Abstract
MicroRNAs (miRNAs) are small RNAs that participate in the regulation of genes associated with the differentiation and proliferation. In this study, 5 novel miRNAs were identified from human mesenchymal stem cells and characterized using various analyses. To investigate the potential functions associated with the regulation of cell differentiation, the differences in miRNA expression were examined in undifferentiated and differentiated human embryonic stem (ES) cells using reverse transcription (RT)-PCR analysis. Specifically, 3 miRNAs exhibited decreased expression levels in human umbilical vein endothelial cells (HUVECs) and endothelial cells derived from human ES cells. Putative target genes related to differentiation or maturation of endothelial cells were predicted by seed sequences of 2 novel miRNAs and analyzed for their expression via miRNA-mediated regulation using a luciferase assay. In HUVECs, CDH5 gene expression was directly repressed by hsa-miR-6086. Similarly, hsa-miR-6087 significantly downregulated endoglin expression. Therefore, the roles of these 2 miRNAs may be to directly suppress their target genes, popularly known as endothelial cell markers. Taken together, our results demonstrate that several novel miRNAs perform critical roles in human endothelial cell development.
Introduction
MicroRNAs (miRNAs) are small, endogenous noncoding RNAs that are involved in various biological processes, including the development, differentiation, and proliferation of cells [1]. Recent studies have demonstrated that miRNAs are involved in gene regulation or can serve as biological markers for diagnosing diseases. Human miR-15-a and miR-16-1 have been shown to be commonly deleted or downregulated in patients with B-cell chronic lymphocytic leukemia, suggesting that they may regulate biological processes, similar to tumor suppressors [2]. miRNAs have also been shown to mediate post-transcriptional regulation of mRNA in eukaryotic cells through the suppression of gene expression by recognizing complementary nucleic acid targets [3]. Mature miRNAs are excised from 60- to 80-nucleotide pre-miRNA, double-stranded RNA (dsRNA) fold-back structures, mimicking a dsRNA hairpin, by the Dicer RNase III endonuclease, resulting in ∼18–24-nucleotide-long oligos [4]. The resulting mature miRNA can directly inhibit the translation of mRNA by binding to the target mRNA, forming an RNA-induced silencing complex (RISC) [5]. Formation of RISC is mediated by the binding of miRNA to complementary sites in 3′-untranslated regions (3′-UTRs) of the target mRNAs in a sequence-dependent manner [6]. A specific miRNA has been predicted to bind a specific recognition sequence within the 3′-UTR of numerous transcripts [7]. Individual mRNAs have also been shown to harbor multiple recognition sequences for various miRNAs [8].
miRNAs have been suggested to regulate up to 30% of the genes within the human genome by binding to 3′-UTRs [9]. Experimental evidence has revealed that miRNAs play important roles in various diseases, such as cancer, diabetes, viral infection, and cardiac dysfunction [10]. Further, miRNAs were shown to be involved in the survival, proliferation, and differentiation of stem cells [11]. Although germ cells are segregated relatively late in mammalian development, at peri-gastrulation, they are restricted after the first fertilized egg cleavage in lower organisms [12]. Mice defective in the Dicer endonuclease gene (Dicer−/−) die at embryonic day 7.5. This study has revealed that Dicer−/− embryonic stem (ES) cells are defective in miRNA maturation, resulting in proliferation and differentiation defects [13]. Recently, pyrosequencing of small RNAs isolated from normal ES and mutant ES (Dicer−/−) cells identified 46 novel miRNAs from 110,000 miRNA transcripts. Further, the loci of 4 miRNAs and their human homologs were demonstrated to participate in the regulation of oncogenesis, suggesting that miRNAs may vitally function in ES cells as cell cycle regulators [14].
Human ES cells have the potential to be a valuable resource for regenerative medicine because of their unlimited proliferation and differentiation potentials [15]. Although ES cells are immortal and pluripotent, the specific gene expression patterns that are responsible for unique physiologic states are not well understood [16]. Analyses of ES cells revealed various aspects of the RNAi-mediated control of cellular differentiation and epigenetic reprogramming in mammals [17].
In this study, 5 novel miRNAs were identified from human mesenchymal stem cells, and their expression patterns were examined in undifferentiated human ES cells and differentiated endothelial cells. The results demonstrated differential expression of novel miRNAs during ES cell differentiation, suggesting that they may play critical roles in human embryonic development.
Materials and Methods
Culture of human mesenchymal stem cells
Human bone marrow-derived mesenchymal stem cells were purchased from Cambrex Bio Science. Cells were cultured in α-MEM (Invitrogen) supplemented with 10% fetal bovine serum (FBS; HyClone) and 20 μg/mL gentamicin (Invitrogen). Cells were routinely maintained according to the manufacturer's instructions.
Maintenance and differentiation of human ES cells
Human ES cells (CHA3 human ES cells and H9 human ES cells) used in a previous study [18] were cultured on mitotically inactivated STO cells (CRL-1503) (ATCC) in ES cell medium consisting of DMEM/F12 (1:1) supplemented with 100 mM MEM nonessential amino acids, 100 U/mL penicillin, 0.1 mg/mL streptomycin, 55 mM β-mercaptoethanol, 20% Knockout Serum Replacement, and 4 ng/mL of recombinant human basic fibroblast growth factor (bFGF; R&D Systems). Feeder cells were cultured in DMEM supplemented with 100 mM MEM nonessential amino acids, 100 U/mL penicillin, 100 μg/mL streptomycin, 55 mM β-mercaptoethanol, and 10% FBS (HyClone). Media were changed daily. Human ES cells were manually subcultured using fire-thrown Pasteur pipettes on freshly prepared STO cells every 7 days. All cultures were maintained at 37°C and 5% CO2. All reagents were purchased from Gibco BRL unless otherwise indicated. The CHA3 human ES cell and H9 human ES cell lines were provided by the Stem Cell Research Laboratory of CHA University and the WiCell Research Institute, respectively.
