholar]
- 20.Park I. Lee JL. Ryu MH. Kim TW. Sook Lee S. Hyun Park D. Soo Lee S. Wan Seo D. Koo Lee S. Kim MH. Prognostic factors and predictive model in patients with advanced biliary tract adenocarcinoma receiving first-line palliative chemotherapy. Cancer. 2009;115:4148–4155. doi: 10.1002/cncr.24472. [DOI] [PubMed] [Google Scholar]
- 21.Penel N. Vanseymortier M. Bonneterre ME. Clisant S. Dansin E. Vendel Y. Beuscart R. Bonneterre J. Prognostic factors among cancer patients with good performance status screened for phase I trials. Invest New Drugs. 2008;26:53–58. doi: 10.1007/s10637-007-9088-x. [DOI] [PubMed] [Google Scholar]
- 22.Roxburgh CS. McMillan DC. Role of systemic inflammatory response in predicting survival in patients with primary operable cancer. Future Oncol. 2010;6:149–163. doi: 10.2217/fon.09.136. [DOI] [PubMed] [Google Scholar]
- 23.Proctor MJ. Morrison DS. Talwar D. Balmer SM. O'Reilly DS. Foulis AK. Horgan PG. McMillan DC. An inflammation-based prognostic score (mGPS) predicts cancer survival independent of tumour site: A Glasgow Inflammation Outcome Study. Br J Cancer. 2011;104:726–734. doi: 10.1038/sj.bjc.6606087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hauser CA. Stockler MR. Tattersall MH. Prognostic factors in patients with recently diagnosed incurable cancer: A systematic review. Support Care Cancer. 2006;14:999–1011. doi: 10.1007/s00520-006-0079-9. [DOI] [PubMed] [Google Scholar]
- 25.Bachelot T. Ray-Coquard I. Catimel G. Ardiet C. Guastalla JP. Dumortier A. Chauvin F. Droz JP. Philip T. Clavel M. Multivariable analysis of prognostic factors for toxicity and survival for patients enrolled in phase I clinical trials. Ann Oncol. 2000;11:151–156. doi: 10.1023/a:1008368319526. [DOI] [PubMed] [Google Scholar]
- 26.Kohne CH. Cunningham D. Di CF. Glimelius B. Blijham G. Aranda E. Scheithauer W. Rougier P. Palmer M. Wils J. Baron B. Pignatti F. Schoffski P. Micheel S. Hecker H. Clinical determinants of survival in patients with 5-fluorouracil-based treatment for metastatic colorectal cancer: results of a multivariate analysis of 3825 patients. Ann Oncol. 2002;13:308–317. doi: 10.1093/annonc/mdf034. [DOI] [PubMed] [Google Scholar]
- 27.Vigano A. Bruera E. Jhangri GS. Newman SC. Fields AL. Suarez-Almazor ME. Clinical survival predictors in patients with advanced cancer. Arch Intern Med. 2000;160:861–868. doi: 10.1001/archinte.160.6.861. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.