Abstract
We have developed FINDSITEX, an extension of FINDSITE, a protein threading based algorithm for the inference of protein binding sites, biochemical function and virtual ligand screening, that removes the limitation that holo protein structures (those containing bound ligands) of a sufficiently large set of distant evolutionarily related proteins to the target be solved; rather, predicted protein structures and experimental ligand binding information are employed. To provide the predicted protein structures, a fast and accurate version of our recently developed TASSERVMT, TASSERVMT-lite, for template-based protein structural modeling applicable up to 1000 residues is developed and tested, with comparable performance to the top CASP9 servers. Then, a hybrid approach that combines structure alignments with an evolutionary similarity score for identifying functional relationships between target and proteins with binding data has been developed. By way of illustration, FINDSITEX is applied to 998 identified human G-protein coupled receptors (GPCRs). First, TASSERVMT-lite provides updates of all human GPCR structures previously modeled in our lab. We then use these structures and the new function similarity detection algorithm to screen all human GPCRs against the ZINC8 non-redundant (TC<0.7) ligand set combined with ligands from the GLIDA database (a total of 88,949 compounds). Testing (excluding GPCRs whose sequence identity > 30% to the target from the binding data library) on a 168 human GPCR set with known binding data, the average enrichment factor in the top 1% of the compound library (EF0.01) is 22.7, whereas EF0.01 by FINDSITE is 7.1. For virtual screening when just the target and its native ligands are excluded, then the average EF0.01 reaches 41.4. We also analyze off-target interactions for the 168 protein test set. All predicted structures, virtual screening data and off-target interactions for the 998 human GPCRs are available at http://cssb.biology.gatech.edu/skolnick/webservice/gpcr/index.html.
Keywords: TASSERVMT, FINDSITE, GPCR modeling, template-based modeling, virtual screening
Introduction
Protein structure can play an important role in the inference and understanding of the biochemical function of proteins1–3. However, in this post genomic era, the number of protein sequences of unknown biochemical function is far larger than the number of experimentally solved protein structures3. This is especially true for certain classes of proteins, such as G-protein coupled receptors (GPCR), whose structures are hard to obtain experimentally4. To catch up with the rapid growth of sequence data, computational methods that can provide appropriately accurate protein structure predictions on a proteome scale are needed. To address this, a number of relatively fast and accurate template based automated protein structure prediction methods have been developed 5–11,12. Then, having a structure in hand, the next issue is to employ the structure to infer the biochemical function of the protein. The knowledge of protein function is useful in the early stages of drug discovery that use the predicted protein structures in virtual ligand screening13–20. Here, ligand homology modeling, LHM, algorithms, the earliest of which was FINDSITE13, are quite promising. All variants share these steps: One identifies a set of holo threading templates (templates with bound ligands), clusters the structures and extracts ligand binding information useful for virtual ligand screening. The disadvantage of LHM is that it requires that a sufficient number of evolutionary related protein structures with bound ligands be solved. Thus, LHM cannot be applied to protein families such as GPCRs where the number of solved structures (either apo or holo) is very small. To remove this limitation, in this paper, we describe the development of FINDSITEX, which replaces the experimentally solved structure library with predicted structures and incorporates known ligand binding information to create a set of virtual holo structures. By way of illustration, we apply FINDSITEX to the biomedically important GPCR protein family21.
In order to apply FINDSITEX to proteomes, we require a rapid and accurate protein structure prediction method. To address this need, we developed TASSERVMT-lite. Then, we require a library of proteins with known binding ligands. We describe the modifications of our traditional FINDSITE approach to accommodate predicted structures and ligand binding information without the three dimensional poses of the ligand-protein complex for the protein template. In what follows, we give the necessary background information for each aspect of FINDSITEX.
Structures of targets and library proteins with known binding data are modeled by a template-based approach. By the term template, we mean a protein with a solved structure that provides the intial coordinates on which the target structure is modeled. Template-based structure prediction methods involve: (1) identification of structural templates by threading; (2) alignment of the target sequence to the template structures; (3) building a full-length model and refinement of the target structure from the initial template-based model. Each of these steps plays a role in determining the ultimate accuracy of the target structure. Sometimes, a compromise is required between accuracy and speed. For example, in the model building and refinement stage, one could use a fast method such as MODELLER6 which has moderate accuracy or a slower but better method, such as TASSER 7, 22.