The isolation and characterization of endothelial cells of both human ES cell lines were performed using the procedures from our previous study [15,19]. For differentiating human ES cells into endothelial cells, embryoid bodies (EBs) were formed. Briefly, human ES cells were detached from feeder cells and transferred into suspension ES culture medium in the absence of bFGF for 9 days. EBs were allowed to attach and grow on gelatinized culture dishes in differentiation medium (Gibco BRL) for 7 to 9 days. The centers of the differentiating EBs were isolated and cultured in a defined medium (EGM-2; LONZA). Mouse anti-human monoclonal von Willebrand factor (vWF) antibodies were used to purify vWF–positive (vWF+) cells from the isolated cells (Chemicon) using a FACS Vantage flow cytometer (BD Bioscience). The sorted vWF+ cells were cultured in EGM-2.
Isolation and cloning of novel miRNAs
Total RNA was extracted from human mesenchymal stem cells with 3 different donors using TRI reagent (Molecular Research Center, Inc.). To enrich for RNAs smaller than 200 nucleotides (nts) prior to novel miRNA cloning, the mirVana RNA Isolation Kit (Ambion) was used according to the manufacturer's instructions. Small RNAs were cloned into vectors using a DynaExpress miRNA Cloning Kit (BioDynamics Laboratory, Inc.) according to the manufacturer's instructions with modifications. Total RNA was prepared as a mixture of RNAs isolated from mesenchymal stem cells with 3 different donors and enriched twice by RNA precipitation. A total of 447.3 and 404.2 μg of RNA was acquired and separated on a denaturing polyacrylamide gel to isolate small RNAs of 18–28 nts in length. Purified 18–28-nt-long RNAs were dephosphorylated by alkaline phosphatase and subjected to phenol/chloroform extraction with ethanol precipitation. The 3′-linker was ligated to the dephosphorylated RNAs of 18–28 nts. The ligated products were blocked at the 3′-end to prevent circularization via 5′-linker ligation and purified on a polyacrylamide gel. The resulting miRNAs 36 to 46 nts in length were cut and extracted from the gel. The products were ligated with ribonucleotide 5′-linker at the 5′-end after their phosphorylation, followed by polyacrylamide gel electrophoresis purification as previously discussed. The resulting miRNAs with 2 linkers ranging from 53 to 63 nts were reverse-transcribed to make cDNA. After amplification, PCR products were analyzed on a 3% agarose gel and cloned into a T vector (Promega). The isolated putative clones were transformed into DH5α cells using a DokDo Mini-prep Kit (ELPIS-Biotech), sequenced, and analyzed (Macrogen).
Characterization of putative novel miRNAs from total isolated small RNAs
After the sequencing analysis, small RNAs were evaluated for miRNA characteristics using a web-based program. The presence of putative miRNA sequences in the human genome was assessed using NCBI BLAST. Predictions of precursor sequence and a stem-loop structure with surrounding putative miRNA sequence in the genome were confirmed using RNAfold (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi). The novelty of the putative miRNA was assessed with miRBase (http://microrna.sanger.ac.us/sequences/).
Analysis of miRNA expression using the TaqMan miRNA assay
The expression of 2 miRNAs was quantified by real-time PCR analysis using TaqMan miRNA assays (Applied Biosystems). Customized RT primers were synthesized to complementary sequences of mature miRNAs and analyzed with the Custom TaqMan Small RNA assay [20]. Complementary DNA templates were standardized with RNU48 [21] and subjected to PCR analysis at 40 cycles according to the manufacturer's instructions. Data were generated using the CFX Manager™ software (Bio-Rad).
Analysis of miRNA expression using a polyA-tailed RT-PCR assay
Small RNAs were isolated from undifferentiated human ES cells and differentiated endothelial cells using the mirVana miRNA Isolation Kit according to the manufacturer's instructions. For poly(A)-tailed RT-PCR analysis, 1 μg of small RNA was polyadenylated using poly(A) polymerase followed by purification of poly(A)-tailed RNAs with NucAway Spin Columns (Applied Biosystems), as previously described [22], according to the manufacturer's instructions. Purified poly(A)-tailed RNAs were reverse-transcribed into single-strand cDNA using the Superscript III Transcriptase (Invitogen) with the RTQ adapter. The reverse primer was the same tailing sequence for all miRNAs.
Constructs, mimic transfection, and the dual-luciferase assay
The 3′-UTRs of the target genes were cloned into pGL4 RL vectors (Promega) at the XbaI site. Site-directed mutagenesis was performed using a Dokdo™ Site-Specific Mutagenesis Kit (ELPIS-Biotech) to mutate 7 base pairs in the predicted seed region targeted by hms-smR-9 and smR-28 in the target 3′-UTR. HeLa cells (5×104) were plated and transfected with a 1:7 mixture (150 ng) of the Firefly luciferase control plasmid (pGL3-control) (Promega) and each of the 3′-UTRs was conjugated with the Renilla luciferase construct and 20 nM of miRNA mimics (Genolution, Inc.) using Lipofectamine 2000 (Invitrogen) in a 24-well culture plate. After 24 h, cells were lysed and assayed for luciferase activity using the Dual Luciferase Assay System (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity for each culture well. The ratio of luciferase activity was measured using the VICTOR3 luminometer (PerkinElmer). Data represented 3 independent experiments performed on different days.
Western blot analysis
Protein extracts were prepared using a PRO-PREP Kit (Intron, Inc.) according to the manufacturer's instructions. After electrophoresis using the Gradi-Gel II Gradient PAGE Analysis Kit (Elpis Biotech), the separated proteins were blotted onto PVDF transfer membranes (Hybond-P; Amersham Biosciences). The blots were probed with mouse monoclonal endoglin IgG (1:1,000) and mouse monoclonal CDH5 antibodies (1:750) overnight at 4°C. A horseradish peroxidase-conjugated goat anti-mouse secondary IgG was used. Rabbit anti-GAPDH was used for the normalization control. All antibodies used in this study were purchased from Santa Cruz Biotechnology (Santa Cruz Biotechnology). Immunocomplexes were visualized with the western blotting Luminol Reagent (Santa Cruz Biotechnology) using the luminescent image analyzer LAS-3000 (Fujifilm).
