Abstract
The transcription factor DLX3 plays a decisive role in bone development of vertebrates. In neural-crest derived stem cells from the dental follicle (DFCs), DLX3 is differentially expressed during osteogenic differentiation, while other osteogenic transcription factors such as DLX5 or RUNX2 are not highly induced. DLX3 has therefore a decisive role in the differentiation of DFCs, but its actual biological effects and regulation are unknown. This study investigated the DLX3-regulated processes in DFCs. After DLX3 overexpression, DFCs acquired a spindle-like cell shape with reorganized actin filaments. Here, marker genes for cell morphology, proliferation, apoptosis, and osteogenic differentiation were significantly regulated as shown in a microarray analysis. Further experiments showed that DFCs viability is directly influenced by the expression of DLX3, for example, the amount of apoptotic cells was increased after DLX3 silencing. This transcription factor stimulates the osteogenic differentiation of DFCs and regulates the BMP/SMAD1-pathway. Interestingly, BMP2 did highly induce DLX3 and reverse the inhibitory effect of DLX3 silencing in osteogenic differentiation. However, after DLX3 overexpression in DFCs, a BMP2 supplementation did not improve the expression of DLX3 and the osteogenic differentiation. In conclusion, DLX3 influences cell viability and regulates osteogenic differentiation of DFCs via a BMP2-dependent pathway and a feedback control.
Introduction
Human dental follicle cells (DFCs) belong to a stem cell population isolated from dental follicles of impacted human wisdom teeth, and are able to differentiate into periodontium-like tissues, such as the periodontal ligament (PDL), the alveolar bone, and the mineralized bone-like cementum [1,2]. These cells are multipotent and can be differentiated, for example, into adipocytes, chondrocytes, and neural cells [2–4]. Although molecular mechanisms in DFCs are not yet completely understood, the differentiation and function of dental follicular cells are known to be controlled by a network of regulatory molecules including transcription factors, growth factors, and cytokines [5,6].
DLX3, an essential transcription factor for embryonic development [7], plays a decisive role in bone development and can be detected in craniofacial bone [8,9]. This transcription-factor contains a homeodomain, which is related to the distal-less domain of Drosophila and was detected even in structures involving epithelial-mesenchymal interaction, such as tooth germs and hair follicles. It is also expressed in otic and olfatory placodes, epidermis, the limb bud, branchial arches, the placenta, and osteoblasts [8–10]. A 4-bp deletion in the DLX3 gene is responsible for the autosomal dominant tricho-dento-osseous (TDO) syndrome. The major phenotypic characteristics of TDO are increased bone mineral density and thickness in the craniofacial bones, enamel hypoplasia, severe taurodontism, and unique kinky/curly hair [11–13]. Analyses of normal and mutant DLX3 in diverse secretory cells of mineralized tissues, such as odontoblasts, ameloblasts, osteoblasts, and chondrocytes, revealed that the influence of DLX3 expression on mineralized tissue development vary specifically according to the terminal differentiation for each cell type [9,14,15].
Previous studies have shown a direct involvement of DLX3 in the regulation of bone differentiation markers [9,16]. In osteoblasts, DLX3 together with DLX5 plays a major role in the transcriptional activity of BGLAP (osteocalcin) and in the induction of BMP2-mediated RUNX2 expression [17,18].
Recently, an increased DLX3 expression was also shown during the osteogenic differentiation of DFCs in vitro [19]. However, additional osteogenic transcription factors such as MSX2, RUNX2, DLX5, or OSTERIX were not regulated in DFCs during osteogenic differentiation, suggesting an important role for DLX3 in the progression of differentiation. To evaluate regulatory mechanisms of osteogenic differentiation in DFCs, we investigated the influence of DLX3 on the differentiation of DFCs. Therefore, we compared cell proliferation, cell morphology, apoptosis, and osteogenic differentiation after overexpression or silencing of DLX3 in DFCs. Moreover, we analyzed the relation of BMP2 and DLX3 with respect to the differentiation of DFCs.
Materials and Methods
Cell Culture
DFCs were isolated as described previously [2,20]. Briefly, impacted human third molars were surgically removed and collected from patients with informed consent. The attached dental follicle was separated from the mineralized tooth. The follicle tissues were cleaned and then digested in a solution of collagenase type I, hyaluronidase (Sigma-Aldrich, Munich, Germany), and DNAse I (Roche, Mannheim, Germany) for 1 h at 37°C. Digested tissues were seeded into T25 flasks in Mesenchym Stem Medium (PAA, Pasching, Austria) at 37°C in 5% CO2. Non-adherent cells were removed after medium-change. The standard basal medium was Dulbecco's modified Eagle's medium (DMEM; PAA), supplemented with 10% fetal bovine serum (FBS; PAA), and 100 μg/mL penicillin/streptomycin. In experiments, DFCs were used at cell passage 6.
DFCs at passage 4 were analyzed for stem cell associated markers with flow cytometry for characterization. The following antibodies were used: anti-CD44-FITC, anti-Nestin-PE, anti-CD105-APC, and anti-CD146-FITC (Miltenyi Biotec, Bergisch Gladbach, Germany). Single-cell suspensions of DFCs were incubated with monoclonal antibodies for 45 min at 4°C, washed once in PBS with 2nM ethylenediaminetetraacetic acid (EDTA) and 0.5% BSA. Immunoglobulin G (IgG) isotype-matched control (Miltenyi Biotec) and immunoglobulin M (IgM) isotype-matched control (BioLegend) were used as negative control. Flow cytometry analyses were done using the fluorescence-activated cell sorter (FACS) Canto II (Becton Dickinson, Heidelberg, Germany). DFCs express CD44, CD105, and Nestin. However, DFCs were negative for CD146 (Supplementary Fig. S1; Supplementary materials are available online at http://www.liebertpub.com/scd).
Transfection of DFCs with DLX3 expression plasmid
Transient transfection was performed using the FuGENE®HD Transfection Reagent (Roche). The DLX3 expression plasmid pCMV-V5DLX3 (pDLX3) (kindly provided by Dr. M I. Morasso National Institute of Arthritis and Musculoskeletal and Skin Diseases, National Institutes of Health, Bethesda, Maryland), expresses DLX3 under the control of a constitutive CMV promoter [21]. As control, an empty vector without an insert (pEV) was used. A constitutive green fluorescence protein expressing plasmid (pCMV-AC-GFP) was purchased from Origene (Rockville, Maryland). Two days before transfection, DFCs were seeded at a cell density of 7×103 cells per cm2. For transfection, the cells were grown until subconfluence and cultivated in standard basal medium without antibiotics. To form the transfection complex, the FuGENE®HD (Roche) reagent and DNA, diluted in DMEM, were combined in a ratio of 5:2 (5 μL FuGENE®HD per 2 μg DNA) and incubated at room temperature for 15 min. Then the preincubated transfection complex was added to the cultures. For a DLX3 dose-dependent test, dilutions of transfection complexes were used. After 24 h of transfection, standard basal medium containing antibiotics was changed into the plates. The expression of DLX3 in DFCs was determined by quantitative reverse transcription-polymerase chain reaction (qRT-PCR) or western blot after 48 h.
The transfection efficiency was determined before. DFCs were transfected with the plasmid pCMV-AC-GFP. Here, more than 50% of the investigated cells were fluorescent (unpublished data).
Transfection of DFCs with DLX3siRNA
The efficiency of the small interfering RNA (siRNA) transfection procedure in DFCs was successfully established with the RNA interference (RNAi) Human/Mouse Starter Kit (Qiagen, Hilden, Germany) previously. Four different nucleotides (20–23 nt) targeting human DLX3 mRNA (GenBank No. NM_005220) were tested for silencing. DLX3 siRNA(6) and DLX3 siRNA(7) showed the highest inhibition efficiencies and were selected for silencing experiments. A negative nonspecific siRNA (NS siRNA; Qiagen) was used for control. Two days before transfection, DFCs were seeded at a cell density of 7×103 cells per cm2. For transfection, the cells were grown until subconfluence and cultivated in standard basal medium containing antibiotics. DLX3 siRNA or NS siRNA (20 μM) were diluted in 50–100 μL DMEM at a final concentration of 50 nM and combined with the HiPerFect Transfection reagent (Qiagen) and incubated at room temperature for 10 min to form the transfection complexes. Subsequently, the latter were added to culture plates. After 48 h, the inhibitory effect of DLX3 siRNA in DFCs was determined by qRT-PCR and western blots.
Osteogenic differentiation
For osteogenic differentiation, DFCs were stimulated with dexamethasone or BMP2. The dexamethasone supplemented standard osteogenic differentiation medium (ODM) or BMP2-containing medium (BMP2) comprised of DMEM (PAA) supplemented with 1% FBS (PAA), 100 μmol/L ascorbic acid 2-phosphate, 4-(2-hydroxyethyl)-1-piperazine ethanesulfonic acid (HEPES) (20 mmol/L) and 2.8 mmol/L β-glycerophosphat, 1×10−7 mol/L dexamethasone sodium phosphate (Sigma-Aldrich) or 50 ng/mL BMP2 (Biomol, Hamburg, Germany). Long-term cultures were cultivated in ODM containing 10% FBS. Calcium staining of differentiated cells was performed with alizarin red S. The quantitation of alizarin staining was done as previously described [20]. For BMP2 neutralization, 15 μg/mL of BMP2 antibody (R&D Systems, Minneapolis, Minnesota) was added to the cell culture medium. DFCs were treated with BMP2 antibody or control IgG (15 μg/mL) for 30 min at 4°C prior to seeding. The inhibition of BMP signaling with this BMP2 specific antibody was verified in BMP2 induced DFCs prior to treatment as part of procedure (data not shown).
