Abstract
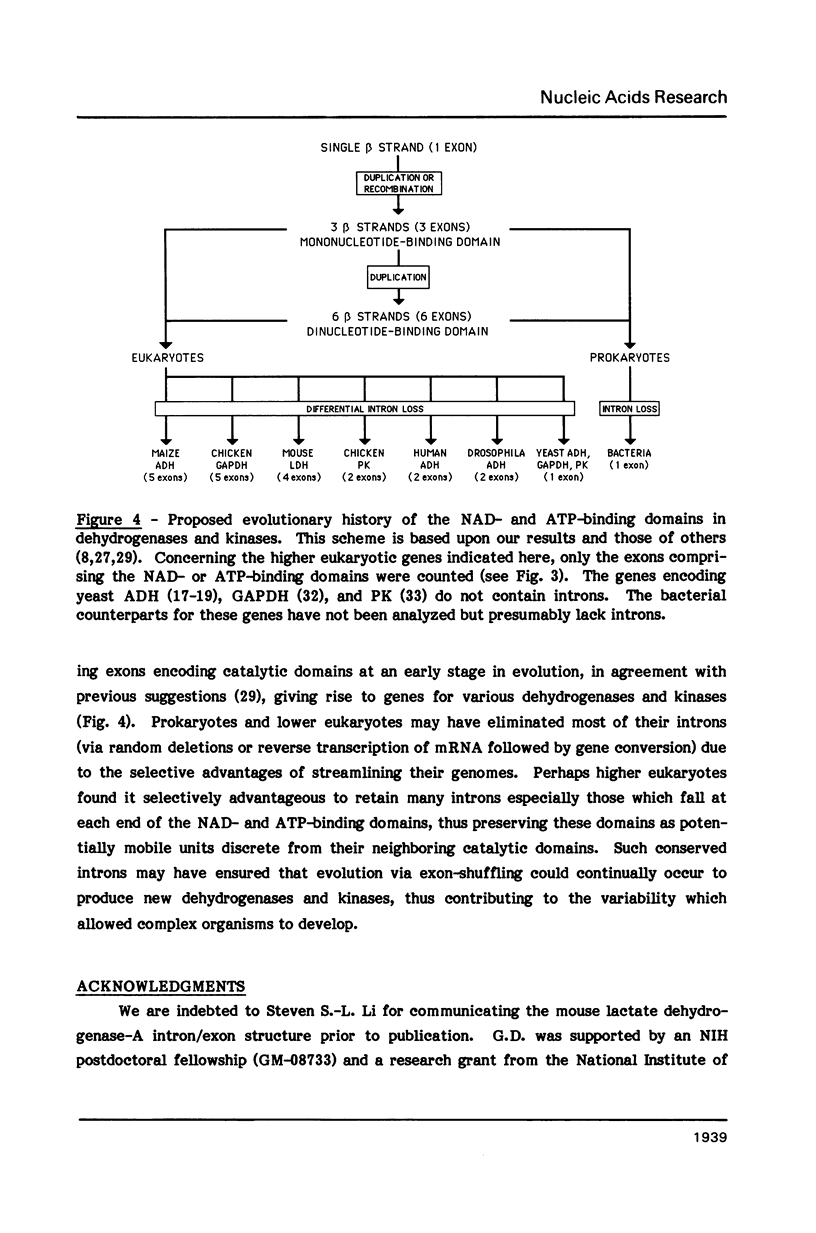
It has been suggested that the intron/exon structure of a gene corresponds to its evolutionary history. Accordingly, early in evolution DNA segments encoding short functional polypeptides may have been rearranged (exon-shuffling) to create full-length genes and RNA splicing may have been developed to remove intervening sequences (introns) in order to preserve translational reading frames. A conflicting viewpoint would be that introns were randomly inserted into previously uninterrupted genes after their initial evolutionary development. If so, the sites of introns would be unlikely to consistently reflect the domain structure of the protein. To address this question, the intron/exon structure of the gene encoding human alcohol dehydrogenase (ADH) was determined and compared to the gene structures for other ADHs and related proteins, all of which possess nucleotide-binding domains. Our results indicate that the introns in the nucleotide-binding domains of all the genes examined do indeed fall at positions which separate the short functional polypeptides (i.e. beta strands) which are believed to comprise this domain. We argue that our data is most easily explained by the hypothesis that introns were present in an ancestral nucleotide-binding domain which was later rearranged by exon-shuffling to form the various dehydrogenases and kinases which utilize such a domain.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bennetzen J. L., Hall B. D. The primary structure of the Saccharomyces cerevisiae gene for alcohol dehydrogenase. J Biol Chem. 1982 Mar 25;257(6):3018–3025. [PubMed] [Google Scholar]
- Benyajati C., Place A. R., Powers D. A., Sofer W. Alcohol dehydrogenase gene of Drosophila melanogaster: relationship of intervening sequences to functional domains in the protein. Proc Natl Acad Sci U S A. 1981 May;78(5):2717–2721. doi: 10.1073/pnas.78.5.2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benyajati C., Spoerel N., Haymerle H., Ashburner M. The messenger RNA for alcohol dehydrogenase in Drosophila melanogaster differs in its 5' end in different developmental stages. Cell. 1983 May;33(1):125–133. doi: 10.1016/0092-8674(83)90341-0. [DOI] [PubMed] [Google Scholar]
- Brändén C. I., Eklund H., Cambillau C., Pryor A. J. Correlation of exons with structural domains in alcohol dehydrogenase. EMBO J. 1984 Jun;3(6):1307–1310. doi: 10.1002/j.1460-2075.1984.tb01967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke R. L., Tekamp-Olson P., Najarian R. The isolation, characterization, and sequence of the pyruvate kinase gene of Saccharomyces cerevisiae. J Biol Chem. 1983 Feb 25;258(4):2193–2201. [PubMed] [Google Scholar]