Results
Cloning of small RNAs from human mesenchymal stem cells
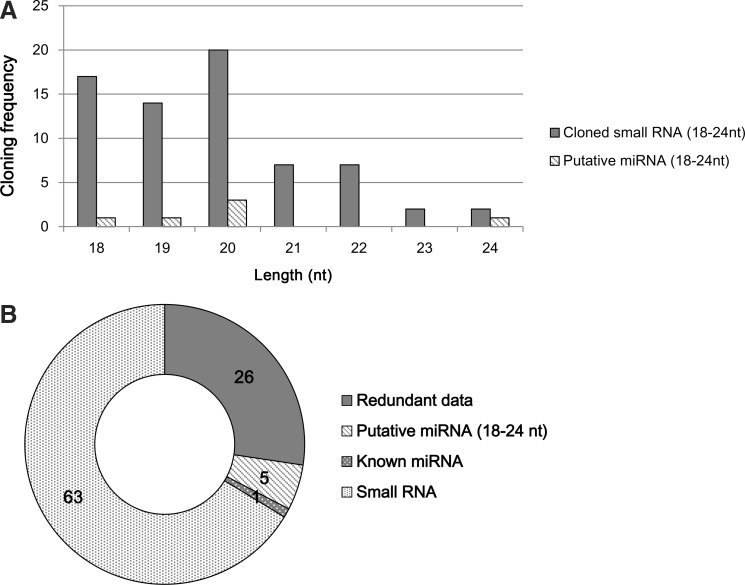
Human mesenchymal stem cells from 3 different donors were individually cultured and harvested for total RNA isolation. To isolate putative small RNAs, RNAs between 18 and 28 nts, which correspond to the average length of mature miRNAs (ie, cloned small RNA), were purified from total precipitated RNA. Approximately 95 small RNA sequences were obtained from the total RNA of human mesenchymal stem cells. After the sequencing analysis, 95 putative small RNA clones between 18 and 24 nts were identified using NCBI blast searches (Fig. 1A). Among them, the sequences of 26 clones overlapped (Fig. 1B). All putative small RNAs mapped to human chromosomes (Supplementary Table S1; Supplementary Data are available online at www.liebertonline.com/scd).
FIG. 1.
Characterization of small RNAs. Five novel miRNAs were isolated from hMSCs. (A) The size distributions of small RNAs cloned from hMSCs (B) The results of small RNAs depicted in a pie diagram. miRNA, microRNA; hMSCs, human mesenchymal stem cells.
Identification of novel miRNAs from human mesenchymal stem cells
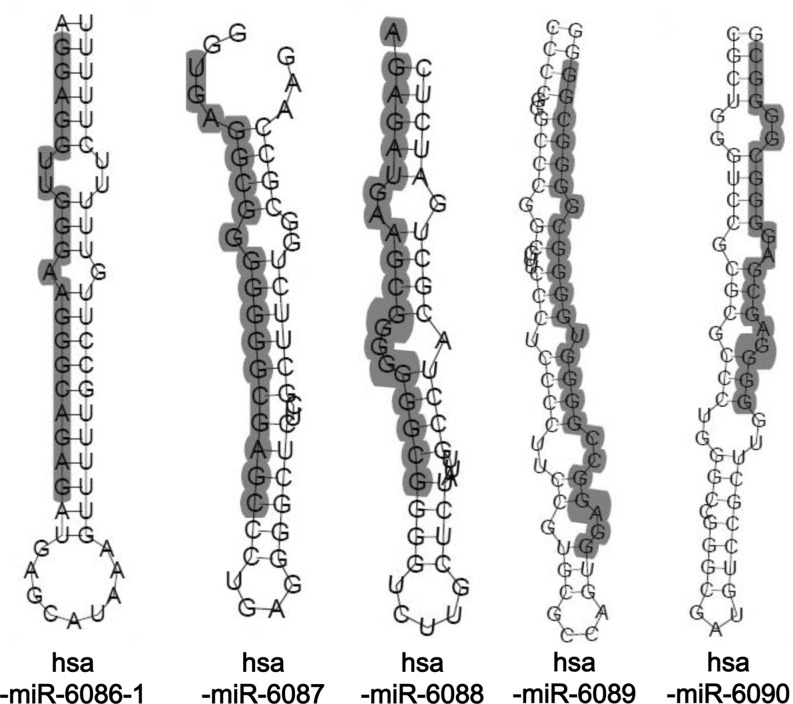
To confirm the identity of putative miRNAs, the following conditions were analyzed: size of small RNAs (18–24 nts), the capacity for the formation of a stem-loop structure (Fig. 2) and the cellular expression [20,23,24]. Among 69 small RNAs, 5 novel miRNAs and one previously known miRNA were identified (Fig. 1B). The novel miRNAs ranged between 18 and 24 nts in length (Table 1). As shown in Fig. 2, the precursor miRNAs could form stem-loop structures when analyzed using an RNA fold program. The sequences of 5 novel miRNAs were conserved among various species (Supplementary Table S2). To map the chromosomal location of the miRNAs, their sequences were compared with the human genome (NCBI blast, Genome browser). As shown in Table 1, the novel miRNAs mapped to the following locations: hsa-miR-6090 on chromosome 11, hsa-miR-6088 on chromosome 19, hsa-miR-6086, hsa-miR-6087, and hsa-miR-6089 on chromosome X.
FIG. 2.
Predicted secondary structure of novel cloned pre-miRNAs. Human genomic sequences upstream and downstream of the novel miRNAs were folded with the computer program RNAfold. Mature miRNA sequences are marked in gray.
Table 1.