Alkaline phosphatase (ALP) activity detection
The osteogenic differentiation potential of DFCs after overexpression or silencing of DLX3 was evaluated after 10 days of cultivation in ODM, BMP2, or standard basal medium. The alkaline phosphatase (ALP) activity was detected with naphthol and fast red violet [19]. Total ALP activity was determined quantitatively by p-nitrophenol phosphate conversion method using the phosphatase assay kit (Jena Bioscience, Jena, Germany). ALP activity values were normalized to total DNA concentration determined by Quant-iT™ PicoGreen® dsDNA Assay (Invitrogen). The ALP activity per cell was normalized (100%) to the mean of the correspondent control cells at day 10 of differentiation with ODM or BMP2. Each experiment was repeated up to 4 times.
Cell Proliferation
For cell proliferation analysis, the standard basal medium was used. The cell proliferation was estimated with the WST-1 assay (Roche) after 24 h and 72 h of transfection with either pDLX3 or DLX3 siRNAs. WST-1 is a colorimetric assay based on the cleavage of the tetrazolium salt WST-1 by mitochondrial dehydrogenase in viable cells. Cell cultures were incubated with the WST-1 reagent for 4 h at 37°C. The optical density was measured at 450-nm wavelength thereafter. After 24 h of transfection, DFCs were used for normalization (= 100%) either with the control vector (pEV) or the control (NS) siRNA. Each experiment was repeated 4 times.
Rhodamine Phalloidin staining of cells
Forty-eight hours after transfection, DFCs, which were cultivated on chamber-slides, were washed with PBS and fixed with 2% formaldehyde for 30 min. Cells were washed with PBS and permeabilized with 0.5% Triton X-100 (Sigma-Aldrich) for 3 min. After washing, DFCs were stained with 300 μL rhodamine phalloidin (14 μM of stock solution dissolved 3:1000 in 1× PBS containing 0.1% BSA) for 30 min. The cell nuclei were stained with Hoechst 33258 for 15 min. Cells were washed twice in PBS and imaged with a fluorescence microscope (Axio Scope, Zeiss, Germany).
Microarray analysis
Total RNA was extracted using the NucleoSpin® RNA II kit (Macherey Nagel, Düren, Germany) and quality controlled using the RNA 6000 Nano LabChip (Agilent Technologies, Santa Clara, California). DNA microarray analyses were carried out with Affymetrix Human Gene 1.0 ST arrays according to the Affymetrix standard protocol. RNAs were used from DFCs after 48 h of transfection with pDLX3 or pEV. Two microarrays were performed with RNAs from independent cultures of the applied conditions. Microarray hybridizations were carried out at the Centre of Excellence for Fluorescent Bioanalytics at the University of Regensburg. Each microarray configuration is deposited at the National Center for Biotechnology Information Gene Expression Omnibus (GEO; accession number GSE29753). Data were analyzed with the NetAffx Analysis Center and the Robust Multi-array Analysis (RMA) algorithm. A fold-change of more than 2 with a maximal p-value of 0.05 was considered significant. The Database for Annotation, Visualization, and Integrated Discovery (DAVID; http://niaid.abcc.ncifcrf.gov) was used for annotations of significant regulated transcripts after differentiation. The data were also analyzed by Ingenuity Pathways Analysis (IPA; Ingenuity® Systems, http://www.ingenuity.com). Genes that met the fold change cut-off were associated with biological functions in Ingenuity's Knowledge Base. Right-tailed Fisher's exact test was used to calculate a p-value determining the probability that each biological function assigned to that data set was due to chance alone.
Quantitative reverse-transcription polymerase chain reaction
Total RNA was isolated from DFCs with NucleoSpin® RNA II (Macherey Nagel). The cDNA synthesis was performed using 400 ng total RNA and RevertAid™ M-MuLV Reverse Transcriptase Kit (Fermentas, St. Leon-Rot, Germany). Quantitative PCR was performed with the Fast Start DNA Master SYBR® Green I kit (Roche). Sequences for primers and probes are listed in Supplementary Table S1 (Supplementary Data available online at www.liebertonline.com/scd). Quantitative RT-PCR was performed with the LightCycler PCR System (Roche). The LightCycler 4.05 software was used for estimation of threshold cycles (Ct value). Glyceraldehyde-3-phosphate-dehydrogenase (GAPDH) was used as a housekeeping gene. Quantification was done with the ΔΔCt calculation method as described by Winer et al. [22]. The total RNA from correspondent controls of experiments was used for calibration (relative gene expression=1).
Western blotting
For protein extraction, DFCs at indicated time points were washed with PBS and harvested by trypsin-EDTA treatment. After extensively washing with basal medium and PBS to eliminate trypsin, DFCs were treated with lysis buffer (1 mM Na-Orthovanadate, 150 mM NaCl, 1 mM EDTA, and 1% NP-40) on ice for 30 min. Solutions for protein extractions contain protease-inhibitor tablets (complete mini, Roche) to minimize protein degradation. An aliquot of 25 μg protein extract was denatured and reduced by heating in sodium dodecyl sulfate (SDS) sample buffer containing dithiothreitol, separated by SDS-polyacrylamide gel electrophoresis in 12% Tris-glycine gels (Invitrogen, Carlsbad, California) and transferred to nitrocellulose membranes. The membranes were blocked with skimmed milk for 60 min followed by the incubation with antigen-specific antibodies overnight at 4°C. Antibodies which detect the following proteins were used: DLX3 (1E9; Chemicon, Billerica, Massachusetts), RUNX2 (M-70; Santa Cruz Biotechnology, Santa Cruz, California), β-Actin (AC-15; Novus Biologicals, Littleton, Colorado), V5-Epitope (Invitrogen); and pSMAD1 (41D10), BAX (D2E11), and BCL2 (D5568; all purchased from Cell Signaling). After having been washed, the membranes were incubated with horseradish peroxidase-conjugated antibodies for 60 min, which were specific either for mouse IgG (Promega) or rabbit IgG (Cell Signaling). Detection was performed by chemiluminescence (GE Healthcare, Amersham).
Apoptosis analysis
Apoptosis in DFCs was determined with Annexin V-FITC/PI assay as described previously [23]. For measurement of apoptosis, the cells were transfected with DLX3 siRNA or pDLX3 (see above). In addition, after 4 h of transfection with pDLX3, 0.5 μM camptothecin (MBL, Woburn, Massachusetts) was added to the cell cultures. After 48 h DFCs were harvested by trypsin-EDTA treatment, washed with ice-cold PBS (2% BSA), and stained with PI and fluorescein isothiocyanate (FITC) conjugated Annexin V using an Annexin V-FITC Apoptosis Detection Kit I (Becton Dickinson). Annexin V-FITC identifies cells in early apoptosis by detecting externalized phosphatidylserine, and PI identifies cells that have lost plasma membrane integrity (i.e., necrotic or late apoptotic cells). The cells were resuspended in 100 μL of 1×binding buffer supplemented with 1 μL of Annexin V-FITC and 5 μL of PI and kept at room temperature in the dark for 15 min according to the manufacturer's instructions. Following the addition of 400 μL of 1× binding buffer, the stained cells were kept on ice and subjected to analysis using a FACS Canto flow cytometer (BD Biosciences, San Jose, California). The FITC fluorescence was measured through a 530/30 band pass filter and the PI fluorescence through 650 nm.
Chromatin immunoprecipitation
DLX3-transfected DFCs cultivated in basal medium for 72 h were used for chromatin immunoprecipitation. Approximately 1×107 cells per immunoprecipitation were incubated with fixation buffer (500 mM HEPES/KOH [pH 7.9]; 100 mM NaCl; 1mM EDTA [pH 8.0]; 0.5 mM EGTA [pH 8.0]; 1% formaldehyde) for 10 min. For neutralization, glycine was added to a final concentration of 125 mM and cells were washed with cold PBS.
Cells were harvested, centrifuged, and lysed in a lysis buffer containing 10 mM HEPES/KOH [pH 7.9], 85 mM KCl, 1 mM EDTA [pH 8.0], and 0.1% NP-40. To isolate the nuclei, cells were lysed with 50 mM Tris/HCl [pH 7.0]; 1% SDS, 0.5% Empigen BB (Fluka); and 10 mM EDTA [pH 8.0]. Samples were sonicated (4×10 s, amplitude 10%) to reduce the DNA length into 0.2-1.0 kb, and cellular debris was removed by centrifugation at 13,000 rpm for 5 min at 4°C.
