Abstract
Background
The development of nonviral gene delivery systems is one of the most intriguing topics in nanomedicine. However, despite the advances made in recent years, several key issues remain unsettled. One of the main problems relates to the difficulty in designing nanodevices for targeted delivery of genes and other drugs to specific anatomic sites. In this study, we describe the development of a novel chitosan nanobubble-based gene delivery system for ultrasound-triggered release.
Methods and results
Chitosan was selected for the nanobubble shell because of its low toxicity, low immunogenicity, and excellent biocompatibility, while the core consisted of perfluoropentane. DNA-loaded chitosan nanobubbles were formed with a mean diameter of less than 300 nm and a positive surface charge. Transmission electron microscopic analysis confirmed composition of the core-shell structure. The ability of the chitosan nanobubbles to complex with and protect DNA was confirmed by agarose gel assay. Chitosan nanobubbles were found to be stable following insonation (2.5 MHz) for up to 3 minutes at 37°C. DNA release was evaluated in vitro in both the presence and absence of ultrasound. The release of chitosan nanobubble-bound plasmid DNA occurred after just one minute of insonation. In vitro transfection experiments were performed by exposing adherent COS7 cells to ultrasound in the presence of different concentrations of plasmid DNA-loaded nanobubbles. In the absence of ultrasound, nanobubbles failed to trigger transfection at all concentrations tested. In contrast, 30 seconds of ultrasound promoted a moderate degree of transfection. Cell viability experiments demonstrated that neither ultrasound nor the nanobubbles affected cell viability under these experimental conditions.
Conclusion
Based on these results, chitosan nanobubbles have the potential to be promising tools for ultrasound-mediated DNA delivery.
Keywords: chitosan, nanobubbles, transfection, DNA, gene carrier, ultrasound
Introduction
Gene therapy technology has the potential to offer novel treatments for cancer and other diseases caused by gene anomalies. However, one of the most significant challenges faced by the field is the development of safe, nontoxic, and efficient nanodelivery systems. Viral vectors represent efficient carriers for gene transduction, but they also come with certain limitations, including toxicity and immunogenicity.1 Nonviral vectors have consequently attracted much interest as gene carriers to overcome these problems, but their transduction efficiency is very low, although many efforts have recently been directed towards improving this aspect.2
The use of ultrasound as an external stimulus for gene transfer has also been the subject of much investigation. As a physical transfer methodology, ultrasound can circumvent many of the problems associated with gene transfer, and its noninvasive nature confers an important advantage over other physical methods.3 Ultrasound can be strategically used to increase release of active substances from microbubbles, ie, gas-filled microspheres. The use of microbubbles as gene delivery vehicles and transfection-enhancing agents is particularly promising.2,4,5 Various methods have been proposed for the delivery of genes using microbubbles, including: direct physical incorporation of DNA into the microbubble shell during fabrication; use of cationic lipids incorporated into the microbubble shell to bind DNA electrostatically; use of single or multiple layers of cationic polymer on the microbubble shell to bind DNA electrostatically; covalent linking of DNA-nanoparticle carriers; and use of complementary DNA strands to load nanoparticles. Most of these microbubble formulations have demonstrated high DNA-loading capacity and/or high transfection efficiency.
Under the action of ultrasound, microbubbles undergo cavitation or produce volumetric oscillations. Microbubble cavitation under a certain level of ultrasound pressure can transiently modify cell permeability and can create pores in the endothelial layer (sonoporation), which aid drug and gene delivery. Sonoporation is being studied as an effective means of promoting extravasation of large macromolecules, such as plasmid DNA, to improve their delivery to tissues and targeted release.6 In particular, much research has been directed towards combined use of ultrasound and microbubbles with the aim of improving transfection efficiency.7,8 Several formulations of microbubbles with shells composed of lipids, proteins, polymers, and surfactants have been designed as DNA carriers.9 The combined use of ultrasound with DNA-bound bubbles has been found to improve DNA transfection in both in vitro and in vivo experiments compared with administration of naked DNA alone.2,10 More recently, submicron-sized bubbles, termed nanobubbles, have been designed and proposed as a prospective tool for gene delivery.11–14
An advantage of nanosized bubbles is their potential extravasation capacity and their potential to accumulate in pathological tissues via the enhanced permeability and retention effect.15,16
In a previous study, we developed DNA-loaded diethylaminoethyl (DEAE)-dextran nanobubbles as gene delivery systems. DEAE-dextran nanobubbles were able to complex and transfect DNA.17 In order to design a nanobubble system that can be administered in vivo without potential toxicity problems related to the presence of DEAE-dextran, here we consider a new nanobubble formulation using the known biocompatible polysaccharide, chitosan. The aim of the current study was to formulate and characterize in vitro new nanobubbles composed of a chitosan shell and a perfluoropentane core as possible DNA carriers for localized ultrasound-mediated gene delivery.
Materials and methods
Materials
Ethanol (96%) was obtained from Carlo Erba (Milan, Italy). Soybean lecithin (Epikuron 200®) was kindly gifted by Degussa (Hamburg, Germany). Perfluoropentane and medium molecular weight chitosan were sourced from Sigma-Aldrich (St Louis, MO). Ultrapure water was obtained using a 1–800 Millipore system (Molsheim, France).
Cell culture
A COS7 cell line (derived from kidney cells of the African green monkey) was cultured in Dulbecco’s Modif ied Eagle’s Medium-High Glucose (PAA, 4061 Pasching, Austria) supplemented with heat-inactivated 10% bovine serum (Gibco/BRL, Gaithersburg, MD) and 1% Zell Shield (Minerva Biolabs, Berlin, Germany).
