Recent development of molecular tools for pathogen detection introduced the possibility of early targeted antimicrobial treatment that, in turn, may improve the outcome of patients with sepsis [1].
We retrospectively evaluated 54 results of the SeptiFast test from patients with solid malignancy admitted to the ICU between June 2009 and August 2011. Specimens from suspected bloodstream infection were analyzed using LightCycler SeptiFast (Roche Molecular Diagnostics, Prague, Czech Republic) according to the manufacturer's instructions and evaluated in comparison with blood culture results obtained from blood sampled no longer than 24 hours before or after sampling for SeptiFast. Blood culturing and identification were performed according to the routine diagnostic procedures. The total number of complementary blood cultures analyzed was 85. Consistently, negative results from SeptiFast and blood culture were obtained in 21 (39%) cases.
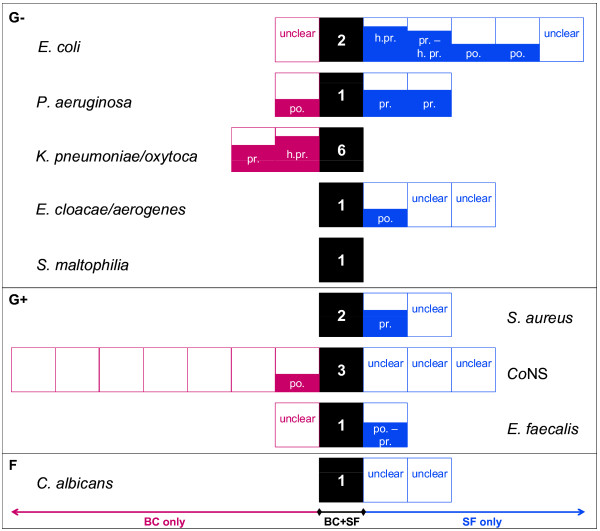
To assess the true positivity of both microbiological methods, discrepant cases were evaluated in the context of clinical and laboratory findings by two independent physicians experienced in critical care (Figure 1). The clinically relevant presence of a pathogen detected by blood culture but not by SeptiFast was recorded for Klebsiella pneumoniae/oxytoca, and in both cases the presence of another member of Enterobacteriaceae was reported by SeptiFast, Escherichia coli and Enterobacter cloacae/aerogenes - thus misidentification of strains with atypical phenotype cannot be excluded [3]. Our results also show that SeptiFast is more efficient in detection of clinically relevant infection by E. coli and Pseudomonas aeruginosa.
Figure 1.
Scheme summarizing detection of pathogens together with clinical evaluation of discrepant findings. Totally 47 pathogens were detected in 29 SeptiFast (SF) and/or 43 blood culture (BC) samples. From 20 pathogens listed on the SF menu [2], nine were detected in the present study and all pathogens detected by blood culture were on the SF menu, 18 microorganisms were detected by both BC and SF, 12 by BC only, and 18 by SF only. The clinical evaluation is as follows: each pathogen detected by either method was classified as possible infection (based on clinical data; for example, benefit from antimicrobial therapy), as probable infection (based on clinical data and cultivation of the pathogen from other site), as highly probable infection (based on clinical data and presence of the pathogen in blood within the relevant time window), as unlike infection (for example, contamination) or as undetermined. To obtain orientation in false positive and false negative results in pathogen detection by SF and cultivation, we assigned a score value to each positive classification as follows: possible infection (score 2), probable infection (score 3), and highly probable infection (score 4). Black boxes represent summary of cases when pathogen was detected by both methods, SF and BC. Red boxes left of concordant events show detection by BC only, and blue objects right of black squares represent events of pathogen detection by SF only. Empty boxes, cases evaluated by both ICU experts as unlike infection; unclear, cases evaluated by ICU experts discrepantly (for example, unlike vs. possible or probable infection). The size of filled objects together with abbreviated classification describe the evaluation of ICU experts for each case: h.pr., highly probable infection; pr., probable infection; po., possible infection. Coagulase-negative staphylococci (CoNS) were detected far more often by BC than SF, probably as a result of contamination of blood culture sample by staphylococci from normal skin flora. Regarding CoNS, SF is able to reflect the amount of CoNS DNA template; if its level is below the discrimination threshold (crossing point (CP) value of amplification growth curve >20), the result is reported as negative. In this context, we retrospectively evaluated all SF raw data files for the CoNS amplification curve and its CP. The CoNS curve with CP >20 was present in 15 out of 44 (34%) SF staphylococci-negative with a lowest CP of 21.4, supporting the necessity for the discrimination threshold in the SF evaluation algorithm for commensal bacteria.