Sepharose CL-4B beads (Sigma) were prepared for preclearing with salmon sperm DNA (Sigma) (10μg/μL in 20 mM Tris/HCl; 100 mM NaCl; 2 mM EDTA; 1% Triton-X 100; 20% BSA) for 1h at 4°C. Approximately 340 μL of cell nuclear extract were transferred to a clean microcentrifuge tube and precleared with 640 μL of preincubated sepharose CL-4B beads for 2 hours at 4°C. Precleared lysate was centrifuged for 5 min at 13 000 rpm. For control, a DLX3 specific antibody or an unspecific immunoglobulin G (IgG) antibody (2.5 μg) were added to 200 μl of supernatant of the precleared lysates and incubated over night at 4°C.
ProteinA/proteinG sepharose beads (Sigma) were blocked with salmon sperm DNA (10μg/μL in 20 mM Tris/HCl; 100 mM NaCl; 2 mM EDTA; 1% Triton-X 100; 20% BSA) for 1h at 4°C. Blocked proteinA/proteinG sepharose beads were added to the samples and incubated for 2 h at 4°C. Subsequently, using Ultrafree MC spin columns (Millipore, Temecula, California), the beads were washed (20 mM Tris/HCl [pH 7.4]; 150 mM NaCl; 0.1% SDS; 1% Triton-X 100), and the precipitated antibody/antigen/DNA was eluted in elution buffer (0.1 mM NaHCO3; 1% SDS). Transcription factor/DNA cross-links were reversed over night at 65°C.
For chromatin purification, the QiaQuick Kit (Qiagen) was used according to the manufacturer. The DNA was analyzed by PCR with specific primers for DLX3 binding sites on promoter-regions of the RUNX2 (primers: forward ‘TTGAAAAGAAGAAAACCATTTGC’; reverse ‘AGCTTGTGGGATTTCCAAAC’) and ZBTB16 (Primers: forward ‘TACTCTCAGCATCCCTGGCT’; reverse ‘TCAACCACTACAACATGCCC’) genes. The promoter binding sites for DLX3 were predicted with the MatInspector/MatBase program of Genomatix software suite (Genomatix, Munich, Germany).
Results
DLX3 overexpression and inhibition in DFCs
For the analyses of molecular function, the overexpression and inhibition of DLX3 in DFCs were established. DLX3 was overexpressed in DFCs after transfection of the pDLX3 plasmid or inhibited with a siRNA complementary for a short sequence of DLX3.
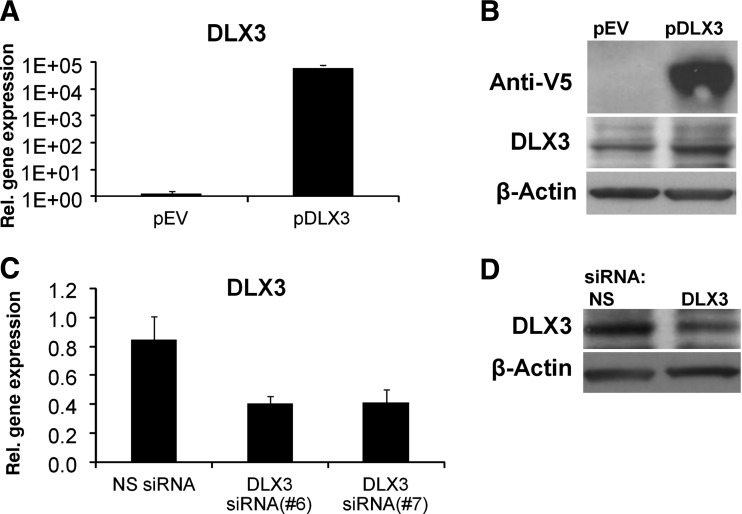
The DLX3 mRNA level was highly increased (1×105) in DFCs transfected with pDLX3 (Fig. 1A). The expression of the recombinant DLX3 protein was detected with an anti-V5 antibody and anti-DLX3 antibody in DFCs (Fig. 1B). DLX3 gene expression was down-regulated after transfection with a DLX3 specific siRNA (Fig. 1C). A western blot analysis with an anti-DLX3 antibody revealed also that protein expression of DLX3 was down-regulated (Fig. 1D).
FIG. 1.
DLX3 overexpression or DLX3 silencing in dental follicle stem cells (DFCs). The gene expression of DLX3 was determined by quantitative reverse- transcription polymerase chain reaction (qRT-PCR) analysis after 48 h of transfection with the expression plasmid (pDLX3) and the empty vector (pEV) (A) or with two independently DLX3-specific small interfering RNA (siRNA), siRNA(6) and siRNA(7), and a nonspecific siRNA, NS siRNA (C). All values are means plus standard error (σ/√n) of three biological replicates (n=3) per group. Gene expression of DFCs after transfection with pDLX3 or DLX3 siRNA was compared with the control groups pEV or NS siRNA, respectively. DLX3 expression were also verified at protein level by western blotting (B,D); recombinant DLX3 was detected with an epitop-V5 specific antibody and the total DLX3 protein was detected with a specific antibody DLX3. The β-Actin antibody was used as a housekeeper standard. Protein lysates after 72 h of transfection (for details, see Materials and Methods) were used for western blot analysis.
Microarray analyses of DFCs after DLX3 overexpression
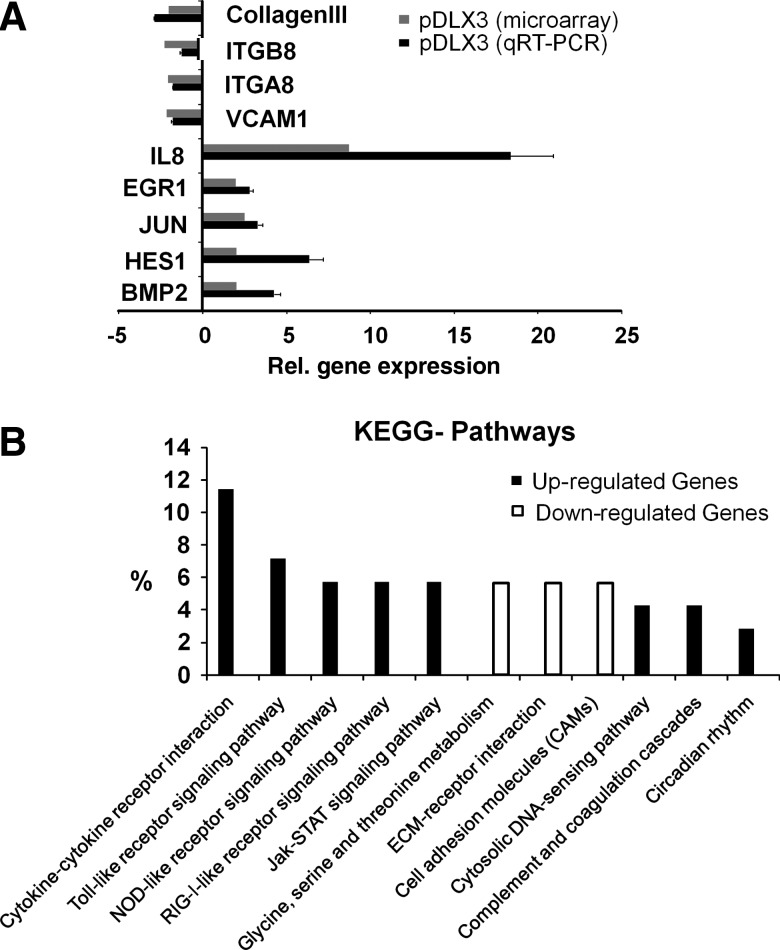
After DLX3 overexpression in DFCs, we identified 73 up-regulated and 55 down-regulated genes (Supplemental Table S2). The highest up-regulated genes were IL8, the chemokines CXCL10 and CXCL11, and MMP1 (Fig. 2A). Moreover, genes such as BMP2, NR4A2, HES1, and ATF3 that were significantly up-regulated are associated with developmental processes. Besides these, connective tissue markers such as Col3A1, ELN (elastin), OMD (osteomodulin), and PLXNC1 (plexin) were significantly down-regulated. Real-time RT-PCR assays of selected regulated genes verified microarray data (Fig. 2A).
FIG. 2.
Microarray analysis after DLX3 overexpression in DFCs. (A) The real-time RT-PCRs confirmed regulated gene expression of selected genes of the DNA microarray analysis. Gray bars represent the mean of gene expression measured by Affymetrix microarray analysis and the black bars of qRT-PCRs. Total RNAs from DFCs transfected with pEV were used for calibration of real-time RT-PCRs (relative gene expression=0). Primers listed in table 1a. (B) Comparative Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis of Affymetrix microarray expression signals from DFCs after DLX3 overexpression (48h). KEGG pathway analysis (p values <0.05: strongly enriched in the annotation categories) of up-regulated genes (black bars) and down-regulated genes (white bars).
The Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis revealed overrepresented pathways such as the Janus kinase, and signal transducer and activator of transcription (Jak-STAT) pathway in the group of up-regulated genes. While in the group of down-regulated genes, for example the glycine, serine, and threonine metabolism, extracellular matrix (ECM)-receptor interaction and cell adhesion molecules (CAMs) pathways are overrepresented (Fig. 2B).
