Abstract
Zinc finger nucleases (ZFNs) are powerful tools of genome engineering but are limited by their inevitable reliance on error-prone nonhomologous end-joining (NHEJ) repair of DNA double-strand breaks (DSBs), which gives rise to randomly generated, unwanted small insertions or deletions (indels) at both on-target and off-target sites. Here, we present programmable DNA-nicking enzymes (nickases) that produce single-strand breaks (SSBs) or nicks, instead of DSBs, which are repaired by error-free homologous recombination (HR) rather than mutagenic NHEJ. Unlike their corresponding nucleases, zinc finger nickases allow site-specific genome modifications only at the on-target site, without the induction of unwanted indels. We propose that programmable nickases will be of broad utility in research, medicine, and biotechnology, enabling precision genome engineering in any cell or organism.
Programmable nucleases, which include ZFNs and Transcription Activator-Like Effector Nucleases (TALENs), are powerful tools of genome engineering that enable targeted mutagenesis (Bibikova et al. 2002; Urnov et al. 2005; Maeder et al. 2008; Kim et al. 2009, 2011b; Miller et al. 2011; Mussolino et al. 2011) and chromosomal rearrangements (Brunet et al. 2009; Lee et al. 2010a,b, 2011) in any cell or organism. ZFNs and TALENs consist of the FokI nuclease domain fused to distinct DNA-binding domains; ZFNs use zinc fingers (Kim et al. 1996), and TALENs use TAL effector repeat domains derived from a plant pathogen, Xanthomonas. These DNA-binding modules allow the creation of custom DNA-binding proteins that target almost any predetermined DNA sequence (Rebar and Pabo 1994; Bae et al. 2003; Boch et al. 2009; Zhang et al. 2011).
ZFNs and TALENs cleave DNA, producing site-specific DSBs in a genome. DSBs—frequently occurring DNA lesions caused by physical, chemical, and biological stresses—are dangerous signals, often leading to cell death and cancer unless properly repaired. Cells are equipped with two competing DSB repair systems: HR and NHEJ (Kanaar et al. 1998). In the presence of homologous DNA, DSBs can be repaired by HR. The HR machinery uses homologous DNA as a template and is an error-free DSB repair system. In contrast, the two end points of a DSB can be efficiently ligated by NHEJ without the use of homologous DNA. Unlike HR, NHEJ is error-prone, often inducing small insertions and deletions (indels) at breakpoint junctions. Repair of nuclease-induced site-specific DSBs by HR or NHEJ gives rise to targeted genome modifications.
Despite the broad utility of these enzymes in basic research, biotechnology, and medicine, genome engineering with programmable nucleases is limited by the inevitable production of DSBs and reliance on error-prone NHEJ. As a result, programmable nucleases often induce randomly generated, unwanted indels at the on-target site even in the presence of homologous donor DNA, because NHEJ is a dominant pathway of DSB repair over HR in higher eukaryotic cells and organisms (Fattah et al. 2010). To make things worse, these enzymes induce off-target mutations at sites that are highly homologous to the intended target site, where they produce off-target DSBs (Gabriel et al. 2011; Mussolino et al. 2011; Pattanayak et al. 2011). In addition, the repair of off-target DSBs via NHEJ can give rise to unwanted chromosomal rearrangements (Brunet et al. 2009; Lee et al. 2010b, 2011). Furthermore, nucleases that induce too many off-target DSBs are toxic to cells, making it difficult, if not impossible, to isolate gene-edited cells (Cornu et al. 2008; Kim et al. 2009).
In principle, it should be possible to overcome these limitations by using an enzyme that either (1) induces a DSB only at the intended target site or (2) does not induce a DSB at the target site but still elicits site-specific mutations. Here, we demonstrate that site-specific DNA-nicking enzymes (nickases)—constructed by engineering the FokI nuclease domain of ZFNs—can induce SSBs in the genome, whose repair via highly accurate HR gives rise to targeted genome modifications. Importantly, SSBs are not repaired by error-prone NHEJ, and, therefore, do not give rise to indels at both on-target and off-target sites. Thus, SSB-inducing zinc finger (ZF) nickases could serve as highly specific mutagens with no or little off-target effects.
