Abstract
Stimulated Raman scattering (SRS) microscopy is a newly developed label-free chemical imaging technique that overcomes the speed limitation of confocal Raman while avoiding the nonresonant-background problem of coherent anti-Stokes Raman scattering (CARS) microscopy. Previous demonstrations were limited to single Raman band measurement. We present a novel modulation multiplexing approach that allows real-time detection of multiple species using the fast Fourier-transform. We demonstrate quantitative determination of chemical concentration of a ternary mixture. Furthermore, two imaging applications are pursued: (1) quantitative determination of oil content, as well as pigment and protein concentration in microalgae cultures; (2) 3D high resolution imaging of blood, lipids, and protein distribution in ex vivo mouse skin tissue. We believe quantitative multiplex SRS uniquely combines the advantage of fast label-free imaging with the fingerprinting capability of Raman spectroscopy and enables numerous applications lipid biology as well as biomedical imaging.
Raman spectroscopy is a powerful technique for noninvasive characterization of both biological and non-biological samples. In order to obtain spatially resolved chemical information, the confocal Raman technique can be employed. However, spontaneous Raman scattering is intrinsically weak and is unsuitable for high speed imaging. Coherent anti-Stokes Raman scattering (CARS) offers orders of magnitude higher sensitivity and video-rate imaging has been achieved 1. However, it has a non-resonant background problem which originates from a four-wave mixing process 2. New developments in CARS microscopy removed the non-resonant background problem by using complicated experimental procedures or post-image data processing 3. Most recently, stimulated Raman scattering (SRS) microscopy has emerged as a alternative to CARS microscopy 4-8. Unlike CARS, SRS microscopy has straightforward image interpretation and quantification, without complications arising from non-resonant background and phase matching condition. Consequently, not only is the signal to noise ratio (SNR) improved, but the Raman spectral fidelity is also preserved. Because SRS has a linear concentration dependence, it has the potential to become a powerful method for label-free quantitative determination of chemical concentration of individual species in a multi-component system.
A major limitation of the current implementation of SRS microscopy is that only a single Raman band can be probed at a time. To quantitatively measure multiple components with overlapping Raman bands, at least the same number of bands needs to be measured. However, as high sensitivity SRS imaging uses a high frequency lock-in detection scheme, the same spectral-resolved detection approach that is commonly employed in fluorescence and multiplex CARS detection is currently impractical to implement due to the difficulty of building a multichannel lock-in that works in the MHz frequency range 9-11. Spectrally tailored excitation is an excellent way of circumventing this problem and can distinguish as many chemical components as the number of spectral elements12. However, this technique is restricted to single species detection per imaging session and it requires all species to be externally calibrated before imaging.
Here we present a novel modulation multiplexing approach that allows multiple Raman bands to be measured simultaneously (Figure 1). Through either in situ calibration or external calibration, multispecies chemical mapping can be implemented at subcellular resolution with speed at least three orders of magnitude faster than confocal Raman imaging. In order to detect multiple Raman bands simultaneously, a broadband Ti:sapphire femtosecond laser is used in place of the picosecond laser as the pump. The femtosecond laser is divided into a number of wavelength bands, with each band corresponding to one Raman shift that is modulated at its own RF frequency. This is achieved with an acousto-optical tunable filter device (AOTF, Crystal Technology). The filtered pump beam is then combined with an electrically synchronized picosecond laser at 1064 nm and sent to a laser scanning microscope (Olympus Fluoview 300). The exact Raman shift is determined by the energy difference between the filtered pump beam and Stokes beam, which is encoded in the modulation frequency. Using a Fourier-transform approach, each Raman band is extracted by demodulating at different frequencies and therefore can be measured simultaneously. The spectral resolution of each band is limited by the AOTF to 33 cm-1.
Figure 1.

(A) Schematic diagram of multiplex SRS setup. HWP: half-wave plate; QWP: quarter-wave plate; EOM: electro-optical modulator; PBS: polarizing beamsplitter; DM: dichroic mirror; PMT: photomultiplier tube; LIA: lock-in amplifier. (B) Workflow of the multiplex Raman encoding and decoding process. (C) Example AOTF filtered spectrum using three channel modulation. Each shaded region corresponds to one channel modulated at a different frequency.