In the past, to address the compromise between speed and accuracy, TASSER-lite7 was developed. The parameters of the structure prediction algorithm TASSER22 were optimized for targets whose sequence identities to the identified templates range from 35% to 90%. The result was a significant speed up in the model building and refinement stage from an average of ~29 hours to 17 minutes for targets of size 40–200 residues. Nevertheless, TASSER-lite retains the accuracy of TASSER22 and shows better performance than MODELLER6. However, 97% of the 998 human GPCRs modeled in this study have a maximal sequence identity to their closest template < 35%. Thus, an approach that removes this limitation is required.
Since publication of our original paper that modeled the structures of all identified human GPCRs23, which was done with the rhodopsins (Sensory, Halo, Bacterior and Bovine) as templates for the majority of GPCR targets, there have been six experimentally determined GPCR structures as well as newly identified GPCR sequences. In a review article 24 on GPCR modeling, it was pointed out that multiple-template based modeling produces better structures than those by single-template based methods; this is true in general and not just for GPCRs25–27. Multiple-template based modeling is applicable to human GPCR targets because threading can identify a set of GPCR and/or rhodopsin templates for the majority of targets. Thus, modeling GPCRs with the multiple-template based approach, TASSERVMT-lite and new GPCR templates should yield more accurate models than in our original GPCR structural database23. Zhang and Zhang28, modeled a subset of all identified human GPCRs using I-TASSER29 with spatial restraints derived from experiments other than X-ray or NMR structure determination. I-TASSER is a variant of TASSER22 based on multiple-templates and is computationally more expensive than TASSERVMT-lite. For a typical GPCR protein that is 350 amino acids in length, full TASSER refinement (which is not more expensive than iterative I-TASSER) takes around 3 days, whereas TASSERVMT-lite needs around 10 hours. Furthermore, spatial restraints are not available for all human GPCRs. Other non-proteome scale modeling approaches for GPCRs include methods that use knowledge-based constraints30; methods for loop modeling31, 32, activated state modeling33, 34, conformational ensemble modeling35 and binding pocket modeling36.
To employ protein models as target receptors for ligand docking in structure-based virtual screening requires approaches that can use binding sites whose inaccuracy is greater than the differences seen in crystal structures of the same protein that binds different ligands, viz. cross-docking. The recently developed FINDSITE/Q-dock ligand homology modeling (LHM) methodology13–15 is an example of an approach that exhibits the desired insensitivity to receptor structure deformation. LHM is designed to extend template-based techniques to model protein-ligand interactions and provides detailed biochemical functional annotation of the target proteins. In practice, LHM consists of three steps: First, functional relationships between proteins are detected by threading methods that are dominated by sequence profile similarity scores to identify functionally important residues, common molecular substructures in binding ligands and the structural conservation of their binding modes. These conserved features are exploited during the initial docking of ligands by a similarity-based approach. Second, a ligand fingerprint profile is constructed from the ligands of identified, potentially functionally similar proteins and used for ligand-based virtual screening to identify small molecules from a compound database that could potentially bind to the target. Finally, the positions of small molecule ligands are placed and adjusted to optimize their interactions with the protein of modeled structure and to rank the predicted poses. These basic ideas have subsequently been applied by a number of groups including those of Zhang, Sternberg and others17, 19, 37, 38. The FINDSITE/Q-dock approach uses FINDSITE as the first step to identify functional relationships and binding ligand substructures from complexes in the PDB39. Due to the scarcity of complexes in the PDB for GPCRs, FINDSITE is inapplicable to the large-scale virtual screening of GPCRs. Successful structure-based virtual screening using homology modeling for few individual GPCR family members can be found elsewhere 38, 40.
In this work, to remove the restriction of FINDSITE that a large set of holo template proteins have solved structures, we developed FINDSITEX. First, the structures of target proteins and proteins with ligand binding information are modeled using TASSERVMT-lite. Functional inference and binding ligand identification is accomplished using the modeled structures and a hybrid sequence/structure based approach that combines the structure alignment method fr-TM-align41 with the BLOSUM62 substitution matrix score42. We show that this simple hybrid approach is better than sequence (BLAST43 & PSI-BLAST5), profile (HHSEARCH8), or structure (fr-TM-align41) based approaches for ligand virtual screening. The average enrichment factor within the top 1% of screened compounds using FINDSITEX is triple that of FINDSITE, even when only remote homologous templates (library protein’s sequence identity < 30% to the target) are used.