Preparation of pDNA
The pEGFP-C3 plasmid (4.7 kb), encoding green fluorescence protein, was transformed in Escherichia coli TOP10 and amplified in Lysogeny broth media at 37°C overnight. The plasmid was purified using the NucleoBond® Xtra plasmid purification system (Macherey-Nagel GmbH and Co, KG, Düren, Germany), according to the manufacturer’s protocol. The purity of pDNA was certified by the absorbance ratio at OD260/OD280, and its integrity was confirmed by gel electrophoresis. Plasmid DNA was stored at −20°C until used.
Synthesis of tetradecylphosphoric acid
To synthesize tetradecylphosphoric acid (C14), 100 mL of benzene and 0.26 mol POCl3 were introduced into a volumetric flask, and 100 mL of benzene and 0.26 mol tetradecanol were then introduced via a funnel and slowly dropped into the flask under magnetic stirring for 8 hours. The mixture was then evaporated using a rotary evaporator; 2 mL of water containing some drops of terz-butilic alcohol, which acts as a catalyst to the reaction, was then added to the dry product obtained. The resulting solution was separated by washing twice with ether in a separating flask. The ether phase was then evaporated off and dried under vacuum overnight to obtain tetradecylphosphoric acid.
Determination of chitosan surface tension
The surface tension of the chitosan solution in water at pH 5.5 was measured by the ring method, using a thin platinum ring (Kruss K10, Germany).
Preparation of chitosan nanobubbles
Chitosan nanobubbles for DNA complexation were obtained using medium molecular weight chitosan (approximately 170,000 Da, degree of deacetylation 75%–85%) for the shell and a perfluorocarbon core. Briefly, to prepare the chitosan nanobubbles, an ethanol solution containing Epikuron 200 (1% w/v) and tetradecylphosphoric acid was added to perfluoropentane and ultrapure water under stirring. To obtain the nanobubbles, a 2.7% w/v chitosan solution at pH 5.0 was added dropwise, whilst the mixture was homogenized using an Ultra-Turrax® homogenizer (IKA, Konigswinter, Germany) for 3 minutes at 13,000 rpm. The nanobubbles were then purified by ultra/diafiltration using a TCF2 instrument (Millipore) with a membrane cutoff at 100,000. DNA-loaded nanobubbles were prepared by adding 300 μL of the nanobubble suspension to 40 μg of pEGFP-C3 plasmid (4.7 kb, pDNA) encoding enhanced green fluorescent protein. The system was incubated for 30 minutes and subsequently characterized.
Characterization of nanobubble formulation
The average diameters and polydispersity indices of the nanobubble formulation were determined by photocorrelation spectroscopy using a 90 Plus instrument (Brookhaven, NY) at a fixed scattering angle of 90° and a temperature of 25°C. Each reported value is the average of ten measurements of three different formulation batches. The polydispersity index indicates the size distribution within a nanobubble population. The electrophoretic mobility and zeta potential of each formulation were determined using the 90 Plus instrument. For zeta potential determination, samples of the formulation were placed into the electrophoretic cell, where an electric field of approximately 15 V/cm was applied. Each sample was analyzed at least in triplicate. The electrophoretic mobility measured was converted into a zeta potential using the Smoluchowski equation.
The morphology of each nanobubble formulation was determined by transmission electron microscopy performed using a Philips CM10 instrument (Philips, Eindhoven, The Netherlands). Nanobubble preparations were dropped onto a Formvar-coated copper grid and air-dried prior to examination. The morphology of the nanobubbles was determined by fluorescent microscopy using a DM2500 Leitz instrument. The viscosity of the nanobubbles was determined at 25°C and 37°C using a capillary viscometer.
Thermal analysis of nanobubbles
Thermal analysis was carried out using a DSC/7 differential scanning calorimeter (Perkin-Elmer, Branford, CT) equipped with a TAC 7/DX instrument controller and the Pyris program. The instrument was calibrated with indium for melting point and heat of fusion before analyses took place. A heating rate of 10°C per minute was used in the 25°C–200°C temperature range. Standard aluminum sample pans for liquids (Perkin- Elmer) were used and about 20 mg of nanobubble aqueous suspension weighed; an empty aluminum pan was used as the reference standard. A chitosan solution was also analyzed for comparison purposes. Analyses were carried out under nitrogen purge; triple runs were made for each sample.
Ultrasound stability of pDNA-loaded nanobubbles
pDNA-loaded nanobubbles were evaluated following their exposure to an ultrasound stimulus of oscillation frequency 2.5 ± 0.1 MHz and an average acoustic pressure distribution value of 2.4 ± 0.2 MPa (nominal frequency 50 Hz; nominal power 30 W). Each formulation was analyzed before and after exposure to ultrasound at 25°C and 37°C for 0.5, 1, 2, 3, and 5 minutes by morphological analysis of the nanobubble to evaluate the integrity of the nanobubble structures.
DNA loading efficiency and complexation capacity of nanobubbles
The loading of pDNA was determined spectrophotometrically at 260 nm using an ultraviolet-visible spectrophotometer (DU 730, Beckman Coulter, Fullerton, CA). Following centrifugation of the DNA-loaded nanobubbles, the amount of free pDNA in the supernatants was determined using a calibration curve created with known amounts of free pDNA (5–30 μg/mL). The pDNA loading efficiency of the nanobubbles was calculated by subtracting the amount of free pDNA from the initial amount added.