We first demonstrate the quantitative determination of chemical concentrations of a ternary solution system. Mixtures of three chemicals: oleic acid, cholesterol, and cyclohexane, with significant Raman band overlap in the CH stretching region, are dissolved in deuterated chloroform. A total of 9 binary and 3 ternary mixtures were prepared. We chose three Raman peaks to probe (2850, 2900, and 2960 cm-1) based on their spontaneous Raman spectra (shown in Figure 2(A)). The power in each channel was 16 mW and the Stokes power was 76 mW at the objective focus. Standard linear algebra operation is used to calculate the binary and ternary mixture concentrations based on measurements on single species. A ternary plot of the calculated concentrations is shown in Figure 2(B). We can see that the concentrations of most solutions are correctly inferred. The residual errors we see in the ternary plot are largely caused by femtosecond laser spectral drift during measurement, synchronization timing jitter, and the inaccuracy of matrix calibration due to the cross-talk of the three channels caused by AOTF diffraction sidelobes. The measurement accuracy can be significantly improved if timing-jitter and cross-talk can be eliminated with better modulation technology.
Figure 2.
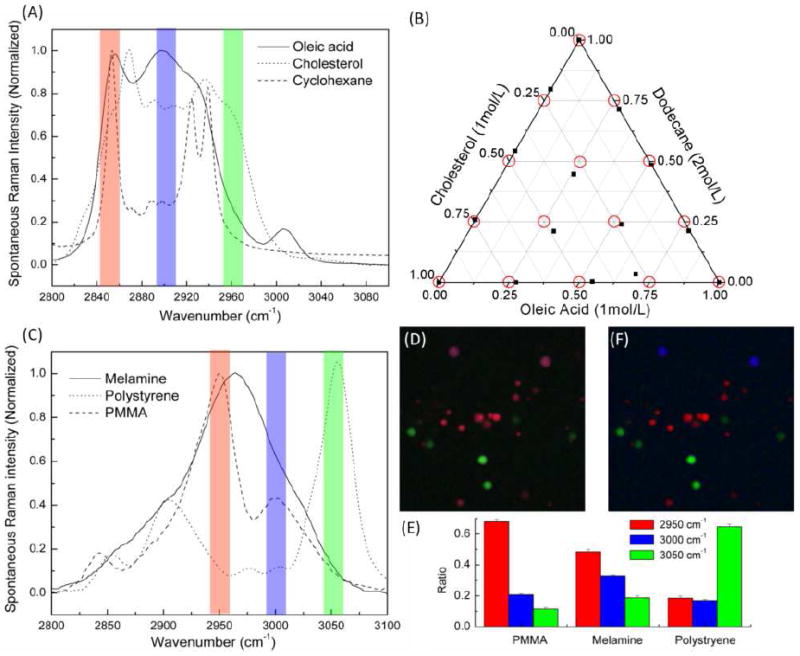
(A) Spontaneous Raman spectra of oleic acid, cholesterol, and cyclohexane. The red, blue and green lines indicate the location of the three Raman bands used for multiplex SRS imaging. (B) Ternary plot of the calculated concentrations of three component mixture based on multiplex SRS measurement. (C). Spontaneous Raman spectra of melamine, PMMA, and polystyrene beads. (D) Raw composite SRS image: red represents image at 2950 cm-1, blue represents image at 3000 cm-1 and green represents image at 3050cm-1. (E) Calibration bar graph of the normalized Raman intensities of each kind of bead at the designated three Raman bands. (F) Reconstructed Raman image (red: PMMA, blue: melamine, and green: polystyrene).
We note that the multiplex data acquisition can be carried out at a rate > 5 kHz, at least two orders of magnitude faster than either conventional Raman imaging or multiplex CARS imaging. Moreover, for spatially segregated samples, calibration can be obtained in situ using the three channel images. To demonstrate this capability, we imaged mixtures of three different polymer beads embedded in agarose gel: 2 μm polystyrene (PS), 1-10 μm polymethyl methacrylate (PMMA) and 2 μm melamine. Three images (512×512 pixels with 200 μs pixel dwell time) are generated simultaneously. Figure 2 (D) shows the composite image of three channels at 2950, 3000 and 3050 cm-1, represented in red, blue and green color respectively. The red and blue channels exhibit small differences, due to the similar Raman spectra of melamine and PMMA. To distinguish them clearly, we first constructed the calibration matrix using in situ imaging data from three different kinds of beads and then applied the inverse matrix to the three channel image. Figure 2 (E) shows the normalized matrix value for the three different kinds of beads in bar graph. It agrees very well with that of spontaneous Raman spectra of the three species shown in Figure 2(C). The calculated concentration images are recompiled into a RGB composite image with each color showing one species (Figure 2 (F)). Different beads are now readily distinguished.