The outline of the remainder of this paper is as follows: In the Methods section, we give details on the functional relationship identification and virtual screening methodology of FINDSITEX. In the Results and Discussion, we present the validation and prediction results of modeling and virtual screening for all human GPCRs and discuss current and future work. The details of TASSERVMT-lite can be found in Supplementary Materials.
Methods
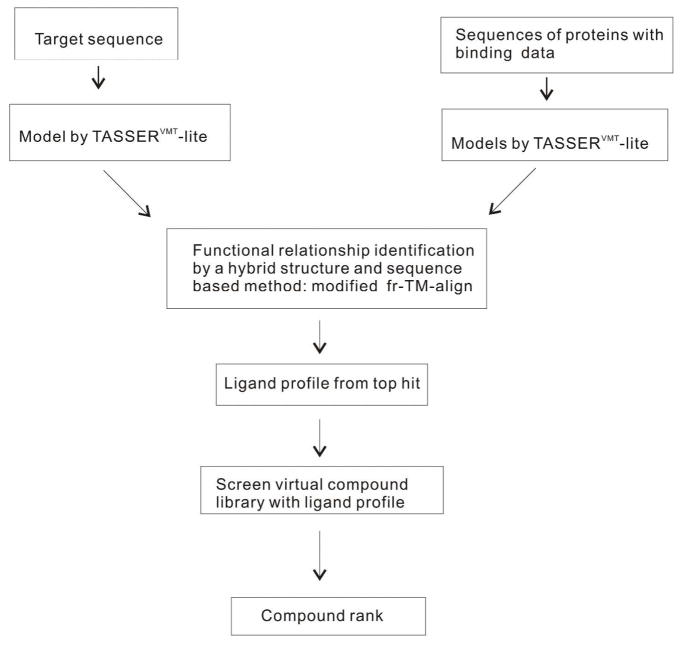
The flowchart for FINDSITEX is given in Figure 1. Models of both target and proteins with binding ligands are modeled with TASSERVMT-lite. Then, the structure and sequence of the target are used to search the binding data library for evolutionarily related proteins. The binding ligands of the top first ranked protein from the library are used to build a molecular fingerprint profile. Subsequently, the fingerprint profile is used to search the compound library for ligands that potentially bind to the target. Here, no attempt is made to predict the binding pose of the target’s ligand other than the ligand structure and likelihood of binding (indicated by ligand ranks). Details of each step are given below.
Figure 1.
FINDSITEX Flowchart.
Modified fr-TM-align for functional similarity identification
In order to use modeled structures for ligand virtual screening across protein family members in general and distant protein family members in particular, a method to rank and select those family members likely to bind the same ligands as the target is needed. One simple way is to use sequence similarity alone to identify the evolutionarily related proteins by BLAST5. In this case, protein structures are unnecessary. Another way is to use a structural alignment method such as the fr-TM-align41, an update of TM-align44. Purely structural alignment methods tend to include many false positives (proteins that are structurally similar, but which have no evolutionary or functional relationship45). A third way of ranking proteins likely to bind similar ligands to those of the target involves two steps: threading and then structural alignment on the threading selected subset as was done in classic FINDSITE13, 46. In this work, we introduce another way of ranking related family members that works better than purely sequence-based or structure-based methods and is simpler than the two-step method of FINDSITE. We modify the fr-TM-align to use an evolutionary score in the final output (the evolutionary score does not affect the structural alignment) to reduce the false positives that typically result from structure comparison. The output score is the summation of the BLOSUM62 substitution matrix 42 values over the aligned residues provided by fr-TM-align and is normalized by target length. In other words, fr-TM-align is used to build the equivalent sequence alignment and BLOSUM62 is used to calculate the sequence alignment score (without gap penalties and is normalized by target length):
| (1) |
This score will be used to rank library proteins. The larger the score is, the closer is the library protein’s function to the target.