Cellular growth and differentiation, cellular development, cellular movement, cell death (apoptosis), and cell-to-cell signaling and interaction were the most overrepresented molecular and cellular functions found after an analysis with Ingenuity (Supplementary Table S3). Organismal survival and skeletal system development and function were associated in the group of regulated genes (Supplementary Table S4). Interestingly, the most overrepresented network of associated functions contains more specific terms such as inhibition of apoptosis, proliferation of normal cells, morphology of cells, and chemotaxis of bone cell lines or binding of osteoblasts (Supplementary Figure S2).
Cell proliferation, morphology, and apoptosis
Amongst others, most overrepresented biological functions in DFCs after DLX3 overexpression were cell proliferation, morphology of cells, and inhibition of apoptosis.
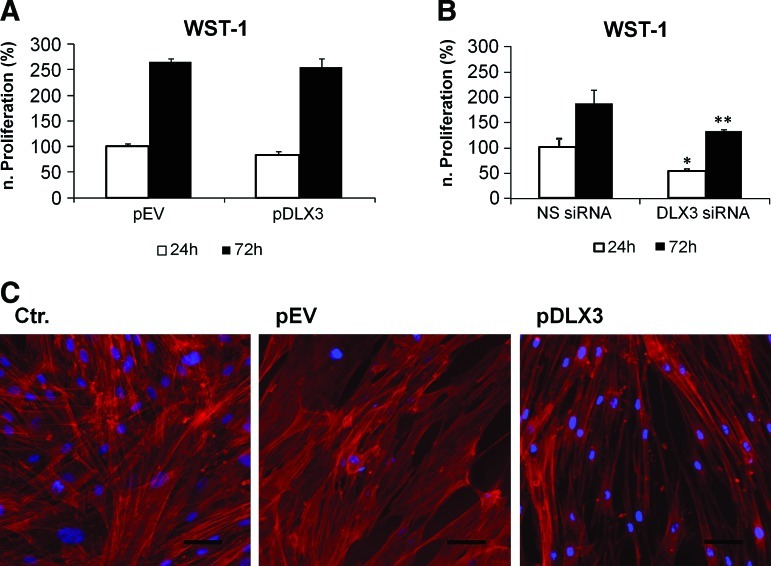
Cell proliferation was maintained after DLX3 overexpression, but it was significantly reduced after silencing of DLX3 (Fig. 3A, B). The morphology of DFCs after DLX3-overexpression or -silencing was characterized using fluorescent staining. Interestingly, DFCs transfected with pDLX3 had a more spindle like cell shape with reorganized actin filaments (Fig. 3C); however, DFCs after DLX3 silencing were similar to controls (data not shown).
FIG. 3.
Influence of DLX3 on proliferation and morphology of DFCs. For evaluation of cell proliferation, the metabolic activity of DFCs was measured with WST-1 after 24 h and 72 h of transfection (A, B). The proliferation was not influenced by DLX3 overexpression (A), but decreased after DLX3 silencing (B). All values are means plus standard error (σ/√n) of 4 biological replicates. DFCs after DLX3 overexpression and silencing were compared with controls after 24 h and 72 h. Statistics were done using the Student's t test. *p<0.05 t test, **p<0.005. (C) Cell morphology changes of DFCs after DLX3 overexpression was visualized by staining of actin filaments and the cell nucleus (for details see Materials and Methods). Standard bars: 50 μm. Color images available online at www.liebertonline.com/scd
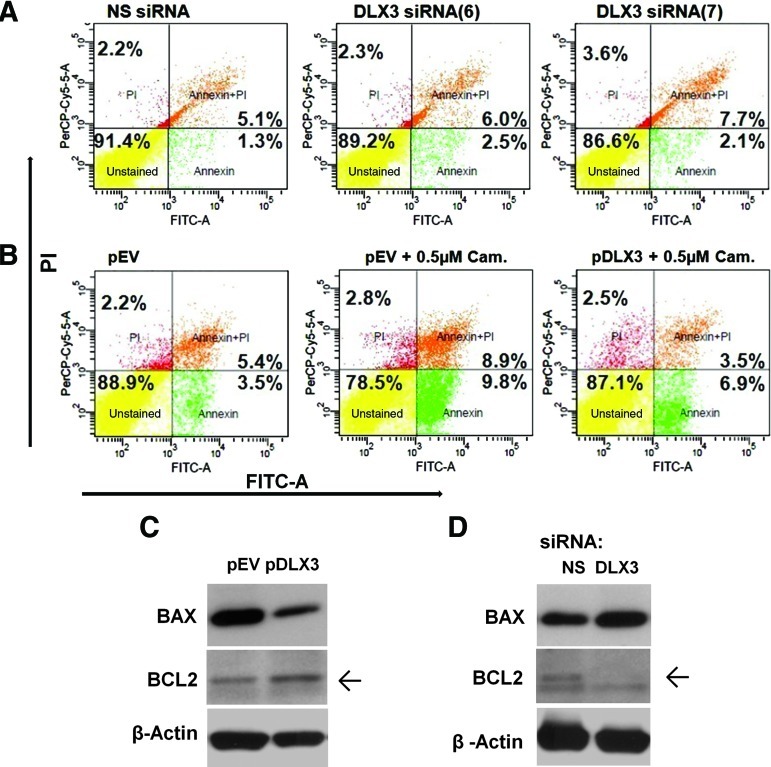
To verify the influence of DLX3 on apoptosis in DFCs, a flow cytometry assay with FITC Annexin V was carried out after gene regulation of DLX3. Results of this investigation suggest that DLX3 silencing exhibits approximately 48% more apoptotic cells than the respective control with a reduction of viable cells from total of 91.4% (NS siRNA) down to 86.6% (DLX3 siRNA7) (Fig. 4A). On the other hand, the number of apoptotic cells of DFCs after transfection with pDLX3 and incubation with the cytotoxic quinoline alkaloid camptothecine was about 7-fold diminished in comparison with the empty vector (Fig. 4B). The FACS analysis was supported by the increased and decreased expression of the pro-apoptotic protein BAX, after silencing and overexpression of DLX3, respectively (Fig. 4C). A western blot analysis with an anti-BCL2 antibody revealed also that the expression of the anti-apoptotic protein was down-regulated and increased, after silencing and overexpression of DLX3, respectively (Fig. 4D).
FIG. 4.
Apoptosis of DFCs. FACS analysis after transfection with (A) DLX3 specific siRNAs (6 and 7) and a NS siRNA, or (B) with pDLX3 and pEV in DFCs after treatment with Camptothecin. For visualization of cell viability the Annexin V/PI assay was applied. Viable cells (annexin V−; PI−) are shown in the lower left quadrant of density plots. Apoptotic cells (annexin V+; PI−) are shown in the lower right quadrant. Cells in the late apoptosis (annexin V+; PI+) are shown in the upper right quadrant and necrotic cells (annexin V−; PI+) are in the upper left quadrant. Western blot analysis with BAX, BCL2, and β-Actin antibodies and protein lysates of DFCs, 72 h after transfection with (C) pDLX3 and pEV or (D) DLX3 siRNA(6) and NS siRNA on basal medium. Color images available online at www.liebertonline.com/scd
DLX3 stimulates directly the osteogenic differentiation of DFCs
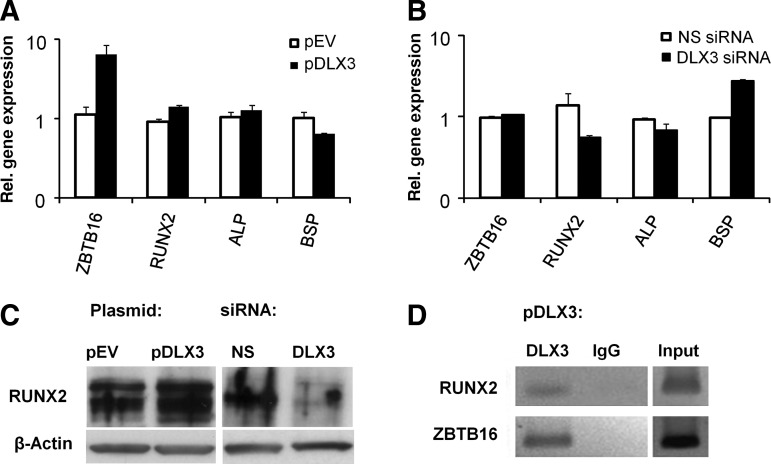
We analyzed the gene expression of osteogenic markers, which were associated with the differentiation of DFCs [20,24]. Here, osteogenic markers such as alkaline phosphatase (ALP) and RUNX2 or transcription factors, which were associated with differentiated DFCs, such as ZBTB16 were highly up-regulated in DLX3-overexpressing cells. In contrast, bone sialoprotein (BSP) was down-regulated after DLX3 overexpression (Fig. 5A). After DLX3 silencing, RUNX2, ALP, and BSP were contrarily regulated (Fig. 5B). The RUNX2 qRT-PCR results were verified by western blot (Fig. 5C). A DLX3-specific chromatin immunoprecipitation (ChIP) assay verified that DLX3 binds directly on promoters of osteogenic marker genes RUNX2 and ZBTB16 after DLX3 overexpression in DFCs (Fig. 5D). Moreover, RUNX2, ZBTB16, and BMP2 were regulated by DLX3 in a dose-dependent manner (Supplementary Fig. S3).