In the first application, we demonstrate the use of multiplex SRS for studying microalgae biochemical composition, especially lipid content. The need for renewable energy sources has sparked growing interest in green algae oil production 13,14. The lipids (mainly triglyceride) stored in the microalgae could be converted into bio-diesel efficiently. Various methods have been employed to evaluate the genetic and environmental factors that affect the oil producing capability of microalgae, but all have significant drawbacks. Fluorescent probes face problems such as nonspecific labeling, inaccuracy in quantification, photobleaching and phototoxicity 15. Traditional biochemical methods such as chromatography and mass spectrometry cannot analyze single cells and do not allow continuous in situ monitoring of oil accumulation. Most recently, Raman microspectroscopy has been used to study algal lipid composition but with very low throughput 16. CARS and SRS are excellent label-free alternatives to investigate lipid content because lipids are usually concentrated in droplets and have very strong Raman signal 17,18, but chlorophyll and carotenoids (both are referred to as pigment from here on), which are abundant in green algae, have strong two-color two-photon absorption (TPA) that interferes with Raman imaging. In addition, it is important to quantitatively separate protein from lipids, which is not possible with single Raman band imaging. TPA is a related nonlinear optical process that generates modulation signal in this pump-probe type imaging experiment, but it has much broader spectra response compared to Raman 19. With our new multiplex SRS approach, three channels can be used to separate pigment TPA, lipids SRS and protein SRS.
Microalgae cultures were imaged at three different bands: 2780, 2850, and 2940 cm-1. TPA contributes to all three channels due to its broad spectra response. At 2780 cm-1, protein and lipid signals are almost negligible. At 2850 cm-1, lipids have large Raman signal, and at 2940 cm-1, both lipids and protein have strong Raman signals. We calibrated the signal distribution in three channels from protein and lipids using two prepared samples: one is 30% bovine serum albumin solution and the other is oleic acid liquid (Sigma). For pigment calibration we used the obtained experimental images, similar to the calibration process used in the bead imaging. Cell cultures of Botryococcus Braunii microalgae are grown in modified bold 3N medium (both from UTEX). We compared two different lighting conditions: under continuous illumination versus a 12:12-h light: dark cycle. The optical power is lowered to 22 mW for the pump (all three channels) and 40 mW for the Stokes in order to minimize photodamage due to TPA. Figures 3(A) shows an example raw composite image of the SRS+TPA signal of microalgae. We can see that the pigment signal dominates all three channels, but is mostly located at the periphery of the cell in a clamshell shape, whereas lipid droplets are concentrated in the center of the cell. After spectra unmixing, the image clearly distinguishes the three major components (Figure 3(B)). We took a total of 15 frames for each sample (see Supporting information). With thresholding methods, the sizes of the cells were calculated. The average cell size for continuous illumination sample is much larger than that of 12:12h light: dark cycle sample lighting (Data not shown). It is known that algal cells under intense illumination could attain higher biomass compared with cells that had been adapted to low-level irradiance 20. To be more quantitative, we compared the cell averaged biomass for the two different types. Figure 3(C) shows the bar graph of the result. There is an increase in cellular concentration for all three components when microalgae are grown under continuous illumination. In particular, pigment increases by 95%, lipids by 68% and protein by 56%. The oil content can be gauged by the fraction of lipids mass in the total cell mass. We used the ratio of lipids to protein as a semi-quantitative measure of oil content. Clearly under continuous lighting, the lipids/protein ratio in the cell increases. This result is corroborated by other studies 21,22. Interestingly, the pigment content increases the most. This is probably due to the adaptation of algal cells to light to increase photosynthetic activities.
Figure 3.

multiplex SRS imaging of microalgal cells under different illumination conditions. (A) Raw composite images of microalgal cells at three bands: 2780 cm-1 (red), 2850cm-1 (green) and 2940 cm-1 (blue); (B) corresponding unmixed images: chlorophyll and carotenoids (red), lipids (green) and protein (blue); (C) bar graph of cell averaged concentration of pigment, lipids and protein for the two different samples. Scale bar: 10 μm.