Finally, pDNA complexation was evaluated by electrophoresis in an agarose gel. Different volumes of nanobubble aqueous suspension (6, 8, 10, 12 μL) were incubated with 4 μg of DNA. DNA-loaded chitosan nanobubbles were then loaded into 1% w/v agarose gel and electrophoresis run in TAE buffer (40 mM Tris base, 20 mM acetic acid, and 1 mM EDTA; pH 8.0) at 60 V for one hour. The gel was then stained in a 0.5 μg/mL ethidium bromide solution. For the positive control, a solution of pDNA (0.1 μg/μL) was used. The banding pattern was visualized using an ultraviolet transil-luminator and photographed with a Polaroid camera.
Sodium dodecyl sulfate test
A sodium dodecyl sulfate (SDS) test was carried out to evaluate the strength of the pDNA electrostatic interaction, whereby the SDS concentration required to displace the pDNA from the nanobubbles was assessed. The nanobubble formulations were incubated for 3 minutes in increasing concentrations of SDS solution (1, 2, 3, 4, and 5%, w/v) and then evaluated by electrophoresis analysis: 3% SDS was required to displace pDNA completely from the nanobubbles. Further experiments established that SDS does not affect nanobubble structure (data not shown). A 3% SDS solution was consequently used in subsequent experiments evaluating the capacity of the nanobubbles to protect pDNA from DNase activity.
DNase stability
The capacity of the nanobubbles to protect pDNA from DNase activity was evaluated. pDNA (0.12 mg ml) and the pDNA-loaded nanobubbles were incubated with DNase I (1 U/mL) at 25°C for 15 minutes. Following incubation of nanobubbles in 3% SDS solution (w/v) for 3 minutes, the nanobubbles were then analyzed by gel electrophoresis for the presence of ethidium bromide-bound DNA.
In vitro release of plasmid DNA in absence and presence of ultrasound
A chitosan-DNA nanobubble aqueous suspension (1 mL) was diluted in 4 mL of phosphate-buffered saline (pH 7.4) at 37°C. At defined time intervals, the supernatant was collected by centrifugation and chitosan nanobubbles were resuspended in 4 mL of fresh buffer. Plasmid DNA released into the supernatants was quantified spectrophotometrically at 260 nm using an ultraviolet-visible spectrophotometer. To evaluate the effect of ultrasound on DNA release, the same release experiment was carried out following insonation of DNA-loaded chitosan nanobubbles for 10, 30, and 60 seconds, and for 2, 3 and 5 minutes using an ultrasound probe with an oscillation frequency of 2.5 ± 0.1 MHz, and the amount of released DNA was quantified (as described above).
Determination of hemolytic activity
The hemolytic activity of the chitosan nanobubbles was evaluated in human blood. Different percentages (v/v) of nanobubble formulations (1.5, 3, 6, 8, and 10%) were added to a suspension of erythrocytes (30%, v/v) in phosphate buffer at pH 7.4. A solution containing a suspension of erythrocytes (30%, v/v) in phosphate buffer pH 7.4 was used as the blank condition, to which an excess of ammonium chloride was added to obtain complete hemolysis for the hemolytic control. Following 90 minutes of incubation at 37°C, the samples were centrifuged at 2000 rpm for 10 minutes and the supernatant was analyzed using a Lambda 2 Perkin-Elmer spectrophotometer at a wavelength of 543 nm. The percentage of hemolysis was calculated with respect to the 100% hemolysis control.
Transfection experiments
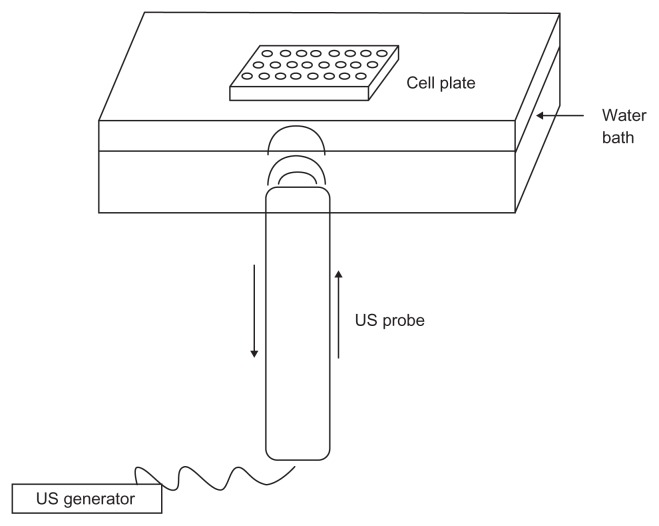
To test the ability of DEAE:DNA nanobubbles to transfect cells, COS7 were seeded at a density of 1 × 104/well in 96-well plates and grown overnight at 37°C in a humidified 5% CO2 incubator. Immediately prior to transfection, the medium was removed and each well received 100 μL of the nanobubble-pDNA complex containing 10, 2, 0.4, or 0.08 μg/mL pDNA diluted in fresh medium. The wells corresponding to the untreated controls received 100 μL of fresh medium. At 24 and 48 hours post-treatment, enhanced green fluorescent protein expression was qualitatively evaluated by confocal laser scanning microscopy using an inverted Zeiss LSM510 fluorescence microscope (Zeiss, Oberkochen, Germany). The same experiments were performed after insonation of the cells transfected with nanobubbles for 5, 10, 15, 30, 60, 90 seconds using an ultrasound probe of oscillation frequency 2.5 ± 0.1 MHz. The schematic apparatus used to insonate the cell plates in the presence of nanobubbles, designed in our laboratory, is reported in Figure 1.
Figure 1.
Schematic apparatus used for transfection experiment in the presence of ultrasound.