Summary of the Characteristics of 6 Novel MicroRNAs
| ID | Sequence of mature miRNA | Size | Chromosomal location | Start | End | Stem-loop | Strand | Genomic location | AT | Evidence |
|---|---|---|---|---|---|---|---|---|---|---|
| hsa-miR-6086-1a | GGAGGUUGGGAAGGGCAGAG | 20 | Xp22.2 | 13608412 | 13608431 | O | + | Intron | 60 | TMA |
| hsa-miR-6086-2 | 3q21.2 | 124914709 | 124914728 | X | − | Intron | ||||
| hsa-miR-6087 | UGAGGCGGGGGGGCGAGC | 18 | Xq22.3 | 108297774 | 108297791 | O | + | Intergenic | 60 | TMA |
| hsa-miR-6088 | AGAGAUGAAGCGGGGGGGCG | 20 | 19q13.32 | 45939912 | 45939931 | O | + | Intergenic | 57 | RT-PCR |
| hsa-miR-6089 | GGAGGCCGGGGUGGGGCGGGGCGG | 24 | Xp22.33 | 2527270 | 2527293 | O | + | Intergenic | 62 | RT-PCR |
| hsa-miR-6090 | GGGGAGCGAGGGGCGGGGC | 19 | 11q24.3 | 128392325 | 128392343 | O | + | Intron | 62 | RT-PCR |
Multiple hit miRNAs in several chromosome.
AT, annealing temperature; TMA, Taqman miRNA assay; miRNA, microRNA; RT-PCR, reverse transcription-PCR.
Among the 5 novel miRNAs, hsa-miR-6086 mapped to multiple chromosomes (Table 1). The sequence of hsa-miR-6086 was observed on both chromosomes 3 and X but did not form a stem-loop structure on chromosome 3. The expression of all 5 miRNAs was examined in human mesenchymal stem cells of 3 different donors using a TaqMan miRNA Assay or poly(A)-tailed RT-PCR analysis (data not shown).
Expression of miRNAs during differentiation of ES cells
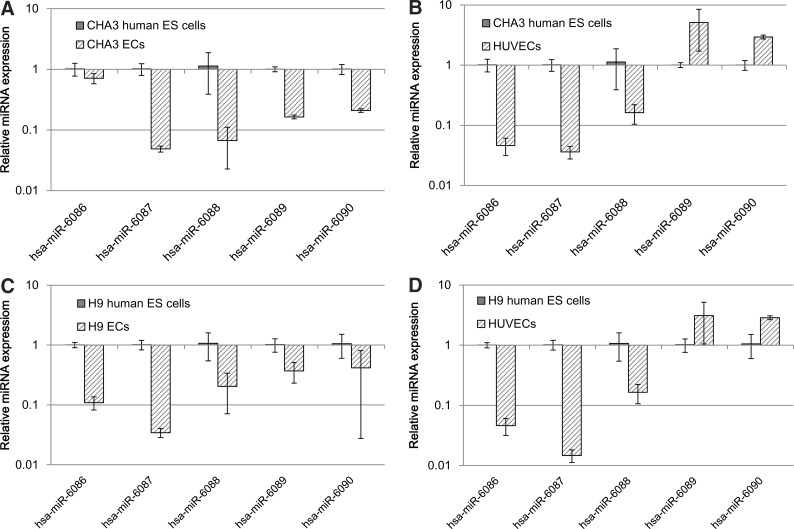
Two TaqMan probes [20] and 3 poly(A)-tailed RT-PCR primers (Supplementary Table S3) were designed to quantify mature miRNAs in cells. In this study, miRNA expression was examined in 2 human ES cell lines, CHA3 human ES cells, and H9 human ES cells. miRNA expression was examined while human ES cells differentiated into endothelial cells. Most of the novel miRNAs were expressed in undifferentiated ES cells and the differentiated endothelial cells and human umbilical vein endothelial cells (HUVECs). The expression of 5 miRNAs, specifically hsa-miR-6086, hsa-miR-6087, hsa-miR-6088, hsa-miR-6089 and hsa-miR-6090, were specifically downregulated during endothelial differentiation of human ES cells (Fig. 3A, C). When analyzed in HUVECs, hsa-miR-6086, hsa-miR-6087, and hsa-miR-6088 expression decreased compared with the expression in undifferentiated human ES cells. In contrast, hsa-miR-6089, and hsa-miR-6090 expression increased relative to the expression in HUVECs (Fig. 3B, D).
FIG. 3.
The miRNA assay was performed to assess the expression of the novel miRNAs in CHA3 and H9 human ES cells, CHA3 and H9 ECs using the TaqMan miRNA assay and poly(A)-tailed reverse transcription-PCR. The following calculation was used to determine the ratios for expression of the miRNAs in CHA3 and H9 ECs compared with CHA3 and H9 human ES cells compared with an internal control (RNU48): dCt=Ct each novel miRNA−Ct RNU48. These values were used to calculate the novel miRNA's ddCt=dCt differentiated stage of ES cells−dCt undifferentiated ES cells. (A) Expression levels are downregulated in CHA3 ECs compared with CHA3 human ES cells. (B) Expression levels are downregulated in CHA3 ECs and upregulated in HUVECs compared with CHA3 human ES cells. (C) Expression levels are downregulated in H9 ECs compared with H9 human ES cells. (D) Expression levels are downregulated in H9 ECs and upregulated in HUVECs compared with H9 humna ES cells. ES, embryonic stem; ECs, endothelial cells; HUVECs, human umbilical vein endothelial cells.