Results
Redesign of a ZFN pair to make nickases
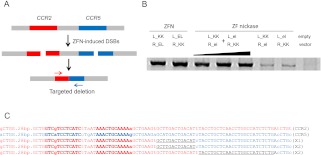
Because two FokI nuclease domains must dimerize on a DNA substrate to cleave DNA (Bitinaite et al. 1998), ZFNs function as dimers rather than monomers. To prevent the formation of an active homodimer, one can use two obligatory heterodimeric FokI variants (Miller et al. 2007; Szczepek et al. 2007) termed “KK” and “EL”: An active nuclease is formed between one KK subunit and the other EL subunit but not between two KK or two EL subunits. We modified ZFN-224 (Perez et al. 2008), which targets the human CCR5 gene, to make nickases. ZFN-224 consists of two ZFN monomers termed L and R, which bind to the left half-site and the right half-site, respectively, separated by a 5-bp spacer (Fig. 1A). Both L_KK/R_EL and L_EL/R_KK are active ZFN pairs. (Hereinafter, L and R refer to zinc finger proteins that bind to the left half-site and the right half-site, respectively, and EL and KK refer to two forms of obligatory heterodimeric FokI domains.) We introduced a mutation (Asp450 to Ala) at the active site of the FokI domain in one subunit to make a catalytically inert monomer (Sanders et al. 2009), which can be paired with a wild-type monomer to yield a nickase. A wild-type monomer alone cannot induce a SSB because two FokI nuclease domains must dimerize to cleave a phosphodiester bond. Catalytically inert monomers, termed “el” and “kk,” carry the Asp450-to-Ala mutation in the EL FokI domain and the KK domain, respectively. Thus, two nickases can be formed, either by pairing L_KK with R_el (designated as L_KK/R_el) or L_kk with R_EL (L_kk/R_EL).
Figure 1.
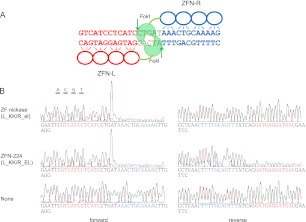
DNA cleavage by ZFNs and ZF nickases. (A) ZFN-224 and its target sequence. ZFN-224 consists of two subunits, L (left, red) and R (right, blue). The two half-site sequences are shown (red and blue) and the 5-bp spacer is shown (black). (Arrows) Cleaved phosphodiester bonds. (B) Run-off DNA sequencing analysis to detect DNA cleavage by ZFN-224 and ZF nickases. A plasmid containing the ZFN-224 target site was incubated with the nuclease pair or the nickase pair and subjected to run-off sequencing. Note that an additional adenine is added at the end by the template-independent terminal transferase activity of AmpliTaq DNA polymerase used for the sequencing reaction.
In vitro and in cellular assays of nickase activities
We tested whether nickases could induce site-specific SSBs in vitro using recombinant proteins expressed in and purified from Escherichia coli. A plasmid containing the ZFN-224 target sequence was digested with the L_KK/R_el nickase or the L_KK/R_EL nuclease (ZFN-224) and subjected to run-off DNA sequencing (Fig. 1B). ZFN-224 cleaved both strands, as evidenced by shrinking peaks after the spacer sequence. In contrast, the nickase cleaved one strand but not the other.
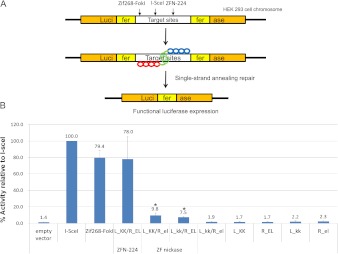
Next, we tested whether nickases could induce targeted mutagenesis using a single-strand annealing (SSA) DNA-repair system in mammalian cells (Fig. 2; Kim et al. 2009). Plasmids that encode nickases were transfected into human embryonic kidney (HEK) 293 cells whose genome contained a stably integrated, partially duplicated firefly luciferase gene that was disrupted by insertion of the CCR5 sequence. Effective ZFNs or nickases would generate a DSB or a SSB, respectively, in the CCR5 sequence, whose repair via SSA allows the restoration of the functional luciferase gene. The efficiency of DNA cleavage by these enzymes can be estimated by measuring luciferase enzyme activity. The highly efficient meganuclease, I-SceI, and Zif268-FokI were used as positive controls. We found that the two nickases, the L_KK/R_el and L_kk/R_EL pairs, were active, restoring the reporter activity partially. Thus, the activity of these nickases (8%–10%, relative to the I-SceI control) was less than that of the nuclease pair (78%). In contrast, all of the monomers alone—L_KK, R_el, L_kk, and R_EL—were inactive. The pair of two mutant forms, L_kk/R_el, also showed no activity in this assay.
Figure 2.