Cell viability assay
To test the cytotoxic effects of exposure to pDNA-loaded nanobubbles and ultrasound, the viability of COS7 cells was determined 48 hours post-transfection using the CellTiter 96 proliferation assay kit (Promega, Madison, WI), according to the manufacturer’s instructions. The effect on cell viability of both parameters was expressed as a percentage, by comparing the absorbances of samples with the respective controls.
Results
Nanobubbles consisting mainly of a chitosan shell and a perfluoropentane core were developed as novel DNA carriers. Chitosan showed a critical micellar concentration of approximately 0.8 mg/mL, indicating its capacity to localize to the interface between the nanobubble perfluoropentane core and the aqueous phase. The viscosity of the nanobubble aqueous suspension was 4.7 centipoise, which, together with the small size of the nanobubbles, renders it suitable for parenteral administration.
The average diameters, polydispersity indices, and zeta potentials of the nanobubble formulations, before and after loading with DNA, are reported in Table 1. The unloaded nanobubbles had sizes in the order of magnitude of nanometers, with an average diameter of less than 500 nm. After incubation of chitosan nanobubbles with DNA, a marked decrease in size was observed, indicating that the presence of DNA condensed the polymer chains via electrostatic interactions, thus confirming plasmid localization.
Table 1.
Physicochemical characteristics of chitosan nanobubbles
| Nanobubble preparation | Diameter mean ± SD (nm) | PI | ZP mean ± SD (mV) |
|---|---|---|---|
| Chitosan nanobubbles without pDNA | 412.0 ± 13.8 | 0.11 | +40.9 ± 2.3 |
| Chitosan nanobubbles with pDNA | 284.2 ± 25.8 | 0.1 | +28.3 ± 1.8 |
Abbreviations: SD, standard deviation; PI, polydispersity index; ZP, zeta potential.
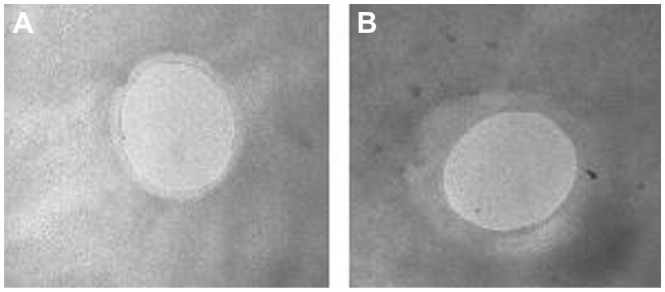
The positive zeta potential of the chitosan nanobubbles decreased after incubation with DNA, indicating the presence of electrostatic interactions between the positive amino chitosan groups and the negative phosphate groups of DNA. A further decrease in surface charge could be obtained by addition of a greater amount of DNA, but lower zeta potentials might favor the aggregation of nanobubbles and affect the physical stability of the nanosuspension. A representative transmission electron microscopic image of DNA-loaded chitosan nanobubbles is reported in Figure 2, showing a well defined core-shell structure with a shell thickness of about 60 nm.
Figure 2.
Transmission electron microscopy image of (A) unloaded and (B) DNA-loaded chitosan nanobubbles.
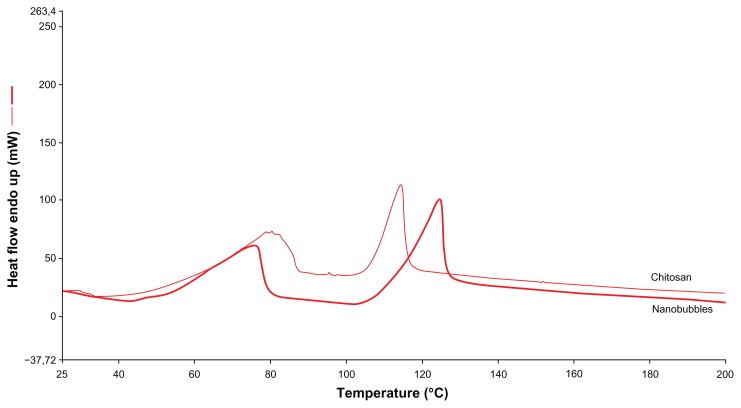
Differential scanning calorimetry thermograms of chitosan nanobubbles are reported in Figure 3. Two main endothermic peaks are present in the differential scanning calorimetry profile. The first broad peak at about 70°C is related to water evaporation, while the second endothermic peak in the 110°C–130°C range is connected to the glass transition temperature of water-plasticized chitosan macromolecules. 18 Chitosan nanobubbles showed a Tpeak at 124.6°C, while the chitosan solution showed a Tpeak at 114.7°C. The melting temperatures differed from those of a pH 5.0 reference chitosan solution (of the same polymer concentration), reflecting a change in the polysaccharide matrix in the nanobubble structure.
Figure 3.
Differential scanning calorimetry profile of chitosan solution and chitosan nanobubbles.
The stability of chitosan nanobubbles after exposure to ultrasound was evaluated. At 25°C, up to 5 minutes of sonication (2.5 MHz) affected neither nanobubble morphology nor structure. On the contrary, at 37°C, nanobubbles begin to rarefy after 3 minutes and completely disappeared after 5 minutes of insonation, indicating a decrease in their stability following exposure to ultrasound that was related to the temperature increase. Considering the low boiling point of perfluoropentane (approximately 30°C), this component might exist as a gas at 37°C; however, the transition might be shifted to higher temperature values due to the small sizes of the nanobubbles. Furthermore, in the presence of ultrasound, the gas core undergoes a conversion from nanodroplet to bubble via a mechanism known as acoustic droplet vaporization. The presence of a central gas core after exposure of the chitosan nanobubbles to ultrasound was verified using a Visualsonics B-mode imaging instrument at 40 MHz. Indeed, chitosan nanobubbles showed good echogenicity (data not shown). The pDNA loading efficiency of the nanobubbles (determined spectrophotometrically) was approximately 75% for a mass of DNA equal to 3.9 × 10−3 μg/μm2. The complexation of DNA with chitosan nanobubbles was confirmed by electrophoresis on an agarose gel (Figure 4). It was possible to observe a fading of the DNA band, indicating complete complexation of the plasmid with nanobubbles at the tested ratio. The amount of DNA loaded onto the nanobubbles was not sufficient to saturate all the positive charges presented on the bubble surface as confirmed by electrophoresis and positive zeta potential values.