Ligand-based virtual screening using the GPCR library with experimental binding data
In this work, we focus on GPCRs, but the methodology can be applied to any distantly related protein family such as ion channels47, kinases48, proteases49, phosphatases50, 51, etc. Ligand-based virtual screening is often the first step in structure based virtual screening14, 15, 38. The structures of target and library GPCRs (GPCR-lib) with experimentally identified binding ligands are built using TASSERVMT-lite. The top ranked GPCR from GPCR-lib is selected using the above modified fr-TM-align approach with the BLOSUM62 score. Ligands binding to the selected GPCR are then filtered so that their pairwise Tanimoto coefficient52 is <0.95 to each other and that the library protein structure has a predicted TM-score ≥ 0.453 to the predicted target structure. If no library protein satisfies these conditions, virtual screening will not be done. Otherwise, we calculate a fingerprint (http://www.daylight.com/dayhtml/doc/theory/theory.finger.html) profile using all the ligands that pass the filtration step. The profile is the summation of the individual fingerprints and is normalized by the number of ligands (equivalent to the average fingerprint of all selected ligands). The normalized profile is used in a continuous Tanimoto coefficient (TC) calculation 54 that ranks the compound fingerprint library:
| (2) |
where x is the target fingerprint profile, y is the fingerprint of the library compound and the summations are over the 1024 fingerprint bits.
Virtual screening results are evaluated by the Enrichment Factor within the top 1% of the compound library defined as:
| (3) |
A true positive is defined as an experimentally known binding ligand or one that has a TC=1 to an experimentally validated binding ligand. EF0.01 ranges from 0 to 100 (100 means that all true positives are within the top 1% of the library).
Human GPCR sequences, structural templates and ligand binding data
The 907 human GPCRs used in our original paper23 are updated from the same sources: http://www.gpcr.org/7tm/ and http://www.expasy.org/cgi-bin/lists?7tmrlist.txt. The new sequences are filtered to be less than 500 residues in length. This results in a set of 998 human GPCR sequences.
All six available experimental GPCR structures (2vt4, 2rh1, 2ydo, 3pbl, 3odu, 3rze) are included in the template library for threading regardless of their sequence identity to each other.
GPCRs with experimental binding data and ligand structures are obtained from the GLIDA database55 (GPCR-lib). These are 168 Human, 98 Mouse and 114 Rat GPCRs and a total of 21,078 non-redundant ligands. The largest protein MGR5_HUMAN in the GPCR-lib has 1,212 residues. There are 7 human GPCRs in the GPCR-lib having > 1000 binding ligands, with the AG2R having the most, 2,205 ligands.
Results
Comparison of predicted GPCR structures with experimental structures
Here, we compare the accuracy of current predictions using TASSERVMT-lite and previous predictions23 for the five human GPCRs with experimental structures. For each target, only the target itself is excluded from the threading template library. Our results are compiled in Table 1. The overall average TM-score is 0.728 for TASSERVMT-lite compared to our previous prediction23 of 0.689 or the SP3 threading prediction of 0.708. The main reason for the increase in accuracy over our original 2006 calculation could be due to the increase in the number of GPCR templates24. Nevertheless, TASSERVMT-lite is 3% better than SP3 threading. For the transmembrane helical portion, the new prediction, whose average RMSD is 2.00 Å, is much better than our 2006 prediction, with an average RMSD of 2.78 Å. However, for the binding pocket, the difference between new and old predictions is less than 0.1Å (3.01 vs. 3.08 Å). As to the extracellular loop 2 (L2), critical for the ligand’s entrance into the binding pocket, both sets of predictions are unsatisfactory (the RMSD to native of the new and 2006 predictions are 10.96 and 13.88 Å, respectively).
Table 1.
Comparison of average model quality for the five known GPCR structures♣
| SP3 threading | Previous model23 | TASSERVMT-lite | Predicted RMSD(TM-score) to native♥ | |
|---|---|---|---|---|
| Overall average RMSD (TM-score) | 10.57Å (0.708) | 13.59Å (0.689) | 8.98Å (0.728) | 7.74Å (0.676) |
| Helical average RMSD (TM-score) | 2.60Å (0.890) | 2.78Å (0.837) | 2.00Å (0.913) | |
| Pocket◆ average RMSD | 3.64Å | 3.08Å | 3.01Å | |
| Extracellular L2 average global RMSD | 13.08Å | 13.88Å | 10.96Å |
They are 2rh1, 2ydo, 3odu, 3pbl, 3rze.
Defined as residues having heavy atom within 5 Å of ligand atom in the crystal structure.
TM-score is given by Eq.(1) from TASSERVMT-lite modeling. A similar equation for RMSD conversion from C-score is used for the RMSD prediction.