Comparison of activities of ZFNs and ZF nickases using a cell-based reporter system. (A) Schematic overview of a single-strand annealing (SSA) system. (B) Measurement of nuclease and nickase activities using the cell-based SSA system. Means and standard deviations (error bars) from at least three independent experiments are shown. P-values were calculated with the Student's t-test; (*) P < 0.05 (empty vector vs. nickase).
Genome editing with zinc finger nickases
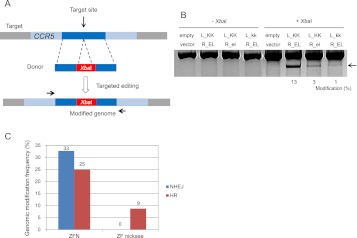
We then tested whether these nickases can induce targeted genome modifications at the endogenous chromosomal site via HR. We transfected human K562 cells with the nickase plasmids and a homologous donor DNA that contained an XbaI site not present in the homologous chromosomal region (Fig. 3A). PCR amplicons of this chromosomal region were partially digested by XbaI, demonstrating 1%–3% genome-editing efficiency by the nickases (Fig. 3B). In agreement with the SSA reporter assay, these SSB-inducing nickases were less active than was the corresponding DSB-inducing nuclease, which showed 13% efficiency.
Figure 3.
Targeted genome editing via HR in human cells with ZF nickases. (A) Schematic overview of HR-mediated genome editing. HR donor DNA consists of two 800-bp homology arms (left and right) and an XbaI site. K562 cells were transfected with plasmids encoding nucleases or ZF nickases plus HR donor plasmid. After 4 d of incubation, genomic DNA was isolated, and the target locus was amplified with primers (arrows) that bind outside of the homology arm sequences. PCR amplicons were digested with XbaI. (B) XbaI-treated and untreated DNA samples were analyzed on a 1% agarose gel. (Arrow) The expected position of XbaI-digested PCR products. Modification frequencies (percentages) were calculated by measuring the band intensity. (C) Comparison of patterns of genomic modifications induced by ZFNs and ZF nickases. PCR products corresponding to genomic modifications were cloned, sequenced, and classified according to their mutation patterns. The DNA sequence of each clone is shown in Supplemental Figure 1.
We also measured genome-editing frequencies by cloning and sequencing the PCR products. We found that the L_KK/R_EL nuclease induced both randomly produced indels (17 clones/total 52 clones, 33%) and homology-directed incorporation of the XbaI site (13/52, 25%) (Fig. 3C; Supplemental Fig. 1). In contrast, the L_KK/R_el nickase did not induce indels at all (0/149 clones) but induced HR-mediated modification at a frequency of 9% (= 13/149). These results show that ZF nickases induce bona fide genome modification in mammalian cells, albeit less efficiently than do ZFNs, and that genome editing with nickases is not accompanied by unwanted indels, demonstrating a critical advantage of nickases over nucleases.
Two pairs of nickases produce a DSB
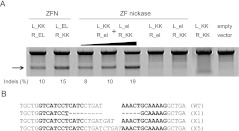
To confirm that nickases induce an SSB at the genomic target site, we introduced two nickases into cultured human cells. Two nickases that generate a SSB on opposite strands may induce a DSB when the two SSBs occur close to each other. This composite DSB could be efficiently repaired by NHEJ. To test this idea, we transfected K562 cells with plasmids that encode two nickases: L_KK/R_el and L_el/R_KK. (We avoided using the combination of L_KK/R_el and L_kk/R_EL because this combination gives rise to the formation of the active ZFN pair, L_KK/R_EL.) We used mismatch-sensitive T7 endonuclease I (T7E1) to detect indels induced by error-prone NHEJ (Fig. 4; Kim et al. 2009). PCR amplicons from cells cotransfected with plasmids encoding the two nickases were partially cleaved at the expected position, indicating the presence of indels at the CCR5 site. DNA sequencing analysis confirmed the induction of indels at the spacer region (Fig. 4B). The mutation frequency induced by these two pairs of nickases was dose-dependent, ranging from 8% to 19%, comparable to that of the original ZFN pairs (10%–15%). In sharp contrast, each nickase alone (L_KK/R_el and L_el/R_KK) did not induce any mutations (assay sensitivity, ∼1%). As expected, the L_KK/R_KK pair (a mismatched combination of obligatory heterodimeric FokI domains) did not show any evidence of indel formation.
Figure 4.