Figure 4.
Electrophoresis of DNA-loaded chitosan nanobubbles on agarose gel.
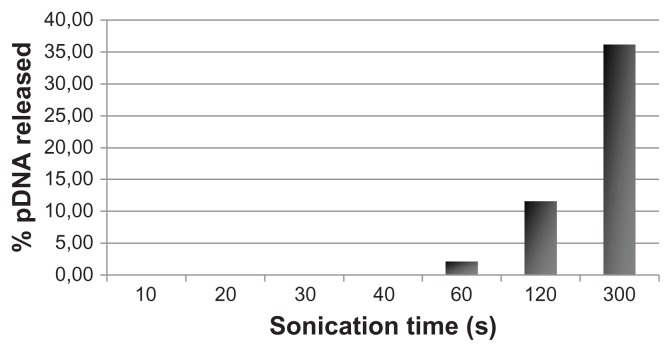
Chitosan nanobubbles were able to protect their loaded DNA from DNase activity. pDNA could then be displaced from the nanobubbles by addition of a 3% SDS solution, as shown by a gel retardation assay (data not shown). In the in vitro release study, no pDNA release from the nanobubbles was observed in the absence of ultrasound, indicating strong interaction between the chitosan shell and the pDNA phosphate groups. In the presence of ultrasound, no pDNA release was observed for sonication times less than one minute (Figure 5). For longer sonication times, pDNA started to be released from the formulation, as shown in the graph.
Figure 5.
In vitro pDNA release from DNA-loaded chitosan nanobubbles after exposure to ultrasound.
Chitosan nanobubbles demonstrated no hemolytic activity when tested in vitro using red blood cells after 90 minutes of incubation at 37°C. For the cell transfection study, the experimental setup was tuned using an ultrasound probe of oscillation frequency 2.5 ± 0.1 MHz and a cell plate-probe distance of 8 cm; 100 μL of nanobubbles was incubated in each well in a 96-well plate.
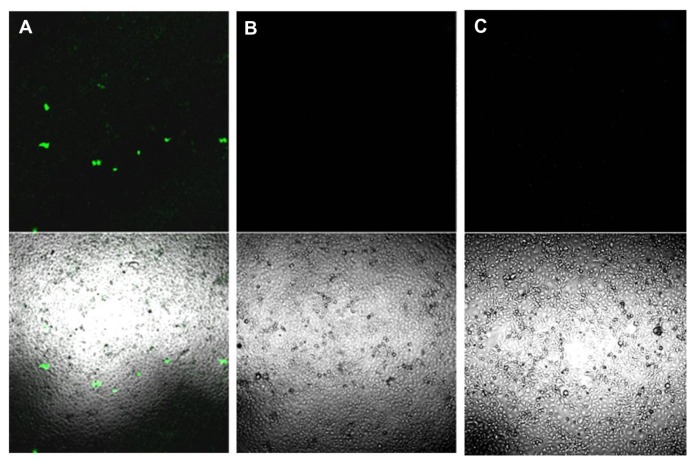
Nonsonified COS7 cells exposed to pDNA-loaded nanobubbles failed to show any green fluorescence protein expression at any of the pDNA concentrations tested (Figure 6B). In contrast, triggering pDNA-loaded nanobubbles (carrying 10 μg/mL of pDNA) with ultrasound for an insonation time of 30 seconds improved the transfection efficiency of the nanobubbles (Figure 6A). The fluorescence signal obtained was not related to autofluorescence phenomena, because no fluorescence signal could be detected in nonsonified cells in the absence of nanobubbles (Figure 6C). Further experiments are in progress to increase the transfection capacity of the formulation in the presence of ultrasound.
Figure 6.
(A) COS7 cells exposed to 30 seconds of insonation in presence of pDNA-loaded nanobubbles carrying 10 μg/mL of pDNA and examined 24 hours post transfection by confocal laser scanning microscopy without fixation. (B) COS7 cells treated as in (A) but not sonified. (C) COS7 cells neither exposed to ultrasound nor to DNA-loaded nanobubbles.
Note: The upper panels show fluorescence images while the lower panels show merged phase-contrast and fluorescence images.
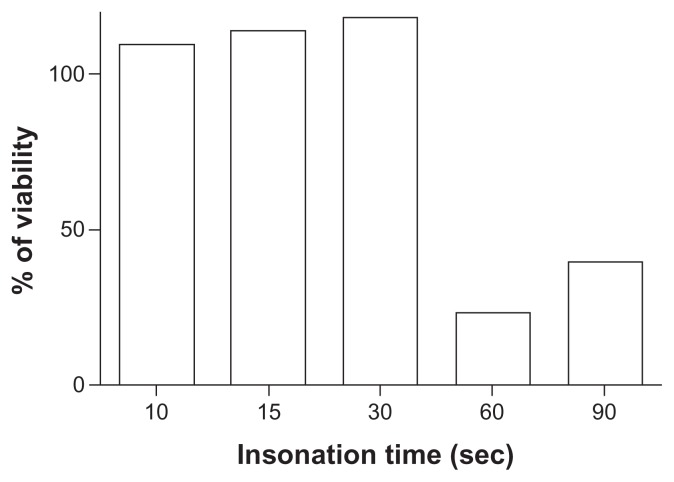
Ultrasound did not affect cell viability under these experimental conditions, as demonstrated in the cell viability assay. In contrast, insonation times longer than 30 seconds resulted in a loss of cell viability in COS7 cells transfected with pDNA-loaded nanobubbles carrying 10 μg/mL of DNA (Figure 7).