FIG. 5.
DLX3 directly regulates expression of osteogenic markers in DFCs. Relative gene expression of ostegenic markers in DFCs was determined 48 h after transfection with (A) pDLX3 and pEV or (B) DLX3 siRNA(6) and NS siRNA by real-time PCR analysis. Sequences for primers and probes are listed in Table 1A. The gene expression of DFCs transfected with the respective controls (pEV or NS siRNA) was used for calibration. All values are means plus standard error (σ/√n) of 3 biological replicates. (C) Western blot analyses were done with RUNX2 and β-Actin specific antibodies and protein lysates of DFCs, 72 h after transfection with pDLX3 and pEV or DLX3 siRNA and NS siRNA in basal medium. (D) Chromatin immunoprecipitation analysis with DFCs (after DLX3 transfection) cultivated 72 h in basal medium, Dulbecco's modified Eagle's medium. The precipitations were done with either a DLX3 specific antibody or an unspecific antibody (immunoglobulin G, IgG) for control. PCRs for DLX3-binding sites on promoters of the genes RUNX2 and ZBTB16 were made with precipitated genomic DNA. ZBTB16, zinc finger and BTB domain containing 16; RUNX2, runt-related transcription factor 2; ALP, alkaline phosphatase; BSP, bone sialoprotein.
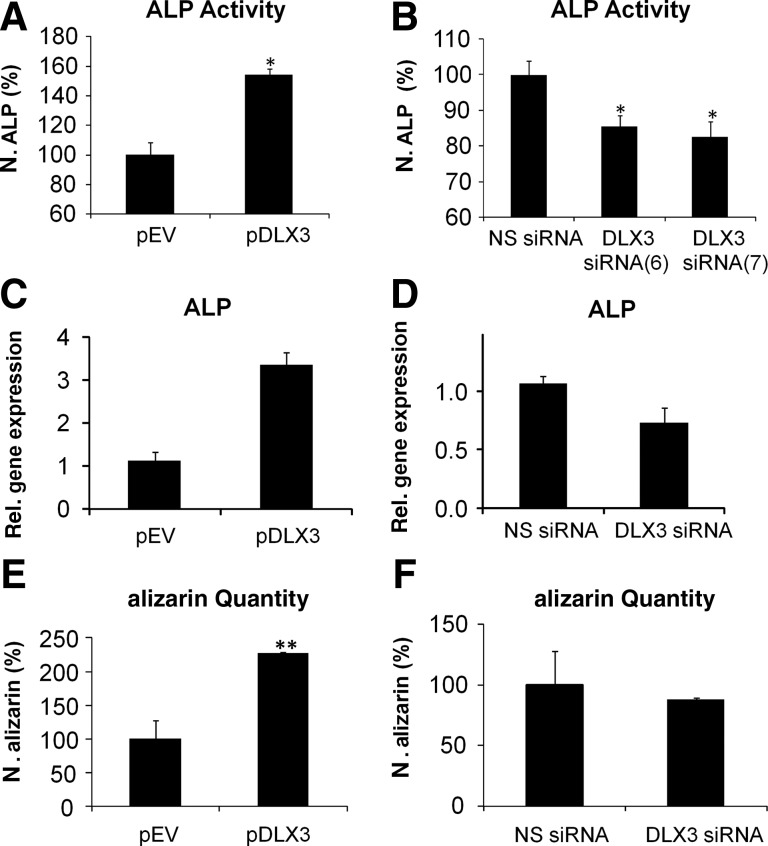
In DFCs the ALP activity was significantly increased after osteogenic differentiation and DLX3 overexpression, but down-regulated after DLX3 silencing (Fig. 6A, B). qRT-PCR analyses revealed that the expression of ALP was also increased or decreased in DFCs after differentiation in ODM and overexpression or silencing of DLX3, respectively (Fig. 6C, D). While DLX3 overexpression induced the highest mineralization of DFCs, they showed a slightly decreased mineralization in comparison to control cells after inhibition of DLX3 (Fig. 6E, F). A measurable ALP activity and mineralization was only observed in cultures with ODM.
FIG. 6.
Influence of DLX3 on osteogenic differentiation of DFCs. The differentiation was evaluated by measurement of the ALP activity (A, B). ALP activity was quantified in DFCs 10 days after induction with osteogenic differentiation medium (ODM) and transfection with (A) pDLX3 and pEV or (B) DLX3 specific siRNAs (6 and 7). ALP activities were calibrated to the activity of the respective control cultures, pEV or NS siRNA, with ODM. All values are means plus standard error (σ/√n) of 4 biological replicates per group. Significant differences are shown with the Student's t test (n=4; *P<0.05). Relative gene expression of ALP was determined 3 days after induction with ODM and transfection with (C) pDLX3 and pEV or a (D) DLX3 specific siRNA(6). Mineral deposits in DFC cultures were estimated with alizarin red staining after 28 days of culture with ODM and transfection with (E) pDLX3 and pEV or (F) DLX3 siRNA(6) and NS siRNA. Quantification of alizarin staining was normalized to the respective controls, pEV or NS siRNA, cultivated in ODM. All values are means plus standard error (σ/√n) of 3 biological replicates per group. Significant differences are shown with the Student's t test (**: p<0.005).
DLX3 stimulates osteogenic differentiation via BMP2 dependent pathway
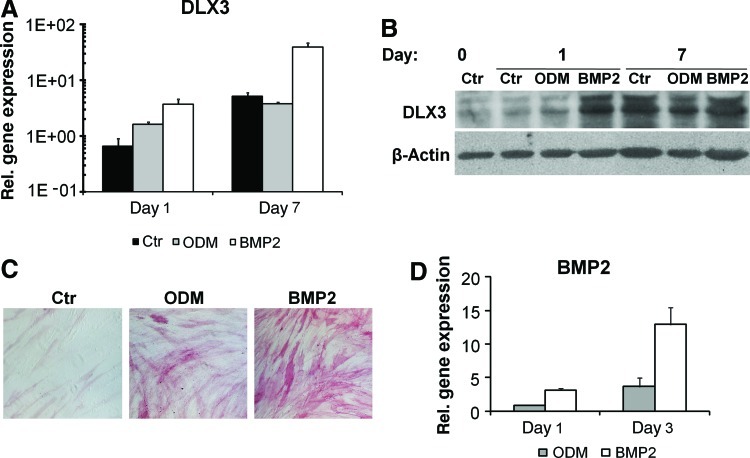
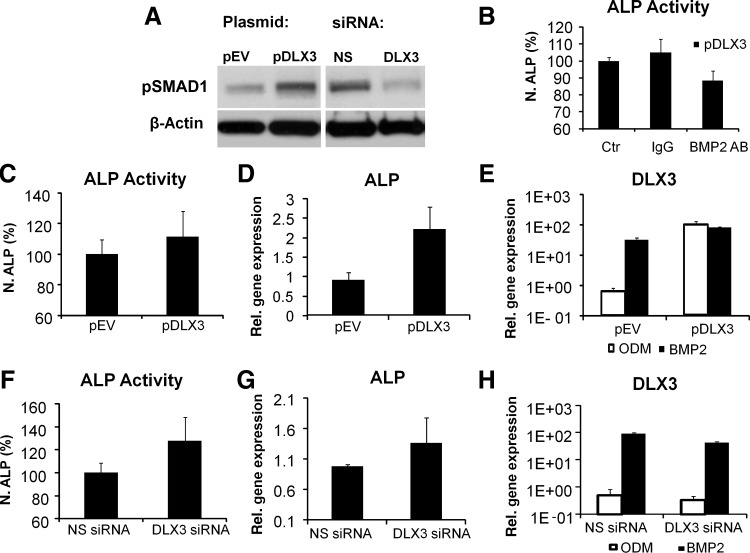
A previous study has demonstrated that DLX3 was up-regulated during osteogenic differentiation with dexamethasone or insulin in DFCs [19]. In addition, we investigated the expression of DLX3 during osteogenic differentiation in BMP2 induced DFCs. Here, DLX3 was up-regulated even at early time points of osteogenic differentiation (Fig. 7A, B). The expression level of DLX3 in DFCs was highly up-regulated in BMP2 in comparison to that in differentiation medium ODM (Fig. 7A, B). Interestingly, BMP2 induces the expression of BMP2 and therefore also the ALP activity in DFCs (Fig. 7D, C). Additional experiments suggest a direct relation between the osteogenic differentiation of DFCs and the expression of BMP2 and DLX3. After DLX3 overexpression BMP2 was significantly up-regulated (Fig. 2A). A western blot analysis for the phosphorylated form of SMAD1 demonstrated the activation and silencing of the BMP pathway after overexpression and silencing of DLX3, respectively (Fig. 8A). Moreover, the inhibition of the BMP pathway with a BMP2 specific antibody diminished the ALP activity in dexamethasone-differentiated DFCs after DLX3 overexpression (Fig. 8B). In contrast to our previous observation with a dexamethasone based protocol (Fig. 6B), the BMP2 induced ALP activity and ALP gene expression was not inhibited after DLX3 silencing (Fig. 8F, G). Moreover, the ALP activity and gene expression was not remarkably increased in DFCs after BMP2 induction and DLX3 overexpression (Fig. 8C, D). The expression of DLX3 was either increased or decreased in BMP2 induced DFCs after overexpression or silencing of DLX3, respectively (Fig. 8 E, H). Interestingly, DLX3 is more highly expressed in DFCs after treatment with BMP2 than in DFCs after treatment with ODM (Fig. 7A, B), but gene expression levels with both treatments were similar after DLX3 overexpression (Fig.8 E, H). These results suggest a feedback mechanism between BMP2 and DLX3.