Two pairs of ZF nickases produce a DSB in the genome. (A) Nuclease or nickase-driven indels detected by T7E1 assay. PCR products amplified using genomic DNA from cells transfected with plasmids (4 μg/monomer) encoding nickases or nucleases were subjected to T7E1 digestion and analyzed by agarose gel electrophoresis. (Arrow) The expected position of the resulting DNA band. (The black triangle above the gel picture) The increase of the transfected plasmid (4, 8, and 10 μg/each monomer). (B) DNA sequences of the CCR5 wild-type and mutant clones. The two half-sites are shown in boldface letters. Microhomologies are underlined, and inserted bases are shown in italics. Dashes indicate deleted bases. The number of occurrences is shown in parentheses; X1 and X5 are the number of each clone. (WT) Wild type.
Nickase-induced SSBs do not give rise to indels
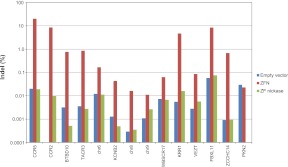
We also performed high-throughput DNA sequencing to confirm that the ZF nickases did not induce indels via NHEJ at either on-target or off-target sites. We measured the frequencies of indels at the CCR5 on-target site and several off-target sites revealed in recent studies (Gabriel et al. 2011; Pattanayak et al. 2011) by analyzing genomic DNA isolated from K562 cells transfected with plasmids encoding ZFN-224 or the L_KK/R_el nickase. As expected, ZFN-224 induced indels at frequencies up to 20% at these sites (Fig. 5; Supplemental Table 1). In sharp contrast, cells expressing the nickase did not show any evidence of indel formation, compared with those containing the empty vector control, at these sites, including the CCR5 site. Apparently, nickase-induced SSBs were faithfully repaired by the endogenous base-excision repair (BER) system, leaving no footprints (Caldecott 2008). Taken together, both the T7E1 assay and the deep sequencing analysis show that ZF nickases, unlike ZFNs, do not trigger error-prone NHEJ to repair DNA breaks.
Figure 5.
Mutagenic NHEJ frequencies at on-target and off-target sites. K562 cells were transfected with plasmids encoding ZFN-224, the ZF nickase (L_KK/R_el), or an empty vector used as a negative control. PCR amplicons corresponding to the CCR5 on-target site and 13 off-target sites were subjected to high-throughput sequencing. Sequences that contained indels within the spacer region were considered to be NHEJ-mediated modifications.
Large chromosomal deletions induced by nickases
Next, we tested whether nickases can induce chromosomal rearrangements at endogenous sites. ZFN-224 recognizes two highly homologous sites, one at the CCR5 locus and the other at the CCR2 locus, and efficiently induces targeted deletions, inversions, and duplications of the intervening 15-kb DNA segments between the two sites (Lee et al. 2010b, 2011). We used PCR to detect the induction of chromosomal deletions in the cells transfected with plasmids encoding ZFNs and ZF nickases (Fig. 6). As expected, the expression of two nickases gave rise to 15-kb deletions. Interestingly, both single nickases also induced 15-kb deletions. We cloned and sequenced PCR products, which confirmed specific deletions of 15-kb DNA segments between the CCR2 and CCR5 sites induced by these nickases (Fig. 6C). However, the patterns of deletion-specific DNA sequences induced by the nickases were quite different from those of DNA sequences induced by ZFNs. No indels were observed at the target site, and no breakpoint junctions could be specified with nickase-mediated deletions. Thus, the CCR2 and CCR5 sequences were fused at variable positions without any insertion or deletion. This suggests that, unlike nuclease-induced DSBs, nickase-induced SSBs are repaired by nonallelic homologous recombination (NAHR) but not by NHEJ (Gu et al. 2008).
Figure 6.
Nickase-mediated genomic deletions in human cells. (A) Schematic representation of ZFN-mediated genomic deletions. (B) PCR products corresponding to the 15-kb genomic deletions in cells transfected with plasmids (4 μg/monomer) encoding nucleases or nickases. (The black triangle above the gel picture) The increase of the transfected plasmid (4, 8, and 10 μg/each monomer). (C) DNA sequences of deletion PCR products that were amplified from genomic DNA isolated from cells transfected with a single nickase pair. Nuclease target sites are shown in boldface letters. Mismatched bases between the CCR2 and CCR5 loci are indicated as lowercase letters. Mutated bases are shown in italics. The regions in which recombination between the CCR2 and CCR5 loci occurred are underlined.
We next measured the frequencies of nickase-mediated genomic deletions using digital PCR analysis (Lee et al. 2010b). The two nickases induced 15-kb deletions of DNA segments between the CCR5 on-target site and the CCR2 off-target site at frequencies of 0.01% (L_KK/R_el) and 0.04% (L_el/R_KK) (Supplemental Table 2). These frequencies were at least 200-fold lower than those of deletions that were generated using the ZFN-224 nuclease (8%). Apparently, the induction of nickase-mediated chromosomal deletions via NAHR repair of SSBs is at least two orders of magnitude less efficient than the induction of deletions via NHEJ repair of DSBs.