- Cavalier-Smith T. Nuclear volume control by nucleoskeletal DNA, selection for cell volume and cell growth rate, and the solution of the DNA C-value paradox. J Cell Sci. 1978 Dec;34:247–278. doi: 10.1242/jcs.34.1.247. [DOI] [PubMed] [Google Scholar]
- Chu F. K., Maley G. F., Maley F., Belfort M. Intervening sequence in the thymidylate synthase gene of bacteriophage T4. Proc Natl Acad Sci U S A. 1984 May;81(10):3049–3053. doi: 10.1073/pnas.81.10.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F. Split genes and RNA splicing. Science. 1979 Apr 20;204(4390):264–271. doi: 10.1126/science.373120. [DOI] [PubMed] [Google Scholar]
- Darnell J. E., Jr Implications of RNA-RNA splicing in evolution of eukaryotic cells. Science. 1978 Dec 22;202(4374):1257–1260. doi: 10.1126/science.364651. [DOI] [PubMed] [Google Scholar]
- Dennis E. S., Gerlach W. L., Pryor A. J., Bennetzen J. L., Inglis A., Llewellyn D., Sachs M. M., Ferl R. J., Peacock W. J. Molecular analysis of the alcohol dehydrogenase (Adh1) gene of maize. Nucleic Acids Res. 1984 May 11;12(9):3983–4000. doi: 10.1093/nar/12.9.3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis E. S., Sachs M. M., Gerlach W. L., Finnegan E. J., Peacock W. J. Molecular analysis of the alcohol dehydrogenase 2 (Adh2) gene of maize. Nucleic Acids Res. 1985 Feb 11;13(3):727–743. doi: 10.1093/nar/13.3.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund H., Nordström B., Zeppezauer E., Söderlund G., Ohlsson I., Boiwe T., Söderberg B. O., Tapia O., Brändén C. I., Akeson A. Three-dimensional structure of horse liver alcohol dehydrogenase at 2-4 A resolution. J Mol Biol. 1976 Mar 25;102(1):27–59. doi: 10.1016/0022-2836(76)90072-3. [DOI] [PubMed] [Google Scholar]
- Gilbert W. Why genes in pieces? Nature. 1978 Feb 9;271(5645):501–501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]
- Hempel J., Bühler R., Kaiser R., Holmquist B., de Zalenski C., von Wartburg J. P., Vallee B., Jörnvall H. Human liver alcohol dehydrogenase. 1. The primary structure of the beta 1 beta 1 isoenzyme. Eur J Biochem. 1984 Dec 17;145(3):437–445. doi: 10.1111/j.1432-1033.1984.tb08573.x. [DOI] [PubMed] [Google Scholar]
- Hempel J., Holmquist B., Fleetwood L., Kaiser R., Barros-Söderling J., Bühler R., Vallee B. L., Jörnvall H. Structural relationships among class I isozymes of human liver alcohol dehydrogenase. Biochemistry. 1985 Sep 24;24(20):5303–5307. doi: 10.1021/bi00341a005. [DOI] [PubMed] [Google Scholar]
- Holland J. P., Holland M. J. Structural comparison of two nontandemly repeated yeast glyceraldehyde-3-phosphate dehydrogenase genes. J Biol Chem. 1980 Mar 25;255(6):2596–2605. [PubMed] [Google Scholar]