In the second application, we show that multiplex SRS could be used to study complicated biological samples where calibration of individual components is not easily available. In those cases, it is useful to use the spatial features of a sample to create pseudo-basis sets (assuming selected spatial locations have only one of the three major components) and use the images themselves to construct the calibration matrix. Consequently, the calculated images may not reflect the relative concentrations of each individual chemical component, but should still reflect the contributions of the major components. To demonstrate this, we imaged the 3D structure of mouse skin tissue. Skin has very complicated structure and compositions, providing both barrier and transport functions. These functions are intimately associated with vasculature and lipids. As discussed previously, the 2850 cm-1 channel mainly has contribution from lipids, while the 2940 cm-1 channel has contributions from both lipids and proteins 8. The 2780 cm-1 channel is off Raman resonance and provides contrast for two-color TPA of hemoglobin, a key species that allows visualization of blood vessels 19,23. By using an independent channel to characterize the contribution from blood, together with lipids and protein SRS signals, we can disentangle major biochemical composition of different skin layers. The calibration matrix is obtained by picking specific regions with features corresponding to protein layer, blood vessels and subcutaneous fat layers based on raw composite images at different imaging depth. After reconstruction, Figure 4 shows spectrally separated blood (red), lipids (green) and protein (blue) images at increasing imaging depths with 6 μm depth increment. Qualitatively, we can observe spatially distinctive features for blood vessels at a depth of around 15-30 μm. The lipid layer on the surface and the fat layer in the hypodermis can be clearly identified. We can also see that in the epidermis, there are many large cell nuclei (with characteristic low lipid concentration), and some have visible nucleoli as marked by higher protein content. Within the fat cell layer, some fat cells are surrounded by large numbers of much smaller lipid droplets. Those exquisite features are indications of the high sensitivity and spatial resolution of our multiplex SRS method. This capability offers the potential of label-free real-time digital pathology.
Figure 4.

Multiplex SRS z stack images of freshly excised mouse ear skin tissue: red - blood contrast, green - lipid contrast, and blue - protein contrast. Each image is 512×512 pixels.
In conclusion, we present multiplex SRS imaging using a novel modulation multiplex approach. Double modulation and double demodulation are employed to achieve high speed and high sensitivity. Combining the advantage of high speed label-free imaging and the chemical fingerprinting power of Raman spectroscopy, multiplex SRS is a powerful and quantitative method for fast label-free chemical composition analysis in biological systems. Compared to sequential wavelength tuning based SRS, multiplex SRS is faster and more reliable. In addition, any sample movement on the subsecond scale will create difficulties in image registration and render quantitative analysis invalid for sequential imaging, but only distorts images in multiplex SRS. The speed of multiplex SRS is currently limited by the modulation speed of AOTF, but it can be further improved by two orders of magnitude (up to 500 kHz) with other technologies such as a multichannel acousto-optical modulator. Using such a modulator, the sensitivity and measurement accuracy could also be significantly improved as a result of better resolution (possibly <15 cm-1) and larger number of channels (up to eight or sixteen, depending on models). The number of components that can be analyzed increases linearly with the number of spectral channels.
Our demonstrated applications are focused on the C-H stretching region because C-H stretching offers strong SRS signal and it is critical to have multiplex SRS for discriminating different species in this strongly congested vibrational region. Eight-channel or sixteen-channel SRS could enable quantification of different lipid species such as triglycerides and cholesterol esters. Extending multiplex modulation to the fingerprint region is straightforward by tuning the Ti: sapphire laser to >900 nm. The SNR of multiplex SRS is only slightly degraded in comparison to narrowband SRS due to cross-talk. Therefore, all previous coherent Raman applications demonstrated in the fingerprint region can be better implemented by multiplex SRS with better chemical specificity and quantitative multi-component analysis. It is worth mentioning that there is a delicate balance between laser bandwidth, number of spectral channels and spectra information. Ideally the larger the spectral bandwidth and the number of channels, the better selectivity can be achieved. However, the laser power at each individual band will be limited, resulting in corresponding degradation of SNR. With proper design, we believe that multiplex SRS has enormous potential as a label-free chemical imaging approach for studying complex systems. It will find important applications in lipid biology studies and biomedical imaging.
Supplementary Material
Acknowledgments
We thank Dr. Xiaohui Ni in providing the mouse skin sample. This work was supported by the US. Department of Energy’s Basic Energy Sciences Program (DE-SC001548) and the NIH T-R01 (1R01EB010244-01) award to X.S.X.