FIG. 7.
DLX3 expression during osteogenic differentiation in DFCs. (A, B) DLX3 expression after 1 and 7 days of induction with either ODM or BMP2. (A) The relative gene expression of DLX3 was determined by qRT-PCR analysis and calibrated to the gene expression of control cells (Ctr) before differentiation. All values are means plus standard error (σ/√n) of 3 biological replicates. (B) Western blot analysis with DLX3 and β-Actin specific antibodies and protein lysates of DFCs cultivated in DMEM (Ctr) before (day 0) and after 1 and 7 days of osteogenic differentiation in ODM or BMP2. (C) ALP activity staining in DFCs after 10 days of induction with ODM or BMP2 and without induction (Ctr, basal medium). All figures have the same magnification. (D) BMP2 expression after 1 and 3 days of induction with either ODM or BMP2. All values are means plus standard error (σ/√n) of 3 biological replicates. Color images available online at www.liebertonline.com/scd
FIG. 8.
Relation between DLX3 and BMP2 expression during osteogenic differentiation in DFCs. (A) Western Blot analysis with pSMAD1 and β-Actin specific antibodies and protein lysates of DFCs, 72 h after transfection with either pDLX3 or DLX3 siRNA in basal medium. (B) ALP activity (day 10 of differentiation in ODM) in DFCs after DLX3 overexpression and selectively inhibition of the BMP-2 pathway with a specific anti-BMP2 neutralizing antibody. The effect was compared with DLX3- transfected DFCs in ODM and an unspecific antibody (IgG) and with DLX3-transfected DFCs in ODM (Ctr.) without antibody treatment. Results are relative ALP activities to the mean activity of the control culture. All values are means plus standard error (σ/√n) of 4 biological replicates per group. (C, F) The ALP activity was quantified in DFCs after 10 days of BMP2 induction and after transfection with either pDLX3 (C) or DLX3 siRNA(6) (F). All values are means plus standard error (σ/√n) of 3 biological replicates per group; the differences were not significant. Relative gene expression of ALP was determined 3 days after induction with BMP2 and transfection with (D) pDLX3 and pEV or a (G) DLX3 specific siRNA(6). DLX3 expression was determined in DFCs after 3 days osteogenic differentiation, induced with BMP2 or ODM, and transfection with pDLX3 and pEV (E) or DLX3 siRNA(6) and NS siRNA (H). All values are means plus standard error (σ/√n) of 3 biological replicates.
Discussion
The mammalian homologues of Drosophila distal-less (DLX) homeobox proteins have functions in skeletal morphogenesis and are involved in early vertebrate morphogenesis of the head and dentition [25,26]. DLX3, for example, is expressed in structures involving epithelial-mesenchymal interaction such as skin, hair, and teeth [8,10]. It is expressed in mineralizing cells such as odontoblast, ameloblast, and osteoblast cells, although a differential expression pattern or function has been reported [14–16]. For example, a 4-bp mutation in DLX3 is related with the TDO syndrome that is characterized with increased bone volume and mineral density but a decreased thickness of dentin [11,12,27,28]. DFCs reside in the dental follicle and are precursor cells for cementoblasts, periodontal ligament fibroblasts, and alveolar osteoblasts [1,2,29]. In this work we characterized DLX3 as an important transcription factor for the differentiation of DFCs into mineralized tissue cells such as alveolar bone osteoblasts.
For molecular investigations of the gene function, we successfully manipulated the DLX3 expression in DFCs by chemical transfection methods. Previous studies have shown that eukaryotic expression systems and antisense oligonucleotide techniques can successfully applied in DFCs [30–32]. For example, Wise et al. achieved a stable silencing of CSF1 for up to 72 h with siRNA in DFCs, while we have reported a high inhibition of DLX3 with siRNA in DFCs even after 3 days of osteogenic differentiation.
Gene expression profiles after DLX3 overexpression were estimated with microarray analyses to evaluate regulated processes in DFCs down-stream of DLX3. Microarray data revealed that the most overrepresented pathway was the cytokine–cytokine receptor interaction pathway with the highly up-regulated gene IL8. IL8 has specific neutrophil and lymphocyte chemotactic activity and is involved in the induction of angiogenesis [33]. Moreover, IL8 is expressed by human osteoblast-like cells, osteosarcoma cell line MG-63, bone marrow stromal cells, and osteoclasts, suggesting that IL8 plays a role in the regulation of cellular functions in bone [34,35]. In our study IL8 is highly up-regulated and participates in most of the regulated pathways after DLX3 overexpression. However, in preliminary experiments IL8 did not modulate the osteogenic differentiation of DFCs (unpublished data). Further experiments have to be done to elucidate the role of IL8 in the process of differentiation of DFCs.
The Jak-STAT pathway including genes sprouty2 (SPRY2) and sprouty4 (SPRY4) are significantly up-regulated after DLX3 overexpression. These genes play an essential role in the bidirectional signal among epithelial and mesenchymal cells, which is also an important process for dental development [36]. Moreover, the Toll-like receptor (TLR) and Nod-like receptor (NLR) signaling pathways were also overrepresented and are well known for triggering innate immune response. Although these pathways are associated with the immune response, new studies showed that both NLRs and TLRs are also involved in the regulation of differentiation in mesenchymal stem cells (MSCs) [37].
Previous studies reported gene expression profiles of DFCs after 1 week of osteogenic differentiation with dexamethasone, BMP-2, IGF-2 and 4 weeks with dexamethasone [20,38]. These studies share significantly down-regulated genes with down-regulated genes after DLX3 overexpression. These genes can be clustered into the amino acid metabolic process, which could be an important biological process for the proliferation of DFCs. Interestingly, 50% of regulated genes after BMP2 induction and after DLX3 overexpression were contrarily regulated in DFCs. These genes can be clustered in functions such as cell adhesion, proliferation, survival, and development process of tissue. Moreover, after DLX3 overexpression DFCs share some regulated genes and biological functions with 4 weeks differentiated DFCs. Here biological functions such as cell activation and viability and genes such as STC1, which are associated with osteogenic differentiation [39], are noteworthy.
An evident regulated process after DLX3 overexpression was the programmed cell death (apoptosis). We showed that silencing of DLX3 induces apoptosis in DFCs, while DLX3 overexpression decreases the number of apoptotic cells. Interestingly, silencing of DLX3 also hampers the cell vitality/proliferation of DFCs. These results suggest that DLX3 participates in the negative regulation mechanism of apoptosis in DFCs. The principal function of apoptosis is the maintenance of tissue homeostasis by balancing cell proliferation and cell death [40]. Moreover, apoptosis is a fundamental component of osteoblast differentiation that contributes to maintaining tissue organization and regulates bone homeostasis [41,42]. It was reported that DLX3 participates in the control of fenretinide-mediated apoptosis in tumor cell lines [43]. Furthermore, Choi et al. reported the influence of a DLX3 mutant for odontoblast apoptosis as a consequence of odontoblast cytodifferentiation disruption in vivo [27].
Another regulated process after DLX3 overexpression was the morphology of cells. Previous studies suggested that cell shape and the alteration of their cytoskeletal components are key regulators of mesenchymal stem cell commitment during differentiation [44,45]. Interestingly, DFCs shape became spindle like with new organized actin filaments after DLX3 overexpression. Genes such as MTSS1, which are important for actin filament morphology regulation [46], were up-regulated in DLX3-overexpressing DFCs. These results propose that DLX3 influences also the cell shape of DFCs.
DLX3 plays an important role in skeletal development and matrix mineralization [9,14]. ALP activity and alizarin red staining showed that DLX3 influences directly the process of osteogenic differentiation and matrix mineralization of DFCs. Furthermore, qRT-PCR analyses showed a regulation of osteogenic marker genes. For example, after DLX3 overexpression in DFCs, gene expression of ZBTB16 was significantly increased. The transcription factor ZBTB16 is a late marker of osteogenic differentiated DFCs. The expression of ZBTB16 was previously reported in DFCs induced with dexamethasone but not in BMP2- or IGF2-differentiated cells [20,24]. ZBTB16 was reported as an inducer of osteoblast-like and osteoblast cell differentiation and mineralization acting upstream of RUNX2 independent but very closely related to the BMP-signaling pathway, [47,48]. Interestingly, ZBTB16, RUNX2, and ALP were up-regulated while BSP was down-regulated after DLX3 overexpression. It is consistent with the knowledge that the expression of RUNX2 and ALP in bone formation has an important role in early and middle stages of differentiation, while the expression of BSP has a significant role in the late stages of osteoblast differentiation. Furthermore, CHIP assays for protein–DNA interactions revealed that DLX3 binds directly the RUNX2 and ZBTB16 promoters and probably regulates their expression (as shown by western blot for RUNX2). Further studies have to evaluate the molecular interactions/functions of DLX3 on promoters of osteogenic marker genes in DFCs. However, our data suggest that DLX3 supports the osteogenic differentiation of DFCs.