Figure 7.
Effect of ultrasound on COS7 viability.
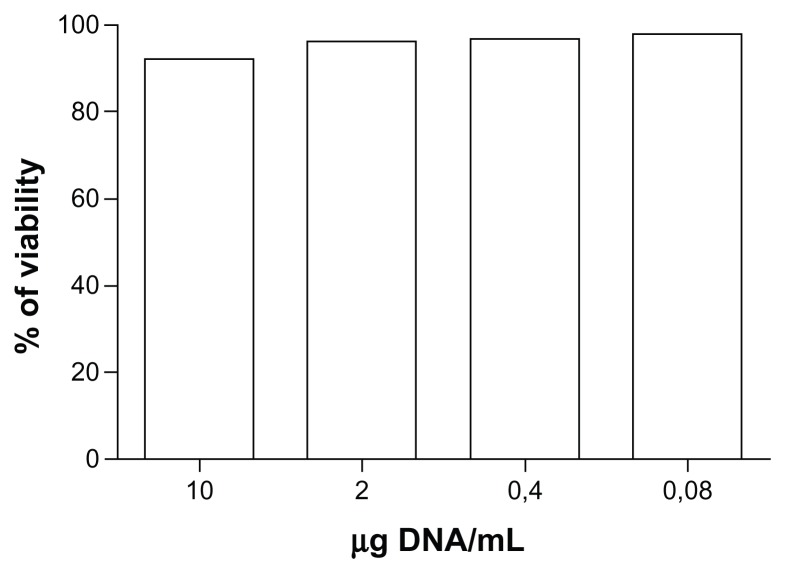
The amount of nanobubbles used to deliver 10 μg/mL of pDNA did not affect cell viability, as shown by the results of the cell viability assay performed on nonsonified COS7 cells treated with different concentrations of nanobubble-delivered pDNA (Figure 8).
Figure 8.
COS7 viability assay after incubation with DNA-loaded chitosan nanobubbles.
Discussion
Recently, several novel microbubble formulations have been developed that aim to improve systemic gene transfection efficiency by loading plasmid DNA or oligonucleotides onto the microbubble surface. However, effective in vivo gene transfection using microbubble carriers requires both a high nucleic acid payload and specific packaging systems to promote intracellular delivery and trafficking to the nucleus.6
Of late, nanobubbles have attracted increasing amounts of attention because of their capacity to load DNA onto their shell; a capacity which holds the potential to enhance circulation times, accumulation in tumor tissues (via the enhanced permeation and retention effect) and cellular trafficking.
In a previous study, we developed nanobubbles consisting of a diethylaminoethyl-dextran (DEAE-dextran) shell and a perfluoropentane core as a DNA carrier. They showed sizes smaller than 500 nm, a positive surface charge, and the ability to complex and transfect DNA. However, a possible drawback of this formulation is its toxic effects at high dosages. To overcome this problem, we designed a new bubble system in the nanometer order of magnitude using chitosan as the polymeric shell.
We selected chitosan for the bubble shell because this polycationic polysaccharide has been increasingly recognized over recent years as providing a safe delivery system for genetic material because it boasts low toxicity, low immunogenicity, excellent biocompatibility, and a high positive charge density.19,20 However, chitosan has the ability to activate macrophages and to stimulate cytokines.21 Chitosan enhances the functions of inflammatory cells such as polymorphonuclear leukocytes, macrophages, and fibroblasts and the stimulation could rely on acetylated residues.22 Consequently, its application for parenteral use should be carefully investigated. With regard to safety of the nanobubbles, no chitosan toxicity was demonstrated in in vivo studies at the doses used to prepare our formulation (1.35 mg/mL). After intravenous administration in rabbits, chitosan levels of 4.5 mg/kg/day caused no effects.21,22 Due to its positive charge, it can easily form polyelectrolyte complexes with negatively charged nucleotides by means of electrostatic interactions. The capacity of chitosan to form a bubble shell has previously been investigated and used for ultrasound-mediated oxygen delivery via the use of gas-filled chitosan nanobubbles.23
In the present study, the formulation was improved to obtain smaller systems. In particular, tetradecylphosphoric acid was intentionally prepared and added to the formulation to produce smaller nanodroplets. This amphiphilic molecule can localize to the perfluoropentane-water interface, lowering the surface tension.
The choice of perfluoropentane for the nanobubble core was due to its already established use in several biomedical applications, and because it is a liquid at room temperature but a vapor at body temperature. This specific feature permitted us to work with a liquid at room temperature using a simple preparation setup. Furthermore, perfluoropentane in nanodroplets can be activated by an external stimulus, like ultrasound, by means of a mechanism called acoustic droplet vaporization, causing the droplet to become a bubble. 24 This means that perfluoropentane can be injected in the form of liquid droplets, dispersed in an aqueous medium, and then converted into bubbles using ultrasound, as shown by Rapoport et al.25 In our work, the chitosan formulation is referred to as “nanobubbles” for the sake of simplicity, but it is important to highlight the fact that before the application of ultrasound it would be more correct to use the term “nanodroplets” when the core is constituted of perfluoropentane for describing the system.