Footnotes
Supporting Information. Detailed descriptions of experimental setup, data analysis and high resolution imaging results. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Evans CL, Xie XS. Annu Rev Anal Chem. 2008;1:883–909. doi: 10.1146/annurev.anchem.1.031207.112754. [DOI] [PubMed] [Google Scholar]
- 2.Cheng JX, Xie XS. J Phys Chem B. 2004;108:827–840. [Google Scholar]
- 3.Day JPR, Domke KF, Rago G, Kano H, Hamaguchi H-o, Vartiainen EM, Bonn M. The Journal of Physical Chemistry B. 2011;115:7713–7725. doi: 10.1021/jp200606e. [DOI] [PubMed] [Google Scholar]
- 4.Ploetz E, Laimgruber S, Berner S, Zinth W, Gilch P. Appl Phys B. 2007;87:389–393. [Google Scholar]
- 5.Freudiger CW, Min W, Saar BG, Lu S, Holtom GR, He C, Tsai JC, Kang JX, Xie XS. Science. 2008;322:1857–1861. doi: 10.1126/science.1165758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ozeki Y, Kitagawa Y, Sumimura K, Nishizawa N, Umemura W, Kajiyama Si, Fukui K, Itoh K. Opt Express. 2010;18:13708–13719. doi: 10.1364/OE.18.013708. [DOI] [PubMed] [Google Scholar]
- 7.Nandakumar P, Kovalev A, Volkmer A. New J Phys. 2009;11:033026. [Google Scholar]
- 8.Saar BG, Freudiger CW, Reichman J, Stanley CM, Holtom GR, Xie XS. Science. 2010;330:1368–1370. doi: 10.1126/science.1197236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muller M, Schins JM. J Phys Chem B. 2002;106:3715–3723. [Google Scholar]
- 10.Cheng J-x, Volkmer A, Book LD, Xie XS. J Phys Chem B. 2002;106:8493–8498. [Google Scholar]
- 11.Kano H, Hamaguchi H. Appl Phys Lett. 2005;86:121113. [Google Scholar]
- 12.Freudiger CW, Min W, Holtom GR, Xu B, Dantus M, Sunney Xie X. Nat Photon. 2011;5:103–109. doi: 10.1038/nphoton.2010.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Horsman M, Wu N, Lan CQ, Dubois-Calero N. Biotechnol Progr. 2008;24:815–820. doi: 10.1021/bp070371k. [DOI] [PubMed] [Google Scholar]
- 14.Mata TM, Martins AA, Caetano NS. Renewable and Sustainable Energy Reviews. 2010;14:217–232. [Google Scholar]
- 15.Elle IC, Olsen LCB, Pultz D, Rodkar SV, Fargeman NJ. FEBS Letters. 2010;584:2183–2193. doi: 10.1016/j.febslet.2010.03.046. [DOI] [PubMed] [Google Scholar]
- 16.Wu H, Volponi JV, Oliver AE, Parikh AN, Simmons BA, Singh S. PNAS. 2011;108:3809–3814. doi: 10.1073/pnas.1009043108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nan X, Potma EO, Xie XS. Biophys J. 2006;91:728–735. doi: 10.1529/biophysj.105.074534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang MC, Min W, Freudiger CW, Ruvkun G, Xie XS. Nat Meth. 2011;8:135–138. doi: 10.1038/nmeth.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu D, Ye T, Matthews TE, Chen BJ, Yurtserver G, Warren WS. Opt Lett. 2007;32:2641–2643. doi: 10.1364/ol.32.002641. [DOI] [PubMed] [Google Scholar]
- 20.Banerjee A, Sharma R, Chisti Y, Banerjee UC. Crit Rev Biotechnol. 2002;22:245–279. doi: 10.1080/07388550290789513. [DOI] [PubMed] [Google Scholar]
- 21.Darzins A, Hu Q, Sommerfeld M, Jarvis E, Ghirardi M, Posewitz M, Seibert M. Plant J. 2008;54:621–639. doi: 10.1111/j.1365-313X.2008.03492.x. [DOI] [PubMed] [Google Scholar]
- 22.Fabregas J, Maseda A, Dominguez A, Ferreira M, Otero A. Biotechnol Lett. 2002;24:1699–1703. [Google Scholar]
- 23.Fu D, Matthews TE, Ye T, Piletic IR, Warren WS. J Biomed Opt. 2008;13:040503. doi: 10.1117/1.2968260. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