A previous study has shown the induction of DLX3 by BMP2 in diverse tissue cells [9]. In this study, we demonstrated a close relationship between the expression of BMP2 and DLX3 during osteogenic differentiation in DFCs. Previous studies have already shown that BMP2 induces the osteogenic differentiation of DFCs [4,20]. In addition we showed that BMP2 induces the expression of DLX3. However, for the first time we could also show that DLX3 induces the expression of BMP2 and therefore activates probably indirectly the BMP pathway, which we could show by the phosphorylation of SMAD1 after the regulation of DLX3. We suggest a close relation between BMP2 (signaling), DLX3, and osteogenic differentiation (ALP activity). The inhibition of BMP signaling with a specific antibody for BMP2 reduces the ALP activity in DLX3 overexpressing DFCs in ODM. In contrast, a BMP2 supplementation in DLX3 overexpressing DFCs did only slightly increase the expression of DLX3 and the ALP activity. In this case, DLX3 expression was high with and without DLX3 overexpression after 3 days of differentiation. Here, the differentiation could not be stimulated with extra BMP2 proteins. Moreover, after DLX3 silencing ALP activity could not be inhibited in BMP2 treated DFCs. We suppose that the inhibition of ALP activity by DLX3 silencing in ODM succeed indirectly via the BMP signaling, which can be reversed by BMP2 supplementation. However, after the induction of the differentiation with BMP2 the expression of DLX3 was about 80 times higher than in DFCs after cultivation in ODM. Alternatively, we can also suppose that DLX3 directly regulates the ALP activity, but DLX3 cannot exceed a definite expression level. This would explain that we cannot find additive DLX3 expression and ALP activity after BMP2 treatment and overexpression of DLX3. Moreover, a reduction of ALP activity was not observed after DLX3 silencing in BMP2 treated DFCs, because the expression of DLX3 was high. We suppose that DLX3 expression level in BMP2 treated DFCs exceed a specific threshold beyond that a regulation of ALP expression was not possible with DLX3 siRNAs. However, further experiments are necessary to prove this hypothesis of DLX3 and ALP expression. Interestingly, 50% of regulated genes were contrarily regulated in DFCs after BMP2 supplementation [20] and DLX3 overexpression. These results suggest that BMP2 (BMP signaling) and the transcription factor DLX3 control each other and the differentiation in DFCs with a feedback control. However, further studies have to evaluate this molecular process in more details.
We find that DLX3 effectively regulates cell proliferation, cell morphology, and apoptosis and enhances dexamethasone-induced osteogenic differentiation and mineralization of DFCs. Moreover, DLX3 and BMP2 induce and control each other during the complex process of osteogenic differentiation in DFCs.
These findings improve our knowledge about the molecular mechanisms of DLX3 and their influence in the osteogenic differentiation of DFCs and therefore in the development of the periodontium.
Supplementary Material
Acknowledgments
This work was supported by a grant of the Deutsche Forschungsgemeinschaft (DFG) MO-1875/2-1 and (DFG) MO-1875/2-2. We thank Prof. Dr. H. Schweikl for his irreplaceable support to the investigation of cellular apoptosis and Mrs. C. Bolay and A. Reck for their technical support.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Ten Cate AR. The development of the periodontium: a largely ectomesenchymally derived unit. Periodontol 2000. 1997;13:9–19. doi: 10.1111/j.1600-0757.1997.tb00093.x. [DOI] [PubMed] [Google Scholar]
- 2.Morsczeck C. Gotz W. Schierholz J. Zeilhofer F. Kuhn U. Mohl C. Sippel C. Hoffmann KH. Isolation of precursor cells (PCs) from human dental follicle of wisdom teeth. Matrix Biol. 2005;24:155–165. doi: 10.1016/j.matbio.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 3.Völlner F. Ernst W. Driemel O. Morsczeck C. A two-step strategy for neuronal differentiation in vitro of human dental follicle cells. Differentiation. 2009;77:433–441. doi: 10.1016/j.diff.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Kemoun P. Laurencin-Dalicieux S. Rue J. Farges JC. Gennero I. Conte-Auriol F. Briand-Mesange F. Gadelorge M. Arzate H. Narayanan AS. Brunel G. Salles JP. Human dental follicle cells acquire cementoblast features under stimulation by BMP-2/-7 and enamel matrix derivatives (EMD) in vitro. Cell Tissue Res. 2007;329:283–294. doi: 10.1007/s00441-007-0397-3. [DOI] [PubMed] [Google Scholar]
- 5.Thesleff I. Mikkola M. The role of growth factors in tooth development. Int Rev Cytol. 2002;217:93–135. doi: 10.1016/s0074-7696(02)17013-6. [DOI] [PubMed] [Google Scholar]
- 6.Sodek J. McKee MD. Molecular and cellular biology of alveolar bone. Periodontol 2000. 2000;24:99–126. doi: 10.1034/j.1600-0757.2000.2240106.x. [DOI] [PubMed] [Google Scholar]
- 7.Morasso MI. Grinberg A. Robinson G. Sargent TD. Mahon KA. Placental failure in mice lacking the homeobox gene Dlx3. Proc Natl Acad Sci U S A. 1999;96:162–167. doi: 10.1073/pnas.96.1.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robinson GW. Mahon KA. Differential and overlapping expression domains of Dlx-2 and Dlx-3 suggest distinct roles for Distal-less homeobox genes in craniofacial development. Mech Dev. 1994;48:199–215. doi: 10.1016/0925-4773(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 9.Hassan MQ. Javed A. Morasso MI. Karlin J. Montecino M. van Wijnen AJ. Stein GS. Stein JL. Lian JB. Dlx3 transcriptional regulation of osteoblast differentiation: temporal recruitment of Msx2, Dlx3, and Dlx5 homeodomain proteins to chromatin of the osteocalcin gene. Mol Cell Biol. 2004;24:9248–9261. doi: 10.1128/MCB.24.20.9248-9261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morasso MI. Mahon KA. Sargent TD. A Xenopus distal-less gene in transgenic mice: conserved regulation in distal limb epidermis and other sites of epithelial-mesenchymal interaction. Proc Natl Acad Sci U S A. 1995;92:3968–3972. doi: 10.1073/pnas.92.9.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kula K. Hall K. Hart T. Wright JT. Craniofacial morphology of the tricho-dento-osseous syndrome. Clin Genet. 1996;50:446–454. doi: 10.1111/j.1399-0004.1996.tb02710.x. [DOI] [PubMed] [Google Scholar]
- 12.Wright JT. Kula K. Hall K. Simmons JH. Hart TC. Analysis of the tricho-dento-osseous syndrome genotype and phenotype. Am J Med Genet. 1997;72:197–204. doi: 10.1002/(sici)1096-8628(19971017)72:2<197::aid-ajmg14>3.3.co;2-0. [DOI] [PubMed] [Google Scholar]
- 13.Islam M. Lurie AG. Reichenberger E. Clinical features of tricho-dento-osseous syndrome and presentation of three new cases: an addition to clinical heterogeneity. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2005;100:736–742. doi: 10.1016/j.tripleo.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 14.Ghoul-Mazgar S. Hotton D. Lézot F. Blin-Wakkach C. Asselin A. Sautier J-M. Berdal A. Expression pattern of Dlx3 during cell differentiation in mineralized tissues. Bone. 2005;37:799–809. doi: 10.1016/j.bone.2005.03.020. [DOI] [PubMed] [Google Scholar]
- 15.Li H. Marijanovic I. Kronenberg MS. Erceg I. Stover ML. Velonis D. Mina M. Heinrich JG. Harris SE, et al. Expression and function of Dlx genes in the osteoblast lineage. Dev Biol. 2008;316:458–470. doi: 10.1016/j.ydbio.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hassan MQ. Tare RS. Lee SH. Mandeville M. Morasso MI. Javed A. van Wijnen AJ. Stein JL. Stein GS. Lian JB. BMP2 commitment to the osteogenic lineage involves activation of Runx2 by DLX3 and a homeodomain transcriptional network. J Biol Chem. 2006;281:40515–40526. doi: 10.1074/jbc.M604508200. [DOI] [PubMed] [Google Scholar]
- 17.Shirakabe K. Terasawa K. Miyama K. Shibuya H. Nishida E. Regulation of the activity of the transcription factor Runx2 by two homeobox proteins, Msx2 and Dlx5. Genes Cells. 2001;6:851–856. doi: 10.1046/j.1365-2443.2001.00466.x. [DOI] [PubMed] [Google Scholar]