Human GPCR threading results
Among the 998 human GPCRs, 988 targets were assigned by SP3 threading as Easy targets (Z-score ≥ 6) that were modeled with the multiple-template based TASSERVMT-lite. The other 10 targets were modeled with a mixed multiple template-based and an ab initio approach, chunk-TASSER56, an upgrade of TASSER for Hard targets (targets having SP3 threading Z-score < 6). Chunk-TASSER is a protein modeling method developed for targets having no identifiable structurally similar templates56. With SP3 threading, 489 targets have 2ydoa (A2A) as their top template, 241 targets have 2ks9a (SUBSTANCE P) as their top template, 158 targets have 3odua (CXCR4 CHEMOKINE) as their top template, 31 targets have 2rh1a (B2-ADRENERGIC) as their top template, 23 targets have 3pbla (DOPAMINE D3) as their top template, 18 targets have 3rzea (HISTAMINE H1) as their top template, 13 targets have 2ziya (SQUID RHODOPSIN) as their top template and 4 targets have 1l9ha (BOVINE RHODOPSIN) as their top template, respectively. The remaining 21 targets do not hit either a GPCR or a Rhodopsin (RH) as the top first template (within the top five, some may hit a GPCR or RH). Some could be falsely annotated as a GPCR or they are too remote to the GPCR and RH templates for SP3 to detect them. For example, targets Q8TDU0, Q16503 and Q16503 have very little PSIPRED57 predicted helical content. Target Q9HB44 has only 145 residues, a size that is too short for a typical GPCR.
Human GPCR modeling results
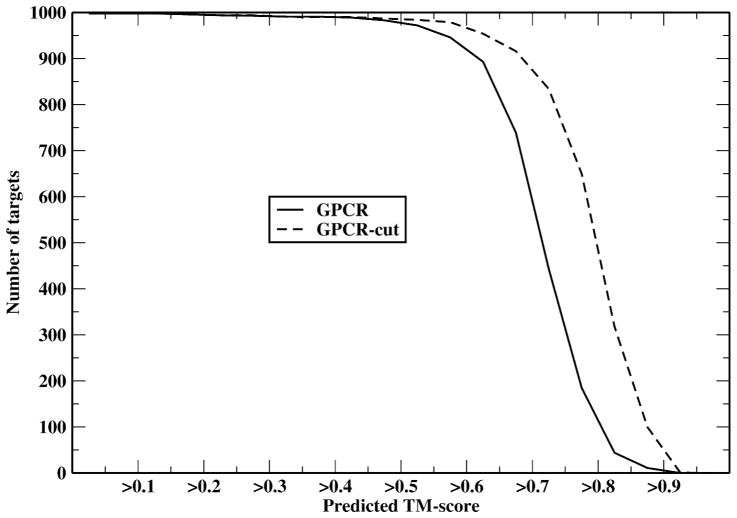
In addition to modeling the full-length 998 target GPCRs using TASSERVMT-lite, we also modeled each sequence with the non-helical tails cut away (GPCR-cut). The average predicted TM-score (see Eq.(1)) for GPCR and GPCR-cut are 0.71 and 0.78, respectively. Thus, without the non-helical tails or for the core part of the GPCR, model quality is likely much better. Figure 2 shows the cumulative distributions of predicted TM-score. Around 97.7% of the full length targets have a predicted TM-score > 0.5. For GPCR-cut, more than 98.6% of targets have a predicted TM-score > 0.5.
Figure 2.
Cumulative distribution of the predicted TM-score for the full length GPCR and the GPCR with non-helical tails removed.
To further check the model quality of our protocol, we compare our models with those of the GPCR-ITASSER28 for a set of 38 common targets in Table 2. The two sets of models on average have a predicted TM-score around 0.7 and their mutual TM-score to each other is also around 0.7, indicating that both sets are likely of the same average quality. GPCR-ITASSER models were modeled using experimentally derived restraints from biochemical data, whereas TASSERVMT-lite models have no such restraints. For reasons that are unclear, the predicted model qualities using GPCR-ITASSER are more diverse (with a predicted TM-score to native in the range 0.42–0.84); some targets have a predicted TM-score greater than 0.8, with a few having a predicted TM-score around 0.45, whereas those of TASSERVMT-lite are more uniform in quality (0.60–0.82).
Table 2.