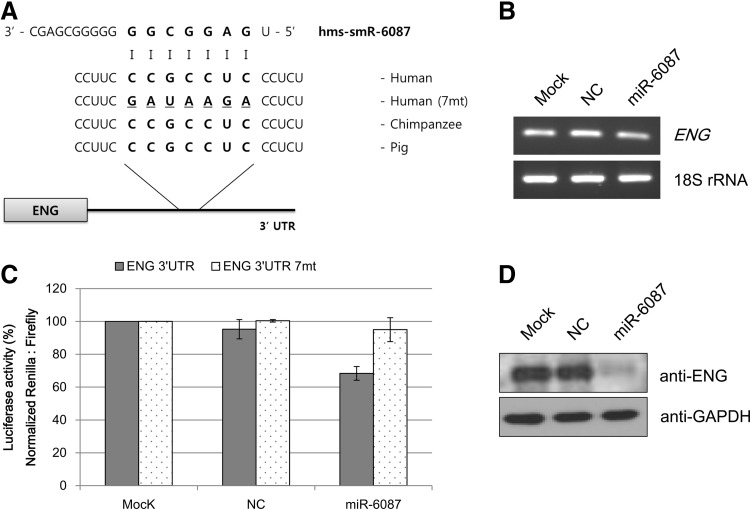
Predicted 3′-UTR of target genes harbors conserved binding site for 3 miRNAs
To characterize the functions of novel miRNAs, the 3′-UTRs of putative target genes were examined to determine the binding site for candidate miRNAs by sequence comparison. Three candidate miRNAs, hsa-miR-6086, hsa-miR-6087, and hsa-miR-6088, were chosen for subsequent analysis. Because their expression gradually decreased during endothelial cell differentiation, the target genes that were upregulated in endothelial cells were examined (data not shown). PECAM, TIE-2, vWF, ICAM1, endoglin, CDH5, and VCAM1 have been shown to act as cellular markers of endothelial cells [15,19,25,26]. Analysis of the predicted target genes was performed by comparing 3′-UTR sequences with those of candidate miRNAs. As shown in Table 2, the analysis revealed that the hsa-miR-6086 sequence matched that of the CDH5 3′-UTR. The results also showed that the 3′-UTR of endoglin has binding sites for hsa-miR-6087. However, hsa-miR-6088 did not bind to any endothelial cell markers in this study. The 3′-UTR sequences of the target genes used in this study were shown to be highly conserved among various mammalian species (Figs. 4A and 5A).
Table 2.
The Putative MicroRNAs Are Matched with 3′- Untranslated Regions of Endothelial Cell Markers
| Roles of genes | Name | NCBI No. | Predicted target-miRNA |
|---|---|---|---|
| Marker of endothelial cell | CDH5 (CD144) | NM_001795.3 | hsa-miR-6086 |
| Endoglin (CD105) | NM_001114753.1 | hsa-miR-6087 |
FIG. 4.
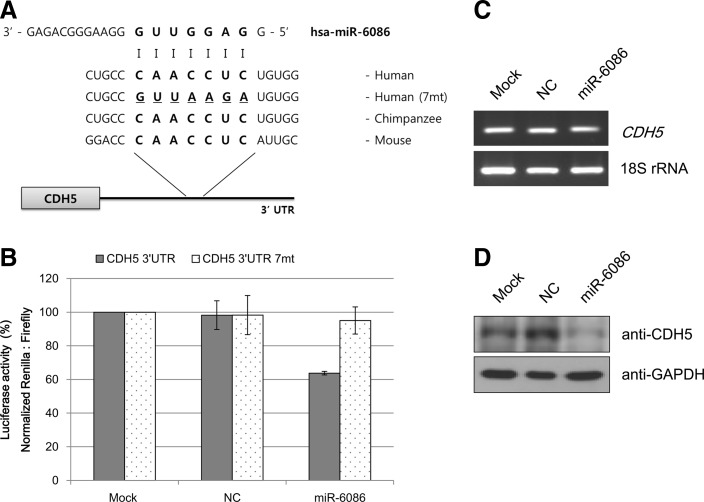
CDH5 expression is negatively regulated by hsa-miR-6086. (A) Sequence alignment between hsa-miR-6086 and the 3′-UTR of the CDH5 mRNA. The binding sites of hsa-miR-6086 in the 3′-UTR of CDH5 are highly conserved in mammals. Dashline: seed mutant region. (B) Luciferase reporter gene assays indicate the effect of hsa-miR-6086 on the activity of the CDH5 3′-UTR reporter. Cotransfection was performed using the plasmids, including the CDH5 3′-UTR and the hsa-miR-6086 binding site mutant (7mt), a hsa-miR-6086 mimic or NC in HeLa cells. All values were normalized to Renilla luciferase activity produced from a cotransfected control plasmid. (C) Semi-quantification of CDH5 mRNA expression indicates that hsa-miR-6086 does not regulate mRNA levels. (D) Immunoblot illustrates that hsa-miR-6086 induces a decrease in CDH5 expression. NC, negative control; UTR, untranslated region.
FIG. 5.
Endoglin expression is directly suppressed by hsa-miR-6087. (A) Sequence alignment between hsa-miR-6087 and the 3′-UTR of ENG mRNA. Binding sites of hsa-miR-6087 in the ENG 3′-UTR are highly conserved in mammals. Dashline: seed mutant region. (B) Luciferase reporter gene assay reveals the effect of hsa-miR-6087 on the activity of the ENG 3′-UTR reporter. Cotransfection was performed using the plasmids with the ENG 3′-UTR or the hsa-miR-6087 binding site mutant (7mt), a hsa-miR-6087 mimic or NC in HeLa cells. All values were normalized to Renilla luciferase activity produced from a cotransfected control plasmid. (C) Semi-quantification of ENG mRNA expression indicates that hsa-miR-6087 does not regulate mRNA levels. (D) Immunoblot illustrates that hsa-miR-6087 induces a decrease in ENG expression.
Two miRNAs directly repressed expression of target genes
Functional analysis of selected miRNA-mRNA interactions was performed using a dual luciferase assay. As shown in Figs. 4B and 5B, the selected miRNAs repressed the activity of Renilla luciferase regulated by the 3′-UTRs of the corresponding genes CDH5 and endoglin. Mutating the modified seed-binding site abrogated the ability of 2 miRNAs to regulate their target genes. Similarly, hsa-miR-6086 repressed the wild type construct by 30%–35% but failed to inhibit the mutant construct (Fig. 4B), and hsa-miR-6087 also failed to suppress the mutant construct (Fig. 5B).
To examine the direct relationship between the selected miRNAs and the expression of their target genes, RT-PCR and western blot analyses were performed in HUVECs. When HUVECs were transfected with mimics of the control and hsa-miR-6086 or hsa-miR-6087, the mRNA expressions of CDH5 and endoglin were not significantly affected (Figs. 4C and 5C). Unlike the mRNA expression, CDH5 protein levels were downregulated in the presence of hsa-miR-6086 in HUVECs (Fig. 4D) compared with the control. Similar results were obtained for endoglin protein expression under the influence of hsa-miR-6087 in HUVECs (Fig. 5D).