Discussion
Our results provide unequivocal evidence that SSB-inducing nickases can be used for targeted genome modifications in higher eukaryotic cells. It has been shown that DNA-nicking enzymes allow homology-directed gene targeting in mammalian cells (Lee et al. 2004; van Nierop et al. 2009) and that nickases can be created by engineering naturally occurring restriction enzymes (Sanders et al. 2009) or meganucleases (McConnell Smith et al. 2009). However, these enzymes are not readily reprogrammed to target any predetermined DNA sequence and thus cannot be used to modify DNA sequences at user-defined genomic sites. In contrast, ZF nickases are programmable, targeting almost any DNA sequence. We provided the first proof-of-principle that these engineered nickases can be used for targeted mutagenesis in cultured human cells.
Programmable nickases have many advantages over nucleases. First, off-target SSBs produced by nickases would be efficiently and faithfully sealed by highly accurate base-excision repair, leaving no footprints at off-target sites. In contrast, nuclease-produced off-target DSBs are repaired by error-prone NHEJ, which gives rise to undesired indels. Furthermore, SSBs occur naturally, and much more frequently than do DSBs, in the genome, and are much less harmful than are DSBs, which often cause cell death and cancer. Many, if not all, nucleases are cytotoxic, making it difficult to isolate clonal populations of gene-edited cells. SSB-producing nickases are likely to be less stressful to cells. In addition, nucleases induce unwanted indels at the on-target site even in the presence of homologous donor DNA. Our deep sequencing analysis indicates that nickases do not induce indels at the on-target site. Last but not least, nickases still can induce unwanted chromosomal rearrangements of DNA segments between on-target and off-target sites via NAHR, but this efficiency is at least two orders of magnitude lower than that of nuclease-mediated rearrangements via NHEJ. Gross chromosomal rearrangements are hallmarks of cancer and are associated with various genetic diseases (Gu et al. 2008). Genome-editing tools that reduce unwanted chromosomal rearrangements would be preferred in many applications such as cell or gene therapy.
We note, however, that nickases come at a price. The efficiency of HR using nickases was a few fold lower than that using corresponding nucleases. It is possible that SSB-triggered HR is intrinsically less efficient than is DSB-triggered HR. Potential users should carefully weigh the pros and cons of nickases and nucleases for specific applications. Because nuclease-mediated HR is more efficient, nucleases may remain the tools of choice for conventional gene knockout and knockin experiments. For applications in stem cell research and gene therapy, in which off-target mutations are of concern, precise genome editing with nickases would be preferred. Fortunately, it takes only a single subcloning step to transform nucleases into nickases or vice versa, and there is no need to start from scratch to make functional genome-editing enzymes. This approach is compatible with pre-characterized ZFNs and TALENs, which can be transformed into the corresponding ZF and TALE nickases, if necessary, by replacing the wild-type FokI domain in one subunit with the catalytically inert FokI domain.
We propose that the efficiency of nickase-mediated genome editing might be enhanced by altering culture conditions (Doyon et al. 2010), using a reporter-based enrichment method (Kim et al. 2011a), or improving the FokI domain via directed evolution (Guo et al. 2010; Doyon et al. 2011), all of which have been useful for enhancing the activity of nucleases. FokI is an enzyme that is evolutionarily programmed to produce DSBs but not SSBs. More efficient SSB-producing enzymes or FokI variants might be used for making improved ZF or TALE nickases.
Recently, independent studies on ZF nickases were reported by two groups. Wang et al. (2012) modified the same CCR5-specific ZFN used in this study to make ZF nickases and showed that these nickases did not induce significant levels of indels at the target site, which is in line with our results. Here, using conventional DNA sequencing, deep sequencing, and the T7E1 assay, we showed that ZF nickases did not induce mutagenic NHEJ at any measurable frequency. In contrast, Joung and colleagues reported that ZF nickases still triggered mutagenic NHEJ, although these enzymes induced SSBs but not DSBs in vitro (Ramirez et al. 2012). It is difficult to reconcile our results with those of Ramirez et al. because they used a reporter system and did not test their nickases at endogenous chromosomal sites. Further studies are warranted to clarify this critical discrepancy.