- Jeck R., Woenckhaus C., Harris J. J., Runswick M. J. Identification of the amino acid residue modified in Bacillus stearothermophilus alcohol dehydrogenase by the NAD+ analogue 4-(3-bromoacetylpyridinio)butyldiphosphoadenosine. Eur J Biochem. 1979 Jan 2;93(1):57–64. doi: 10.1111/j.1432-1033.1979.tb12794.x. [DOI] [PubMed] [Google Scholar]
- Jörnvall H., von Bahr-Lindström H., Jany K. D., Ulmer W., Fröschle M. Extended superfamily of short alcohol-polyol-sugar dehydrogenases: structural similarities between glucose and ribitol dehydrogenases. FEBS Lett. 1984 Jan 9;165(2):190–196. doi: 10.1016/0014-5793(84)80167-2. [DOI] [PubMed] [Google Scholar]
- Jörnvall H., von Bahr-Lindström H., Jeffery J. Extensive variations and basic features in the alcohol dehydrogenase-sorbitol dehydrogenase family. Eur J Biochem. 1984 Apr 2;140(1):17–23. doi: 10.1111/j.1432-1033.1984.tb08061.x. [DOI] [PubMed] [Google Scholar]
- Li S. S., Tiano H. F., Fukasawa K. M., Yagi K., Shimizu M., Sharief F. S., Nakashima Y., Pan Y. E. Protein structure and gene organization of mouse lactate dehydrogenase-A isozyme. Eur J Biochem. 1985 Jun 3;149(2):215–225. doi: 10.1111/j.1432-1033.1985.tb08914.x. [DOI] [PubMed] [Google Scholar]
- Lonberg N., Gilbert W. Intron/exon structure of the chicken pyruvate kinase gene. Cell. 1985 Jan;40(1):81–90. doi: 10.1016/0092-8674(85)90311-3. [DOI] [PubMed] [Google Scholar]
- Russell D. W., Smith M., Williamson V. M., Young E. T. Nucleotide sequence of the yeast alcohol dehydrogenase II gene. J Biol Chem. 1983 Feb 25;258(4):2674–2682. [PubMed] [Google Scholar]
- Russell P. R., Hall B. D. The primary structure of the alcohol dehydrogenase gene from the fission yeast Schizosaccharomyces pombe. J Biol Chem. 1983 Jan 10;258(1):143–149. [PubMed] [Google Scholar]
- Sharp P. A. On the origin of RNA splicing and introns. Cell. 1985 Sep;42(2):397–400. doi: 10.1016/0092-8674(85)90092-3. [DOI] [PubMed] [Google Scholar]
- Stone E. M., Rothblum K. N., Alevy M. C., Kuo T. M., Schwartz R. J. Complete sequence of the chicken glyceraldehyde-3-phosphate dehydrogenase gene. Proc Natl Acad Sci U S A. 1985 Mar;82(6):1628–1632. doi: 10.1073/pnas.82.6.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone E. M., Rothblum K. N., Schwartz R. J. Intron-dependent evolution of chicken glyceraldehyde phosphate dehydrogenase gene. Nature. 1985 Feb 7;313(6002):498–500. doi: 10.1038/313498a0. [DOI] [PubMed] [Google Scholar]
- Stuart D. I., Levine M., Muirhead H., Stammers D. K. Crystal structure of cat muscle pyruvate kinase at a resolution of 2.6 A. J Mol Biol. 1979 Oct 15;134(1):109–142. doi: 10.1016/0022-2836(79)90416-9. [DOI] [PubMed] [Google Scholar]