Indeed, for small droplets stabilized by polymer shells, the Laplace pressure may substantially increase boiling temperature. 24 The Laplace pressure is the pressure difference between the inside and the outside of a droplet or bubble. This effect is caused by the surface tension at the interface between the bulk liquid and the droplet liquid.
The Laplace pressure (ΔP) is given as:
where Pinside is the pressure inside a droplet, Poutside is the pressure outside a droplet, σ is the surface tension, and r is droplet radius.
Excessive pressure inside a droplet would result in increasing the boiling temperature of perfluoropentane. Laplace pressure is reversely proportional to droplet size (as indicated in the equation), thus smaller droplets have higher boiling temperatures than larger droplets.
Complete complexation of DNA with the formulation was achieved, as confirmed by gel retardation assay. Moreover, the strong interaction of DNA with the polycationic nanobubble shell protected the loaded DNA from enzymatic degradation. The in vitro results showing no release of DNA in the absence of ultrasound emphasize the strong interaction between the formulation and its DNA cargo and the high stability of the system.
The precise ultrasound exposure methods used to facilitate gene entry into cells are critical in this regard, and the choice of insonation procedure can dramatically alter the results.25 It is therefore important that the conditions of ultrasound exposure are optimized for their use in ultrasound-mediated gene delivery.26–29 Preliminary studies were carried out to evaluate the experimental parameters to be used in our study. The finely tuned conditions identified for ultrasound-mediated transfection were appropriate for cell safety and did not affect cell viability.
Under the specified conditions, DNA-loaded nanobubbles showed the capacity to transfect COS7 cells without affecting cell viability. A protocol for obtaining ultrasound-mediated gene delivery in vitro was assessed. DNA-loaded chitosan nanobubbles showed moderate transfection capacity following 30 seconds of insonation. This moderate efficiency might be ascribed to possible aggregation phenomena occurring amongst nanobubbles at pH 7.4 when the formulation is incubated in the cell plate. Modifications of the nanobubble formulation are currently being made and tested in order to overcome this drawback. Moreover, fluorescent nanobubbles will be prepared to investigate cell uptake in the presence and absence of ultrasound.
The present study confirmed the feasibility of preparing stable and safe nanobubbles using chitosan as the DNA carrier. This formulation can be considered as a suitable starting point from which improved nanobubble systems comprising a chitosan shell can be developed; for example, cell targeting units could be linked to the polymeric shell. The echogenicity of the formulation might offer the possibility to visualize DNA-loaded nanobubbles released in target tissues. Based on this premise, these new nanobubbles hold the potential to provide a multifunctional platform in nanomedicine.
Conclusion
The present study reports the generation of novel, small-sized, positively charged chitosan nanobubbles. These nanobubbles show the ability to complex with and protect DNA. Their capacity to transfect DNA in vitro was triggered by ultrasound. In the absence of ultrasound, none of the tested DNA-loaded nanobubble concentrations showed any transfection ability. Following 30 seconds of ultrasound treatment, a moderate transfection level was obtained. Shorter sonication times did not result in successful transfection of the DNA cargo into cells, while prolonged sonication times affected cell viability under these test conditions. No formulation-induced cytotoxicity was observed for any of the transfection doses used. Chitosan nanobubbles can be considered as an interesting tool in the development of ultrasound-responsive formulations for targeting DNA delivery.
Acknowledgments
The authors gratefully acknowledge grants from the Miur Ricerca Sanitaria Finalizzata 2006 and 2008 projects to DL.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Patil SD, Rhodes DG, Burgess DJ. DNA-based therapeutics and DNA delivery systems: a comprehensive review. AAPS J. 2007;7:61–77. doi: 10.1208/aapsj070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suzuki R, Oda Y, Utoguchi N, Maruyama K. Progress in the development of ultrasound-mediated gene delivery systems utilizing nano- and microbubbles. J Control Release. 2011;149:36–41. doi: 10.1016/j.jconrel.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 3.Nomikou N, Tiwari P, Trehan T, Gulati K, McHale A. Studies on neutral, cationic and biotinylated cationic microbubbles in enhancing ultrasound-mediated gene delivery in-vitro and in-vivo. Acta Biomater. 2012;8:1273–1280. doi: 10.1016/j.actbio.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 4.Tlaxca J, Anderson C, Klibanov A, et al. Analysis of in-vitro transfection by sonoporation using cationic and neutral microbubbles. Ultrasound Med Biol. 2010;36:1907–1918. doi: 10.1016/j.ultrasmedbio.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Juffermans LJM, Dijkmans PA, Muster RJP, et al. Local drug and gene delivery through microbubbles and ultrasound: a safe and efficient alternative for viral vectors? Neth Heart J. 2004;12:398–403. [PMC free article] [PubMed] [Google Scholar]