Discussion
We identified 5 novel miRNAs from human mesenchymal stem cells. Approximately 95 small RNAs were cloned (Fig. 1A) and sequenced (Supplementary Table S1). Among them, 26 clones were excluded due to their redundancy (Fig. 1B). Sixty-nine positive clones were confirmed as putative miRNAs by sequence analysis using a web-based program. Among the 69 small RNAs, 5 novel miRNAs and 1 previously known miRNA were identified. Formation of a stem-loop structure is a typical characteristic of miRNAs [27]. All of the 5 putative miRNAs identified in this study were predicted to have this unique stem-loop structure (Fig. 2), confirming that they are miRNAs.
Pre-miRNAs are processed into mature miRNAs by several types of RNase III, including Dicer, which then act as regulators of target gene expression [28]. miRNAs were shown to be transcribed from exonic, intronic, and intergenic regions of both protein-coding and noncoding transcripts from RNA polymerase II [29]. Specifically, transcription of miRNA derived from intronic regions depended on the host gene [30]. In this study, novel miRNAs were also confirmed to be transcribed from exonic, intronic, and intergenic regions by transcription genomic analysis using NCBI blast (Table 1).
Although miRNA functions remain poorly understood, miRNAs have been suggested to regulate gene expression in various biological processes, including stress resistance, fat metabolism, cellular proliferation, differentiation, and death [31]. Most novel miRNAs identified in this study exhibit specific expression patterns during human ES cell differentiation. Before characterizing expression profiles of these 5 miRNAs, we selected an endogenous control. For endogenous control expression patterns [32] in human ES and differentiated cells, 4 small nucleolar RNAs were selected for comparison of Ct values expressed in 5 different cells. In this study, RNU48 [21] was more effective than RNU6B, RNU66, and RNU24 as an endogenous control for miRNA expression. The Ct value for RNU6B increased during differentiation compared with undifferentiating human ES cells. However, expression of RNU48 was similar in human ES cells while they were differentiating (Supplementary Fig. S1). CHA3 human ES cell lines [18] and H9 human ES cell lines [33] were provided by the Stem Cell Research Laboratory of CHA University and the WiCell Research Institute, respectively. Two human ES cell lines were successfully differentiated into endothelial cells in our previous study [15]. To examine the patterns of expression associated with cell differentiation, undifferentiated ES and differentiated endothelial cells were subject to 2 different analyses, the TaqMan miRNA assay and poly(A)-tailed RT-PCR [22,34]. When undifferentiated human ES cells were compared with their differentiated endothelial cells, the expression of 5 miRNAs was lower in the differentiated cells than the undifferentiated cells. When ES cells were analyzed with HUVECs, the expression of 3 miRNAs was lower in HUVECs than in undifferentiated ES cells. Conversely, the expression levels of 2 miRNAs, hms-smR-89 and hms-smR-63, were higher in HUVECs than in human ES cells. The differential expression patterns of 2 miRNAs between ES-derived endothelial cells and HUVECs may be a result of their differential endothelial lineage stages. This observation may suggest that both ES-derived endothelial cells and HUVECs are not at the same stage of differentiation [15,26].
To dissect the functions of the novel miRNAs, 3′-UTRs of potential target genes were predicted to determine binding sites for the candidate miRNAs by sequence comparison. The 3 miRNAs, hsa-miR-6086, -6087, and -6088, the expression of which decreased upon endothelial differentiation, were selected. Upon sequence analysis, 2 miRNA sequences matched the 3′-UTRs of the predicted target genes (Figs. 4A and 5A), which have been previously reported as endothelial cell markers by other groups [15,25]. Analysis of target gene regulation by the selected miRNAs was performed using a dual luciferase assay. The luciferase assay revealed that hsa-miR-6086 repressed expression of CDH5 (Fig. 4B), and hsa-miR-6087 downregulated endoglin expression (Fig. 5B). To examine direct relationships between the selected miRNAs and the expression of their target genes and proteins, RT-PCR and western blot analyses were performed in HUVECs. When HUVECs were transfected with mimics of the control or hsa-miR-6086 and hsa-miR-6087, respectively, mRNA levels of the corresponding genes did not significantly decrease in the presence of miRNAs (Figs. 4C and 5C). These findings suggest that the novel miRNAs may not regulate gene expression at the transcriptional level. However, western blot analysis revealed that protein expression was repressed by the presence of the novel miRNAs in this study. There are 2 mechanisms by which miRNAs regulate target genes: (i) the mRNA is degraded when the miRNA exhibits a near-perfect complementary sequence with the target mRNA, and deadenylation and subsequent degradation of the target mRNA occurs (the major mechanism of miRNA action) and (ii) the mRNA is translationally, but not transcriptionally, inhibited when the miRNA sequence is only partially complementary to its target mRNA [35]. Two novel miRNAs have been shown to translationally inhibit target genes [36–38]. After transfection of mimics, formation of cell morphology was not observed (data not shown). In fact, 2 miRNAs did not directly regulate reprogramming factors, where the mRNA levels of reprogramming factors in transfected cells were unchanged (Supplementary Fig. S2). ES-specific miRNAs or iPS-specific miRNAs maintained stemness and undifferentiated ability [39]. In this study, hsa-miR-6086 and -6087 regulated their target genes irrespective of ES- or iPS-specific functions during endothelial differentiation. Specifically, hsa-miR-6086 decreased CDH5 protein expression (Fig. 4D), which was shown to regulate barrier function in endothelial cells [40] and vitally participate in embryonic angiogenesis [41]. In zebra fish and mice, CDH5 deficiency inhibited blood formation and development during embryogenesis [42,43]. Moreover, hsa-miR-6087 suppressed endoglin expression (Fig. 5D), a type of transforming growth factor-β (TGF-β) receptor on endothelial cells [44]. Endoglin has been shown to be expressed in cardiovascular, hematological, reticuloendothelial, and genitourinary systems [45]. Endoglin-deficient mice exhibited defects in angiogenesis and vascular development [46]. These genes were shown to be essential for endothelial differentiation in previous studies. In this study, 2 miRNAs were downregulated in endothelial cells and were shown to negatively regulate their target genes, including endothelial cell markers. Moreover, hsa-miR-6086 and -6087 appeared to function during endothelial biogenesis.