Comparison of predicted model quality of the TASSERVMT-lite model with the GPCR-I-TASSER model for a 38 target subset
| Target ID | Mutual TM-score | GPCR-ITASSER predicted TM-score | TASSERVMT-lite predicted TM-score |
|---|---|---|---|
| O00590 | 0.66 | 0.76 | 0.68 |
| P08172 | 0.61 | 0.79 | 0.67 |
| P08173 | 0.60 | 0.76 | 0.73 |
| P11229 | 0.65 | 0.71 | 0.74 |
| P14416 | 0.80 | 0.73 | 0.73 |
| P21728 | 0.69 | 0.47 | 0.67 |
| P21917 | 0.86 | 0.70 | 0.71 |
| P21918 | 0.67 | 0.45 | 0.62 |
| P25021 | 0.90 | 0.81 | 0.75 |
| P46094 | 0.73 | 0.81 | 0.69 |
| P51677 | 0.71 | 0.83 | 0.68 |
| P51679 | 0.70 | 0.77 | 0.66 |
| P51681 | 0.72 | 0.84 | 0.68 |
| P51684 | 0.86 | 0.72 | 0.74 |
| P51685 | 0.72 | 0.75 | 0.61 |
| P51686 | 0.24 | 0.73 | 0.64 |
| Q99788 | 0.87 | 0.83 | 0.67 |
| Q9H3N8 | 0.72 | 0.69 | 0.65 |
| Q9NPB9 | 0.71 | 0.77 | 0.62 |
| Q9Y5N1 | 0.72 | 0.65 | 0.65 |
| O00421 | 0.72 | 0.81 | 0.67 |
| O00574 | 0.87 | 0.83 | 0.79 |
| P08588 | 0.66 | 0.47 | 0.62 |
| P08913 | 0.63 | 0.72 | 0.82 |
| P13945 | 0.77 | 0.71 | 0.73 |
| P18089 | 0.82 | 0.76 | 0.68 |
| P18825 | 0.62 | 0.62 | 0.80 |
| P25024 | 0.72 | 0.81 | 0.74 |
| P25025 | 0.70 | 0.72 | 0.60 |
| P25106 | 0.69 | 0.74 | 0.73 |
| P32246 | 0.82 | 0.82 | 0.75 |
| P32248 | 0.68 | 0.62 | 0.70 |
| P32302 | 0.69 | 0.69 | 0.71 |
| P35348 | 0.66 | 0.42 | 0.65 |
| P41597 | 0.80 | 0.74 | 0.76 |
| P46092 | 0.70 | 0.80 | 0.69 |
| P49238 | 0.83 | 0.80 | 0.77 |
| P49682 | 0.69 | 0.68 | 0.71 |
| average | 0.72 | 0.72 | 0.70 |
Human GPCR virtual screening results
Virtual screening procedure was tested on the 168 human GPCRs (this 168 set is not identical to a subset of the 998 targets because they are collected from different sources) from the GLIDA database55 that have experimentally determined binding ligands (GPCR-lib). The GLIDA database provides agonist or antagonist information and does not provide binding affinity information. In this work, agonists and antagonists are not distinguished. GPCRs in GPCR-lib having a sequence identity > 30% to a given target were excluded from that target’s ligand profile computation. The computed ligand profile from a potential functionally related library GPCR is then used in continuous TC (Eq.(2)) similarity searching against the ZINC858 non-redundant (TC cutoff 0.7) library containing 67,871 compounds in combination with all the true binding 21,078 non-redundant ligands from the GLIDA database.
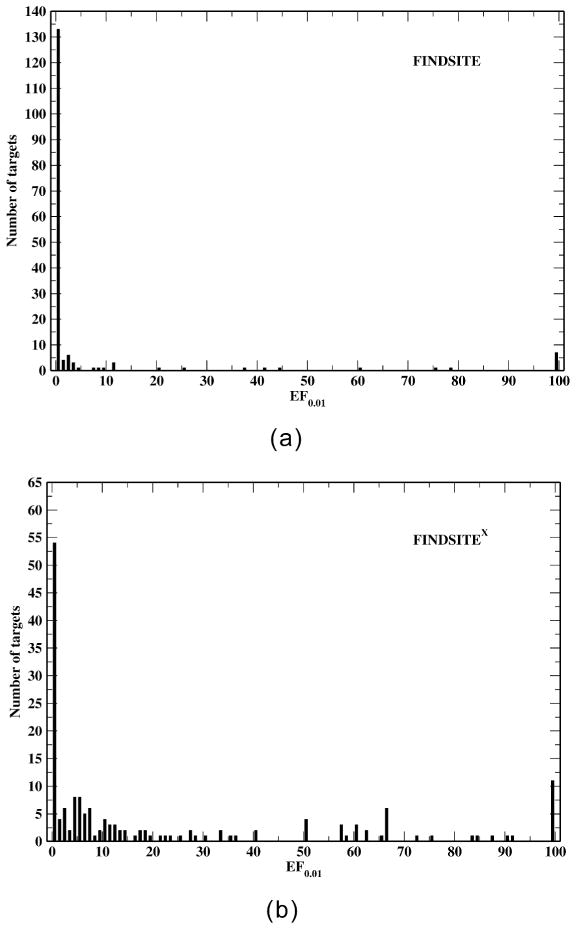
To show the advantage of including biological binding data without holo structures for virtual screening, we compare the EF0.01 distributions of FINDSITEX and of the original FINDSITE in Figure 3. At a 30% sequence identity cutoff, the average EF0.01 by FINDSITEX is 22.7 whereas that from FINDSITE is 7.1. FINDSITEX has 114 targets whose EF0.01 > 1 (i.e. 68% of the targets have an EF0.01 better than random), whereas FINDSITE has only 35 targets whose EF0.01 > 1 (21% of the targets have an EF0.01 better than random). When closely homologous proteins from GPCR-lib are used (Table 3 only excludes the target itself), FINDSITEX gives an average EF0.01 of 41.4, with 91.1% of targets have an EF0.01 better than random, whereas the corresponding values by FINDSITE are 9.6 and 31.5%, respectively.