- 18.Lee M-H. Kim Y-J. Kim H-J. Park H-D. Kang A-R. Kyung H-M. Sung J-H. Wozney JM. Kim H-J. Ryoo H-M. BMP-2-induced Runx2 expression is mediated by Dlx5, and TGF-beta 1 opposes the BMP-2-induced osteoblast differentiation by suppression of Dlx5 expression. J Biol Chem. 2003;278:34387–34394. doi: 10.1074/jbc.M211386200. [DOI] [PubMed] [Google Scholar]
- 19.Morsczeck C. Gene expression of runx2, Osterix, c-fos, DLX-3, DLX-5, and MSX-2 in dental follicle cells during osteogenic differentiation in vitro. Calcif Tissue Int. 2006;78:98–102. doi: 10.1007/s00223-005-0146-0. [DOI] [PubMed] [Google Scholar]
- 20.Saugspier M. Felthaus O. Viale-Bouroncle S. Driemel O. Reichert TE. Schmalz G. Morsczeck C. The differentiation and gene expression profile of human dental follicle cells. Stem Cells Dev. 2010;19:707–717. doi: 10.1089/scd.2010.0027. [DOI] [PubMed] [Google Scholar]
- 21.Duverger O. Lee D. Hassan MQ. Chen SX. Jaisser F. Lian JB. Morasso MI. Molecular consequences of a frameshifted DLX3 mutant leading to Tricho-Dento-Osseous syndrome. J Biol Chem. 2008;283:20198–20208. doi: 10.1074/jbc.M709562200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winer J. Jung CK. Shackel I. Williams PM. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal Biochem. 1999;270:41–49. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
- 23.Krifka S. Petzel C. Bolay C. Hiller K-A. Spagnuolo G. Schmalz G. Schweikl H. Activation of stress-regulated transcription factors by triethylene glycol dimethacrylate monomer. Biomaterials. 2011;32:1787–1795. doi: 10.1016/j.biomaterials.2010.11.031. [DOI] [PubMed] [Google Scholar]
- 24.Morsczeck C. Schmalz G. Reichert TE. Völlner F. Saugspier M. Viale-Bouroncle & O Driemel S. Gene expression profiles of dental follicle cells before and after osteogenic differentiation in vitro. Clin Oral Investig. 2009;13:383–391. doi: 10.1007/s00784-009-0260-x. [DOI] [PubMed] [Google Scholar]
- 25.Qiu M. Bulfone A. Ghattas I. Meneses JJ. Christensen L. Sharpe PT. Presley R. Pedersen RA. Rubenstein JL. Role of the Dlx homeobox genes in proximodistal patterning of the branchial arches: mutations of Dlx-1, Dlx-2, and Dlx-1 and -2 alter morphogenesis of proximal skeletal and soft tissue structures derived from the first and second arches. Dev Biol. 1997;185:165–184. doi: 10.1006/dbio.1997.8556. [DOI] [PubMed] [Google Scholar]
- 26.Bendall AJ. Abate-Shen C. Roles for Msx and Dlx homeoproteins in vertebrate development. Gene. 2000;247:17–31. doi: 10.1016/s0378-1119(00)00081-0. [DOI] [PubMed] [Google Scholar]
- 27.Choi SJ. Song IS. Feng JQ. Gao T. Haruyama N. Gautam P. Robey PG. Hart TC. Mutant DLX 3 disrupts odontoblast polarization and dentin formation. Dev Biol. 2010;344:682–692. doi: 10.1016/j.ydbio.2010.05.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi SJ. Song IS. Ryu OH. Choi SW. Hart PS. Wu WW. Shen R-F. Hart TC. A 4 bp deletion mutation in DLX3 enhances osteoblastic differentiation and bone formation in vitro. Bone. 2008;42:162–171. doi: 10.1016/j.bone.2007.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Handa K. Saito M. Tsunoda A. Yamauchi M. Hattori S. Sato S. Toyoda M. Teranaka T. Narayanan AS. Progenitor cells from dental follicle are able to form cementum matrix in vivo. Connect Tissue Res. 2002;43:406–408. doi: 10.1080/03008200290001023. [DOI] [PubMed] [Google Scholar]
- 30.Wise GE. Yao S. Odgren PR. Pan F. CSF-1 regulation of osteoclastogenesis for tooth eruption. J Dent Res. 2005;84:837–841. doi: 10.1177/154405910508400911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao Z. Liu H. Jin Y. Lingling E. Influence of ADAM28 on biological characteristics of human dental follicle cells. Arch Oral Biol. 2009;54:835–845. doi: 10.1016/j.archoralbio.2009.05.013. [DOI] [PubMed] [Google Scholar]
- 32.Yalvac ME. Ramazanoglu M. Gumru OZ. Sahin F. Palotás A. Rizvanov AA. Comparison and optimisation of transfection of human dental follicle cells, a novel source of stem cells, with different chemical methods and electro-poration. Neurochem Res. 2009;34:1272–1277. doi: 10.1007/s11064-008-9905-4. [DOI] [PubMed] [Google Scholar]
- 33.Rosenkilde MM. Schwartz TW. The chemokine system: a major regulator of angiogenesis in health and disease. APMIS. 2004;112:481–495. doi: 10.1111/j.1600-0463.2004.apm11207-0808.x. [DOI] [PubMed] [Google Scholar]
- 34.Chaudhary LR. Avioli LV. Regulation of interleukin-8 gene expression by interleukin-1beta, osteotropic hormones, and protein kinase inhibitors in normal human bone marrow stromal cells. J Biol Chem. 1996;271:16591–16596. doi: 10.1074/jbc.271.28.16591. [DOI] [PubMed] [Google Scholar]
- 35.Rothe L. Collin-Osdoby P. Chen Y. Sunyer T. Chaudhary L. Tsay A. Goldring S. Avioli L. Osdoby P. Human osteoclasts and osteoclast-like cells synthesize and release high basal and inflammatory stimulated levels of the potent chemokine interleukin-8. Endocrinology. 1998;139:4353–4363. doi: 10.1210/endo.139.10.6247. [DOI] [PubMed] [Google Scholar]
- 36.Klein OD. Minowada G. Peterkova R. Kangas A. Yu BD. Lesot H. Peterka M. Jernvall J. Martin GR. Sprouty genes control diastema tooth development via bidirectional antagonism of epithelial-mesenchymal FGF signaling. Dev Cell. 2006;11:181–190. doi: 10.1016/j.devcel.2006.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim H-S. Shin T-H. Yang S-R. Seo M-S. Kim D-J. Kang S-K. Park J-H. Kang K-S. Implication of NOD1 and NOD2 for the differentiation of multipotent mesenchymal stem cells derived from human umbilical cord blood. PLoS ONE. 2010;5:e15369. doi: 10.1371/journal.pone.0015369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morsczeck C. Petersen J. Völlner F. Driemel O. Reichert T. Beck HC. Proteomic analysis of osteogenic differentiation of dental follicle precursor cells. Electrophoresis. 2009;30:1175–1184. doi: 10.1002/elps.200800796. [DOI] [PubMed] [Google Scholar]
- 39.Yoshiko Y. Maeda N. Aubin JE. Stanniocalcin 1 stimulates osteoblast differentiation in rat calvaria cell cultures. Endocrinology. 2003;144:4134–4143. doi: 10.1210/en.2003-0130. [DOI] [PubMed] [Google Scholar]
- 40.Strasser A. O'Connor L. Dixit VM. Apoptosis signaling. Annu Rev Biochem. 2000;69:217–245. doi: 10.1146/annurev.biochem.69.1.217. [DOI] [PubMed] [Google Scholar]
- 41.Lynch MP. Capparelli C. Stein JL. Stein GS. Lian JB. Apoptosis during bone-like tissue development in vitro. J Cell Biochem. 1998;68:31–49. [PubMed] [Google Scholar]
- 42.Manolagas SC. Birth and death of bone cells: basic regulatory mechanisms and implications for the pathogenesis and treatment of osteoporosis. Endocr Rev. 2000;21:115–137. doi: 10.1210/edrv.21.2.0395. [DOI] [PubMed] [Google Scholar]
- 43.Ferrari N. Paleari L. Palmisano GL. Tammaro P. Levi G. Albini A. Brigati C. Induction of apoptosis by fenretinide in tumor cell lines correlates with DLX2, DLX3 and DLX4 gene expression. Oncol Rep. 2003;10:973–977. [PubMed] [Google Scholar]
- 44.Yourek G. Hussain MA. Mao JJ. Cytoskeletal changes of mesenchymal stem cells during differentiation. ASAIO J. 2007;53:219–228. doi: 10.1097/MAT.0b013e31802deb2d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McBeath R. Pirone DM. Nelson CM. Bhadriraju K. Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y. Zhou K. Zeng X. Lin J. Zhan X. Tyrosine phosphorylation of missing in metastasis protein is implicated in platelet-derived growth factor-mediated cell shape changes. J Biol Chem. 2007;282:7624–7631. doi: 10.1074/jbc.M608448200. [DOI] [PubMed] [Google Scholar]
- 47.Barna M. Hawe N. Niswander L. Pandolfi PP. Plzf regulates limb and axial skeletal patterning. Nat Genet. 2000;25:166–172. doi: 10.1038/76014. [DOI] [PubMed] [Google Scholar]
- 48.Ikeda R. Yoshida K. Tsukahara S. Sakamoto Y. Tanaka H. Furukawa K. Inoue I. The promyelotic leukemia zinc finger promotes osteoblastic differentiation of human mesenchymal stem cells as an upstream regulator of CBFA1. J Biol Chem. 2005;280:8523–8530. doi: 10.1074/jbc.M409442200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.