In summary, this study identified 5 novel miRNAs, which were cloned in human mesenchymal stem cells and differentially expressed during endothelial differentiation of human ES cells. In addition, expression profiles of 3 miRNAs were downregulated in both ES-derived endothelial cells and HUVECs compared with undifferentiated human ES cells. Among them, 2 miRNAs repressed the protein expression of their putative targets genes, known endothelial cell markers, indicative of their involvement in the developmental of human endothelial lineages.
Supplementary Material
Acknowledgments
This work was support by a grant (SC3110) from the Stem Cell Research Center of the 21st Century Frontier Program funded by the Ministry of Education, Science and Technology, Republic of Korea.
Author Disclosure Statement
The authors declare no competing financial interests.
References
- 1.Laurent LC. Chen J. Ulitsky I. Mueller FJ. Lu C. Shamir R. Fan JB. Loring JF. Comprehensive microRNA profiling reveals a unique human embryonic stem cell signature dominated by a single seed sequence. Stem Cells. 2008;26:1506–1516. doi: 10.1634/stemcells.2007-1081. [DOI] [PubMed] [Google Scholar]
- 2.Calin GA. Dumitru CD. Shimizu M. Bichi R. Zupo S. Noch E. Aldler H. Rattan S. Keating M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tolia NH. Joshua-Tor L. Slicer and the argonautes. Nat Chem Biol. 2007;3:36–43. doi: 10.1038/nchembio848. [DOI] [PubMed] [Google Scholar]
- 4.Grishok A. Pasquinelli AE. Conte D. Li N. Parrish S. Ha I. Baillie DL. Fire A. Ruvkun G. Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 5.Nelson KM. Weiss GJ. MicroRNAs and cancer: past, present, and potential future. Mol Cancer Ther. 2008;7:3655–3660. doi: 10.1158/1535-7163.MCT-08-0586. [DOI] [PubMed] [Google Scholar]
- 6.Lagos-Quintana M. Rauhut R. Yalcin A. Meyer J. Lendeckel W. Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 7.Zeng Y. Cullen BR. Sequence requirements for micro RNA processing and function in human cells. RNA. 2003;9:112–123. doi: 10.1261/rna.2780503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wienholds E. Plasterk RH. MicroRNA function in animal development. FEBS Lett. 2005;579:5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- 9.Stark A. Kheradpour P. Parts L. Brennecke J. Hodges E. Hannon GJ. Kellis M. Systematic discovery and characterization of fly microRNAs using 12 Drosophila genomes. Genome Res. 2007;17:1865–1879. doi: 10.1101/gr.6593807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang KM. Dentchev T. Stambolian D. MiRNA expression in the eye. Mamm Genome. 2008;19:510–516. doi: 10.1007/s00335-008-9127-8. [DOI] [PubMed] [Google Scholar]
- 11.Houbaviy HB. Murray MF. Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5:351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 12.Gu P. Reid JG. Gao X. Shaw CA. Creighton C. Tran PL. Zhou X. Drabek RB. Steffen DL, et al. Novel microRNA candidates and miRNA-mRNA pairs in embryonic stem (ES) cells. PLoS One. 2008;3:e2548. doi: 10.1371/journal.pone.0002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatfield SD. Shcherbata HR. Fischer KA. Nakahara K. Carthew RW. Ruohola-Baker H. Stem cell division is regulated by the microRNA pathway. Nature. 2005;435:974–978. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- 14.Calabrese JM. Seila AC. Yeo GW. Sharp PA. RNA sequence analysis defines Dicer's role in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2007;104:18097–18102. doi: 10.1073/pnas.0709193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho SW. Moon SH. Lee SH. Kang SW. Kim J. Lim JM. Kim HS. Kim BS. Chung HM. Improvement of postnatal neovascularization by human embryonic stem cell derived endothelial-like cell transplantation in a mouse model of hindlimb ischemia. Circulation. 2007;116:2409–2419. doi: 10.1161/CIRCULATIONAHA.106.687038. [DOI] [PubMed] [Google Scholar]
- 16.Chen C. Ridzon D. Lee CT. Blake J. Sun Y. Strauss WM. Defining embryonic stem cell identity using differentiation-related microRNAs and their potential targets. Mamm Genome. 2007;18:316–327. doi: 10.1007/s00335-007-9032-6. [DOI] [PubMed] [Google Scholar]
- 17.Kanellopoulou C. Muljo SA. Kung AL. Ganesan S. Drapkin R. Jenuwein T. Livingston DM. Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahn SE. Kim S. Park KH. Moon SH. Lee HJ. Kim GJ. Lee YJ. Cha KY. Chung HM. Primary bone-derived cells induce osteogenic differentiation without exogenous factors in human embryonic stem cells. Biochem Biophys Res Commun. 2006;340:403–408. doi: 10.1016/j.bbrc.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 19.Ahn JS. Moon SH. Kim J. Chung HM. Kim JK. Identification of differentially expressed genes in human embryonic stem cell-derived endothelial cells using suppression subtractive hybridization. Stem Cells Dev. 2010;19:1249–1256. doi: 10.1089/scd.2009.0265. [DOI] [PubMed] [Google Scholar]