Figure 3.
EF0.01 distribution of (a) FINDSITE and (b) FINDSITEX with a sequence cutoff 30% for the 168 human GPCRs that have experimentally identified binding ligands.
Table 3.
Enrichment factor EF0.01 of different methods for functional relationship detection for the 168 testing human GPCRs.
| Method | Sequence cutoff 30% | Exclude target only | No cutoff |
|---|---|---|---|
| fr-TM-align with evolutionary score | 22.7 | 41.4 | 64.2 |
| BLAST | 18.9 | 39.9 | 63.9 |
| PSI-BLAST (5 iterations) | 16.3 | 28.5 | 50.2 |
| HHSEARCH | 13.1 | 33.2 | 64.2 |
| fr-TM-align | 13.3 | 22.3 | 64.2 |
To confirm the usefulness of modeled structures for ligand-based virtual screening, we also used BLAST43, PSI-BLAST5, and HHSEARCH8 that use only sequence information to select the top related library GPCR for ligand profile construction. The results along with the pure structure based fr-TM-align results at different sequence identity cutoffs are given in Table 3. At a sequence identity cutoff of 30%, the average EF0.01 by BLAST, PSI-BLAST (5 iterations) and HHSEARCH are 18.9, 16.3 and 13.1, respectively. Application of modeled structures for the target in combination with the evolutionary score for selection of related GPCRs improves EF0.01 by about 20% (as compared to BLAST) to 22.7. We also examined the effect of applying the evolutionary score in the fr-TM-align’s final score calculation. Without the evolutionary score (i.e. using fr-TM-align’s TM-score), the average EF0.01 is 13.3, a result that is much worse than when the evolutionary score is used (22.7). If only the BLOSSUM62 matrix is used in a Needleman-Wunsch59 alignment, then the resulting EF0.01 will be 19.6, which is slightly better than that of BLAST and ~15% worse than the hybrid method. HHSEARCH and Needleman-Wunsch methods have been used in 60 for a study of GPCR evolution. We note that methods that perform better for structure similarity detection (structure prediction) tend to perform worse for virtual screening at a low sequence cutoff 30%. PSI-BLAST is worse than BLAST, and HHSEARCH and fr-TM-align are worse than PSI-BLAST. This could be due to false positive detection (library structures that are similar to the target but whose functions are dissimilar). When no cutoff is applied (Table 3, No cutoff), all methods except PSI-BLAST give an EF0.01 of around 64. In this case, the true ligands from target itself are used for construction of the ligand profile. An EF0.01 of 64 means that on average, 64% of the true binding ligands are recovered within the 1% of the 88,949 screened compounds if native ligands are used for profile construction. The poorer performance of PSI-BLAST when no sequence cutoff is applied is due to that it cannot distinguish the target native sequence from closely homologous sequences that bind different ligands.