In conclusion, we demonstrated that SSB-producing programmable nickases could be used for targeted genome modifications. ZF nickases, unlike their corresponding nucleases, allowed efficient genome editing without inducing unwanted indels at the target site. Furthermore, nickase-mediated off-target mutations were not detectable even with high-throughput sequencing, demonstrating an unprecedented precision in genome editing. We propose that programmable nickases are novel tools for precision genome engineering, enabling targeted mutagenesis in any cell or organism.
Methods
Plasmids encoding ZFNs
Plasmids that encode the CCR5-targeting ZFNs used in this study were described previously (Kim et al. 2009). To convert FokI nuclease activity to nickase activity, one monomer was made catalytically inactive by introducing the D450A mutation into plasmids encoding obligatory heterodimer FokI variants (KK/EL) by site-directed mutagenesis using the oligonucleotides listed in Supplemental Table 3.
In vitro DNA cleavage assay
DNA sequences that encode ZFN-224 and nickase monomers were amplified and inserted into pET-28b (Novagen) using NheI and XhoI sites. Recombinant proteins were purified using Ni-NTA agarose beads (QIAGEN) from BL21 E. coli transformants. A plasmid DNA (1.6 nM) that contains the ZFN-224 target site was incubated with ZFN or nickase proteins (1.6 nM) in a reaction buffer (20 mM Tris at pH 8.5, 100 μM ZnCl2, 50 μg/mL BSA, 50 mM NaCl, 1 mM DTT, and 40 mM MgCl2) and subjected to dideoxy DNA sequencing.
HR donor construction
The donor plasmid for the CCR5 locus contains a total of 1.6 kb of CCR5 flanking sequences. For the left homology arm, an 800-bp fragment upstream of the ZFN-224 target site was amplified and inserted into pUC18 using the NdeI and XbaI sites. For the right homology arm, the 800-bp fragment downstream from the ZFN-224 target site was amplified and inserted into the vector using the XbaI and SacI sites.
Cell culture
HEK293T/17 (ATCC, CRL-11268) cells and Flp-In T-REx 293 cells (Invitrogen) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 100 units/mL penicillin, 100 μg/mL streptomycin, 0.1 mM nonessential amino acids, and 10% fetal bovine serum (FBS). To maintain stably integrated SSA clones, cells were cultured with 100–150 μg/mL Hygromycin B (Sigma-Aldrich). K562 (ATCC, CCL-243), a human erythroleukemia cell line, was grown in RPMI-1640 with 10% FBS and the penicillin/streptomycin mix (100 U/mL and 100 μg/mL, respectively).
Cell-based SSA reporter system
Flp-In T-REx 293 cells, whose genome contains a stably integrated, partially duplicated firefly luciferase gene that was disrupted by insertion of the CCR5 sequence, were used as previously described (Kim et al. 2009). Each pair of nuclease expression plasmids (500 ng each) was transfected into 2 × 105 reporter cells/well in a 24-well plate using Lipofectamine 2000 (Invitrogen). After 48 h, luciferase gene expression was induced by the addition of 1 μg/mL doxycycline. After 24 h of incubation, the cells were lysed in 20 μL of 1× lysis buffer (Promega), and luciferase activity was determined using 10 μL of luciferase assay reagent (Promega) and 2 μL of cell lysate.
Modified genome detection assay
For the HR assay, 2 × 106 K562 cells were transfected with 10 μg of each nuclease-encoding plasmid and 50 μg of donor plasmid using the 4D-Nucleofector, SF Cell Line 4D-Nucleofector X Kit, Program FF-120 (Lonza) according to the manufacturer's protocol. After 96 h, genomic DNA was isolated, and the target locus was amplified with primers that bind outside of the homology arm sequences (Supplemental Table 3) using HiPi DNA polymerase (Elpisbio). PCR amplicons were digested with XbaI; digested fragments were analyzed on a 1% agarose gel. For detection of local indels, genomic DNA of nuclease-treated cells was analyzed using the T7E1 assay as previously described using target-specific primers (Supplemental Table 3).
DNA sequencing analysis of breakpoint junctions
PCR products corresponding to genomic modifications were purified from agarose gels using the MG Gel Extraction SV system (Macrogen) and cloned into the T-Blunt vector using the T-Blunt PCR Cloning Kit (SolGent). Cloned plasmids were sequenced using M13 primers.
Deep sequencing of on- and off-target sites
2 × 106 K562 cells were nucleofected with 10 μg of each ZFN- or nickase-encoding plasmid and 5 μg of GFP-encoding plasmid using the SF Cell Line 4D-Nucleofector X kit and the FF120/cell line SF program (Lonza) according to the manufacturer's protocol. One-eighth of the cells were subjected to FACS analysis to confirm transfection, and the rest of the cells were harvested and used for genomic DNA isolation 72 h after transfection.