The turnaround time for blood culture analysis is 24 to 48 hours. The workflow in our laboratory allows SeptiFast analysis every working day during a day shift. In such conditions, the turnaround time of SeptiFast results for samples delivered to the laboratory on a workday between 7 am and 2 pm was as follows: 100% of samples, 10 hours; 97% of samples, 8 hours; 59% of samples, 6 hours. The average turnaround time for tests performed with manual DNA isolation was 6 hours 11 minutes; by implementation of automated isolation of DNA using the MagNA Pure Compact System [4], the mean turnaround time was shortened to 5 hours 17 minutes.
In conclusion, detection of pathogen by the LightCycler SeptiFast system is generally not inferior to the results from blood culture. The added value of multiplex DNA amplification-based pathogen detection is the shorter test turnaround time and probably the increased true sensitivity of detection of certain pathogens. Moreover, in clinical settings pathogen detection may be required after an antibiotic administration; in such conditions, cultivation independent methods are of clear benefit [5].
Competing interests
Reimbursement from Roche Diagnostics unrelated to the present work: in 2008, DV and LD for a lecture on a pharmacogenomic and predictive oncology topic, respectively, and in 2010, MV and LD for an expertise work on a protocol for automated DNA isolation from formalin-fixed paraffin-embedded tissues.
Authors' contributions
LD, MV, RT and DV participated in the design of the study. LD and MV carried out the molecular genetics part of the study. DM and PJ evaluated the microbiological findings in the context of clinical status. All authors participated in writing the report.
Contributor Information
Lenka Dubská, Email: dubska@mou.cz.
Martina Vyskočilová, Email: martina.mrkvicova@mou.cz.
Dagmar Minaříková, Email: minarikova@mou.cz.
Petr Jelínek, Email: jelinek@mou.cz.
Renata Tejkalová, Email: renata.tejkalova@fnusa.cz.
Dalibor Valík, Email: valik@mou.cz.
Acknowledgements
The authors received a contribution for the reagents, equipment from Roche Diagnostics for this project, and were supported by the European Regional Development Fund and the State budget of the Czech Republic for RECAMO (CZ.1.05/2.1.00/03.0101).
References
- Garnacho-Montero J, Ortiz-Leyba C, Herrera-Melero I, Aldabó-Pallás T, Cayuela-Dominguez A, Marquez-Vacaro JA, Carbajal-Guerrero J, Garcia-Garmendia JL. Mortality and morbidity attributable to inadequate empirical antimicrobial therapy in patients admitted to the ICU with sepsis: a matched cohort study. J Antimicrob Chemother. 2008;61:436–441. doi: 10.1093/jac/dkm460. [DOI] [PubMed] [Google Scholar]
- Roche Molecular Diagnostics. http://molecular.roche.com/assays/Pages/LightCyclerSeptiFastTestMGRADE.aspx
- Claeys G, De Baere T, Wauters G, Vandecandelaere P, Verschraegen G, Muylaert A, Vaneechoutte M. Extended-spectrum beta-lactamase (ESBL) producing Enterobacter aerogenes phenotypically misidentified as Klebsiella pneumoniae or K. terrigena. BMC Microbiol. 2004;4:49. doi: 10.1186/1471-2180-4-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regueiro BJ, Varela-Ledo E, Martinez-Lamas L, Rodriguez-Calviño J, Aguilera A, Santos A, Gomez-Tato A, Alvarez-Escudero J. Automated extraction improves multiplex molecular detection of infection in septic patients. PLoS One. 2010;5:e13387. doi: 10.1371/journal.pone.0013387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagihara K, Kitagawa Y, Tomonaga M, Tsukasaki K, Kohno S, Seki M, Sugimoto H, Shimazu T, Tasaki O, Matsushima A, Ikeda Y, Okamoto S, Aikawa N, Hori S, Obara H, Ishizaka A, Hasegawa N, Takeda J, Kamihira S, Sugahara K, Asari S, Murata M, Kobayashi Y, Ginba H, Sumiyama Y, Kitajima M. Evaluation of pathogen detection from clinical samples by real-time polymerase chain reaction using a sepsis pathogen DNA detection kit. Crit Care. 2010;14:R159. doi: 10.1186/cc9234. [DOI] [PMC free article] [PubMed] [Google Scholar]