- 6.Sirsi SR, Hernandez S, Zielinski L, et al. Polyplex-microbubble hybrids for ultrasound-guided plasmid DNA delivery to solid tumors. J Control Release. 2012;157:224–234. doi: 10.1016/j.jconrel.2011.09.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hernot S, Klibanov AL. Microbubbles in ultrasound-triggered drug and gene delivery. Adv Drug Deliv Rev. 2008;60:1153–1166. doi: 10.1016/j.addr.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lentacker I, De Geest B, Vandenbroucke R, et al. Ultrasound-responsive polymer-coated microbubbles that bind and protect DNA. Langmuir. 2006;22:7273–7278. doi: 10.1021/la0603828. [DOI] [PubMed] [Google Scholar]
- 9.Sirsi S, Borden M. Microbubble compositions, properties and biomedical applications. Bubble Sci Eng Technol. 2009;1:3–17. doi: 10.1179/175889709X446507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frenkel P, Chen S, Thai T, et al. DNA-loaded albumin microbubbles enhance ultrasound-mediated transfection in-vitro. Ultrasound Med Biol. 2002;28:817–822. doi: 10.1016/s0301-5629(02)00518-5. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Li X, Zhou Y, Huang P, Xu Y. Preparation of nanobubbles for ultrasound imaging and intracellular drug delivery. Int J Pharm. 2010;384:148–152. doi: 10.1016/j.ijpharm.2009.09.027. [DOI] [PubMed] [Google Scholar]
- 12.Lukianova-Hleb E, Samaniego A, Wen J, Metelitsa L, Chang C, Lapotko D. Selective gene transfection of individual cells in vitro with plasmonic nanobubbles. J Control Release. 2011;152:286–293. doi: 10.1016/j.jconrel.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.du Toit LC, Govender T, Pillay V, Choonara Y, Kodama T. Investigating the effect of polymeric approaches on circulation time and physical properties of nanobubbles. Pharm Res. 2011;28:494–504. doi: 10.1007/s11095-010-0247-y. [DOI] [PubMed] [Google Scholar]
- 14.Horie S, Watanabe Y, Chen R, Mori S, Matsumura Y, Kodama T. Development of localized gene delivery using a dual-intensity ultrasound system in the bladder. Ultrasound Med Biol. 2010;36:1867–1875. doi: 10.1016/j.ultrasmedbio.2010.07.015. [DOI] [PubMed] [Google Scholar]
- 15.Iyer A, Khaled G, Fang J, Maeda H. Exploiting the enhanced permeability and retention effect for tumor targeting. Drug Discov Today. 2006;11:812–818. doi: 10.1016/j.drudis.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 16.Cho K, Wang X, Nie S, Chen Z, Shin D. Therapeutic nanoparticles for drug delivery in cancer. Clin Cancer Res. 2008;1:1310. doi: 10.1158/1078-0432.CCR-07-1441. [DOI] [PubMed] [Google Scholar]
- 17.Bisazza A, Civra A, Donalisio M, Lembo D, Cavalli R. The in-vitro characterization of dextran-based nanobubbles as possible DNA transfection agents. Soft Matter. 2011;7:10590–10593. [Google Scholar]
- 18.Princi E, Vincini S, Pedemonte E, et al. Thermal analysis and characterisation of cellulose grafted with acrylic monomers. Thermochim Acta. 2005;426:173–179. [Google Scholar]
- 19.Shu XZ, Zhu KJ. The influence of multivalent phosphate structure on the properties of ionically cross-linked chitosan films for controlled drug release. Eur J Pharm Biopharm. 2002;54:235–243. doi: 10.1016/s0939-6411(02)00052-8. [DOI] [PubMed] [Google Scholar]
- 20.Lee MK, Chun SK, Choi WJ, et al. The use of chitosan as a condensing agent to enhance emulsion mediated gene transfer. Biomaterials. 2005;26:2147–2156. doi: 10.1016/j.biomaterials.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 21.Baldrick P. The safety of chitosan as a pharmaceutical excipient. Regul Toxicol Pharmacol. 2010;56:290–299. doi: 10.1016/j.yrtph.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 22.Porporatto C, Bianco I, Riera C, Correa S. Chitosan induces different L-arginine metabolic pathways in resting and inflammatory macrophages. Biochem Biophys Res Commun. 2003;304:266–272. doi: 10.1016/s0006-291x(03)00579-5. [DOI] [PubMed] [Google Scholar]
- 23.Cavalli R, Bisazza A, Rolfo A, et al. Ultrasound-mediated oxygen delivery from chitosan nanobubbles. Int J Pharm. 2009;378:215–217. doi: 10.1016/j.ijpharm.2009.05.058. [DOI] [PubMed] [Google Scholar]
- 24.Kripfgans O, Fowlkes J, Miller D, Eldevik O, Carson P. Acoustic droplet vaporization for therapeutic and diagnostic applications. Ultrasound Med Biol. 2000;26:1177–1189. doi: 10.1016/s0301-5629(00)00262-3. [DOI] [PubMed] [Google Scholar]
- 25.Rapoport N, Nam KH, Gupta R, et al. Ultrasound-mediated tumor imaging and nanotherapy using drug loaded, block copolymer stabilized perfluorocarbon nanoemulsions. J Control Release. 2011;153:4–15. doi: 10.1016/j.jconrel.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bao S, Thrall BD, Miller DL. Transfection of a reporter plasmid into cultured cells by sonoporation in vitro. Ultrasound Med Biol. 1997;23:953–959. doi: 10.1016/s0301-5629(97)00025-2. [DOI] [PubMed] [Google Scholar]
- 27.Feril LB, Jr, Ogawa R, Tachibana K, Kondo T. Optimized ultrasound- mediated gene transfection in cancer cells. Cancer Sci. 2006;97:1111–1114. doi: 10.1111/j.1349-7006.2006.00286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki R, Takizawa T, Negishi Y, et al. Tumor specific ultrasound enhanced gene transfer in vivo with novel liposomal bubbles. J Control Release. 2008;125:137–144. doi: 10.1016/j.jconrel.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 29.Pislaru SV, Pislaru C, Kinnick RR, et al. Optimization of ultrasound-mediated gene transfer: comparison of contrast agents and ultrasound modalities. Eur Heart J. 2003;24:1690–1698. doi: 10.1016/s0195-668x(03)00469-x. [DOI] [PubMed] [Google Scholar]