The CCR5 on-target site and 13 off-target sites reported in recent studies (Gabriel et al. 2011; Pattanayak et al. 2011) were amplified by PCR with Phusion DNA polymerse (Supplemental Table 2). PCR products were purified with the QIAGEN PCR purification kit and combined into separate equimolar pools for the ZFN, nickase, and empty-vector control samples. A multiplexed Illumina library was prepared according to the manufacturer's specifications. Illumina indices 6 (5′-GCCAAT-3′), 7 (5′-CAGATC-3′), and 8 (5′-ACTTGA-3′) were used for the empty vector-, ZFN-, and nickase-treated libraries, respectively. Equal amounts of the bar-coded libraries were subjected to paired-end read sequencing on an Illumina HiSeq 2000 at the National Instrumentation Center for Environmental Management, College of Agriculture and Life Sciences, Seoul National University. Sequences were identified by searching for exact flanking sequences. Sequences, including insertions and deletions located within the spacer ±1 bp, were considered to be NHEJ-mediated modifications.
Frequencies of chromosomal rearrangements
The frequencies of chromosomal rearrangements were estimated by digital PCR analysis as described (Kim et al. 2010). Genomic DNA samples isolated from cells transfected with plasmids encoding ZFNs were serially diluted in distilled water, and diluted samples were then subjected to nested PCR using appropriate primers (Supplemental Table 3). Critical dilution points that support the amplification of breakpoint junctions were determined. The results were analyzed using the Extreme Limiting Dilution Analysis program (Hu and Smyth 2009) (http://bioinf.wehi.edu.au/software/elda/).
Data access
The deep sequencing data are available at the NCBI Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra) under accession number SRA051467.
Acknowledgments
J.-S.K. is supported by National Research Foundation of Korea (2012-0001225).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.138792.112.
Freely available online through the Genome Research Open Access option.
References
- Bae KH, Kwon YD, Shin HC, Hwang MS, Ryu EH, Park KS, Yang HY, Lee DK, Lee Y, Park J, et al. 2003. Human zinc fingers as building blocks in the construction of artificial transcription factors. Nat Biotechnol 21: 275–280 [DOI] [PubMed] [Google Scholar]
- Bibikova M, Golic M, Golic KG, Carroll D 2002. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics 161: 1169–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitinaite J, Wah DA, Aggarwal AK, Schildkraut I 1998. FokI dimerization is required for DNA cleavage. Proc Natl Acad Sci 95: 10570–10575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U 2009. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326: 1509–1512 [DOI] [PubMed] [Google Scholar]
- Brunet E, Simsek D, Tomishima M, DeKelver R, Choi VM, Gregory P, Urnov F, Weinstock DM, Jasin M 2009. Chromosomal translocations induced at specified loci in human stem cells. Proc Natl Acad Sci 106: 10620–10625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldecott KW 2008. Single-strand break repair and genetic disease. Nat Rev Genet 9: 619–631 [DOI] [PubMed] [Google Scholar]
- Cornu TI, Thibodeau-Beganny S, Guhl E, Alwin S, Eichtinger M, Joung JK, Cathomen T 2008. DNA-binding specificity is a major determinant of the activity and toxicity of zinc-finger nucleases. Mol Ther 16: 352–358 [DOI] [PubMed] [Google Scholar]
- Doyon Y, Choi VM, Xia DF, Vo TD, Gregory PD, Holmes MC 2010. Transient cold shock enhances zinc-finger nuclease-mediated gene disruption. Nat Methods 7: 459–460 [DOI] [PubMed] [Google Scholar]
- Doyon Y, Vo TD, Mendel MC, Greenberg SG, Wang J, Xia DF, Miller JC, Urnov FD, Gregory PD, Holmes MC 2011. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Methods 8: 74–79 [DOI] [PubMed] [Google Scholar]
- Fattah F, Lee EH, Weisensel N, Wang Y, Lichter N, Hendrickson EA 2010. Ku regulates the non-homologous end joining pathway choice of DNA double-strand break repair in human somatic cells. PLoS Genet 6: e1000855 doi: 10.1371/journal.pgen.1000855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel R, Lombardo A, Arens A, Miller JC, Genovese P, Kaeppel C, Nowrouzi A, Bartholomae CC, Wang J, Friedman G, et al. 2011. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat Biotechnol 29: 816–823 [DOI] [PubMed] [Google Scholar]
- Gu W, Zhang F, Lupski JR 2008. Mechanisms for human genomic rearrangements. Pathogenetics 1: 4 doi: 10.1186/1755-8417-1-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Gaj T, Barbas CF III 2010. Directed evolution of an enhanced and highly efficient FokI cleavage domain for zinc finger nucleases. J Mol Biol 400: 96–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Smyth GK 2009. ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods 347: 70–78 [DOI] [PubMed] [Google Scholar]