- 20.Chen C. Ridzon DA. Broomer AJ. Zhou Z. Lee DH. Nguyen JT. Barbisin M. Xu NL. Mahuvakar VR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren J. Jin P. Wang E. Marincola FM. Stroncek DF. MicroRNA and gene expression patterns in the differentiation of human embryonic stem cells. J Transl Med. 2009;7:20. doi: 10.1186/1479-5876-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ro S. Park C. Jin J. Sanders KM. Yan W. A PCR-based method for detection and quantification of small RNAs. Biochem Biophys Res Commun. 2006;351:756–763. doi: 10.1016/j.bbrc.2006.10.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lagos-Quintana M. Rauhut R. Lendeckel W. Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 24.Lai EC. Tomancak P. Williams RW. Rubin GM. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:R42. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nikolova-Krstevski V. Bhasin M. Otu HH. Libermann T. Oettgen P. Gene expression analysis of embryonic stem cells expressing VE-cadherin (CD144) during endothelial differentiation. BMC Genomics. 2008;9:240. doi: 10.1186/1471-2164-9-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J. Moon SH. Lee SH. Lee DR. Koh GY. Chung HM. Effective isolation and culture of endothelial cells in embryoid body differentiated from human embryonic stem cells. Stem Cells Dev. 2007;16:269–280. doi: 10.1089/scd.2006.0108. [DOI] [PubMed] [Google Scholar]
- 27.Afanasyeva EA. Hotz-Wagenblatt A. Glatting KH. Westermann F. New miRNAs cloned from neuroblastoma. BMC Genomics. 2008;9:52. doi: 10.1186/1471-2164-9-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tijsterman M. Plasterk RH. Dicers at RISC; the mechanism of RNAi. Cell. 2004;117:1–3. doi: 10.1016/s0092-8674(04)00293-4. [DOI] [PubMed] [Google Scholar]
- 29.Mattick JS. Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15(Spec No 1):R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 30.Lin SL. Miller JD. Ying SY. Intronic microRNA (miRNA) J Biomed Biotechnol. 2006;2006:26818. doi: 10.1155/JBB/2006/26818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–676. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki T. Higgins PJ. Crawford DR. Control selection for RNA quantitation. Biotechniques. 2000;29:332–337. doi: 10.2144/00292rv02. [DOI] [PubMed] [Google Scholar]
- 33.Amit M. Winkler ME. Menke S. Bruning E. Buscher K. Denner J. Haverich A. Itskovitz-Eldor J. Martin U. No evidence for infection of human embryonic stem cells by feeder cell-derived murine leukemia viruses. Stem Cells. 2005;23:761–771. doi: 10.1634/stemcells.2004-0046. [DOI] [PubMed] [Google Scholar]
- 34.Sdassi N. Silveri L. Laubier J. Tilly G. Costa J. Layani S. Vilotte JL. Le Provost F. Identification and characterization of new miRNAs cloned from normal mouse mammary gland. BMC Genomics. 2009;10:149. doi: 10.1186/1471-2164-10-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brodersen P. Voinnet O. Revisiting the principles of microRNA target recognition and mode of action. Nat Rev Mol Cell Biol. 2009;10:141–148. doi: 10.1038/nrm2619. [DOI] [PubMed] [Google Scholar]
- 36.Poliseno L. Pitto L. Simili M. Mariani L. Riccardi L. Ciucci A. Rizzo M. Evangelista M. Mercatanti A. Pandolfi PP. Rainaldi G. The proto-oncogene LRF is under post-transcriptional control of MiR-20a: implications for senescence. PLoS One. 2008;3:e2542. doi: 10.1371/journal.pone.0002542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doxakis E. Post-transcriptional regulation of alpha-synuclein expression by mir-7 and mir-153. J Biol Chem. 2010;285:12726–12734. doi: 10.1074/jbc.M109.086827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doench JG. Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson KD. Venkatasubrahmanyam S. Jia F. Sun N. Butte AJ. Wu JC. MicroRNA profiling of human-induced pluripotent stem cells. Stem Cells Dev. 2009;18:749–758. doi: 10.1089/scd.2008.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rudini N. Felici A. Giampietro C. Lampugnani M. Corada M. Swirsding K. Garre M. Liebner S. Letarte M. ten Dijke P. Dejana E. VE-cadherin is a critical endothelial regulator of TGF-beta signalling. EMBO J. 2008;27:993–1004. doi: 10.1038/emboj.2008.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vestweber D. VE-cadherin: the major endothelial adhesion molecule controlling cellular junctions and blood vessel formation. Arterioscler Thromb Vasc Biol. 2008;28:223–232. doi: 10.1161/ATVBAHA.107.158014. [DOI] [PubMed] [Google Scholar]
- 42.Larson JD. Wadman SA. Chen E. Kerley L. Clark KJ. Eide M. Lippert S. Nasevicius A. Ekker SC. Hackett PB. Essner JJ. Expression of VE-cadherin in zebrafish embryos: a new tool to evaluate vascular development. Dev Dyn. 2004;231:204–213. doi: 10.1002/dvdy.20102. [DOI] [PubMed] [Google Scholar]
- 43.Gory-Faure S. Prandini MH. Pointu H. Roullot V. Pignot-Paintrand I. Vernet M. Huber P. Role of vascular endothelial-cadherin in vascular morphogenesis. Development. 1999;126:2093–2102. doi: 10.1242/dev.126.10.2093. [DOI] [PubMed] [Google Scholar]
- 44.Lebrin F. Goumans MJ. Jonker L. Carvalho RL. Valdimarsdottir G. Thorikay M. Mummery C. Arthur HM. ten Dijke P. Endoglin promotes endothelial cell proliferation and TGF-beta/ALK1 signal transduction. EMBO J. 2004;23:4018–4028. doi: 10.1038/sj.emboj.7600386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dallas NA. Samuel S. Xia L. Fan F. Gray MJ. Lim SJ. Ellis LM. Endoglin (CD105): a marker of tumor vasculature and potential target for therapy. Clin Cancer Res. 2008;14:1931–1937. doi: 10.1158/1078-0432.CCR-07-4478. [DOI] [PubMed] [Google Scholar]
- 46.Arthur HM. Ure J. Smith AJ. Renforth G. Wilson DI. Torsney E. Charlton R. Parums DV. Jowett T, et al. Endoglin, an ancillary TGFbeta receptor, is required for extraembryonic angiogenesis and plays a key role in heart development. Dev Biol. 2000;217:42–53. doi: 10.1006/dbio.1999.9534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.