- Kanaar R, Hoeijmakers JH, van Gent DC 1998. Molecular mechanisms of DNA double strand break repair. Trends Cell Biol 8: 483–489 [DOI] [PubMed] [Google Scholar]
- Kim YG, Cha J, Chandrasegaran S 1996. Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci 93: 1156–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS 2009. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res 19: 1279–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Lee HJ, Kim E, Kim JS 2010. Analysis of targeted chromosomal deletions induced by zinc finger nucleases. Cold Spring Harb Protoc doi: 10.1101/pdb.prot5477 [DOI] [PubMed] [Google Scholar]
- Kim H, Um E, Cho SR, Jung C, Kim JS 2011a. Surrogate reporters for enrichment of cells with nuclease-induced mutations. Nat Methods 8: 941–943 [DOI] [PubMed] [Google Scholar]
- Kim S, Lee MJ, Kim H, Kang M, Kim JS 2011b. Preassembled zinc-finger arrays for rapid construction of ZFNs. Nat Methods 8: 7 doi: 10.1038/nmeth0111-7a [DOI] [PubMed] [Google Scholar]
- Lee GS, Neiditch MB, Salus SS, Roth DB 2004. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell 117: 171–184 [DOI] [PubMed] [Google Scholar]
- Lee HJ, Kim E, Kim JS 2010a. Site-specific DNA excision via engineered zinc finger nucleases. Trends Biotechnol 28: 445–446 [DOI] [PubMed] [Google Scholar]
- Lee HJ, Kim E, Kim JS 2010b. Targeted chromosomal deletions in human cells using zinc finger nucleases. Genome Res 20: 81–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Kweon J, Kim E, Kim S, Kim JS 2011. Targeted chromosomal duplications and inversions in the human genome using zinc finger nucleases. Genome Res 22: 539–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, et al. 2008. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell 31: 294–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell Smith A, Takeuchi R, Pellenz S, Davis L, Maizels N, Monnat RJ Jr, Stoddard BL 2009. Generation of a nicking enzyme that stimulates site-specific gene conversion from the I-AniI LAGLIDADG homing endonuclease. Proc Natl Acad Sci 106: 5099–5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, et al. 2007. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol 25: 778–785 [DOI] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. 2011. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 29: 143–148 [DOI] [PubMed] [Google Scholar]
- Mussolino C, Morbitzer R, Lutge F, Dannemann N, Lahaye T, Cathomen T 2011. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res 39: 9283–9293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayak V, Ramirez CL, Joung JK, Liu DR 2011. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat Methods 8: 765–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL, et al. 2008. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol 26: 808–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez CL, Certo MT, Mussolino C, Goodwin MJ, Cradick TJ, McCaffrey AP, Cathomen T, Scharenberg AM, Joung JK 2012. Engineered zinc finger nickases induce homology-directed repair with reduced mutagenic effects. Nucleic Acids Res doi: 10.1093/nar/gks179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebar EJ, Pabo CO 1994. Zinc finger phage: Affinity selection of fingers with new DNA-binding specificities. Science 263: 671–673 [DOI] [PubMed] [Google Scholar]
- Sanders KL, Catto LE, Bellamy SR, Halford SE 2009. Targeting individual subunits of the FokI restriction endonuclease to specific DNA strands. Nucleic Acids Res 37: 2105–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepek M, Brondani V, Buchel J, Serrano L, Segal DJ, Cathomen T 2007. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol 25: 786–793 [DOI] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC 2005. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435: 646–651 [DOI] [PubMed] [Google Scholar]
- van Nierop GP, de Vries AA, Holkers M, Vrijsen KR, Goncalves MA 2009. Stimulation of homology-directed gene targeting at an endogenous human locus by a nicking endonuclease. Nucleic Acids Res 37: 5725–5736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Friedman G, Doyon Y, Wang NS, Li CJ, Miller JC, Hua KL, Yan JJ, Babiarz JE, Gregory PD, et al. 2012. Targeted gene addition to a predetermined site in the human genome using a ZFN-based nicking enzyme. Genome Res (this issue) doi: 10.1101/gr.122879.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P 2011. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol 29: 149–153 [DOI] [PMC free article] [PubMed] [Google Scholar]