We then applied FINDSITEX in prediction mode (no GPCR-lib protein is excluded) to all the 998 identified human GPCRs (<500 residues) for ligand-based virtual screening against the combined ZINC8 non-redundant (TC<0.7) compound library and true binding ligands of the GLIDA database (total 88,949 compounds). All predicted structures and virtual screening data are freely available for academic users at http://cssb.biology.gatech.edu/skolnick/webservice/gpcr/index.html. Users can input either the UniProt ID (http://www.uniprot.org/) or a FASTA formatted sequence to search for predictions. Prediction results that can be downloaded from the search are: the top TASSERVMT-lite five all-atom structural models, threading templates and their SP3 Z-scores, template alignments to the target, virtual screening compound rank, target ligand profile (which can be used offline for screening the user’s own compound library), off-target rank (see below), and the five all-atom models modeled when the two end non-helical parts are cut away.
Testing of off-target GPCR predictions
For each human GPCR target, we predict its off-targets (targets that potentially bind to the same ligands). We employ the Kendall τ rank correlation coefficient 61 of the virtual screening compound ranks introduced by Brylinski & Skolnick62. Here, the 168 protein set is used for off-target prediction testing as well. A true off-target for a given target is defined as a target having a maximal TC=1 between all pairs of ligands of the target and the given target (i.e., at least one common ligand in the experimental binding data). A target is predicted as an off-target if its Kendall τ correlation coefficient to the given target is > cutoff. The following quantities are assessed for the off-target prediction.
Target coverage defined as:
Prediction precision is defined for a given target as:
Prediction recall is defined for a given target as:
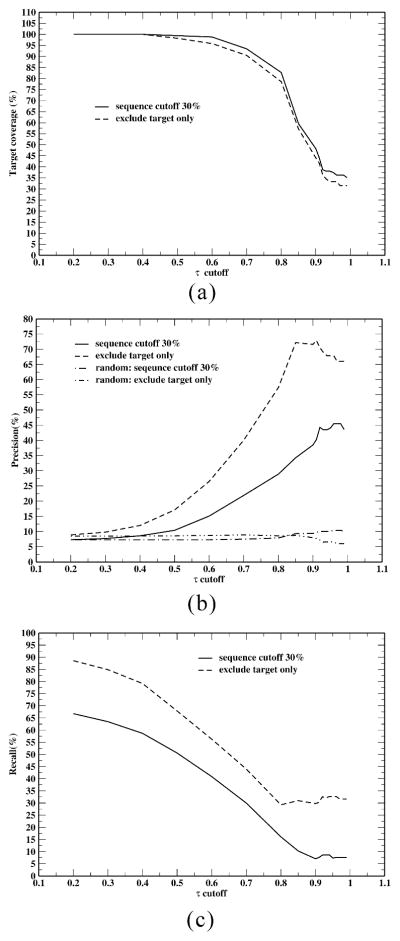
Figure 4a–c shows the dependence of target coverage, prediction precision and recall (averaged per target on the subset of targets having predictions) on the τ cutoff values. A sequence identity cutoff is employed in both ligand profile construction and off-target selection (i.e. the target ligand profile is constructed from ligands of library proteins that have a sequence identity ≤ cutoff to the given target and only off-targets that have a sequence identity ≤ cutoff to the given target are considered). A lower cutoff is used to mimic cases when only evolutionarily remote protein data exists in the binding data library. While the precision and recall depend strongly on the sequence cutoff, the target coverage shows little dependence. With a sequence identity cutoff of 30%, prediction precision reaches a maximal 45.5% at a 0.96 τ cutoff, but only 36% of the 168 targets have predictions and recall is 10.4% for these 36% of the targets. When only the target itself is excluded (i.e. any close homologies are included) and a 0.91 τ cutoff is used, the prediction precision (recall) is 72.9% (30.3%) and 70 targets (or 42%) of the 168 targets have predictions. For both a 30% cutoff and inclusion of close homologues, a prediction with random selection will have a precision of less than 10%. With any τ cutoff > 0.2, precision is better than random.
Figure 4.
Dependences of (a) target coverage, (b) prediction precision and (c) prediction recall on the Kendall τ cutoff value for off-target prediction. Precision and recall are average per target on the subset of targets having predictions.
We next discuss a few examples of identified distantly related off-targets using a τ cutoff of 0.96 when a sequence identity cutoff of 30% is applied. 5HT1A and 5HT2B have a sequence identity of 26% (τ=1 resulted from using the same library protein for ligand profile construction) and share 54 ligands in the GLIDA database; CCR1 and CXCR4 both bind to L000624 (GLIDA ID55) and their sequence identity is 29% (τ=1); DRD2 and 5HT5A have a sequence identity of 25% (τ=1), and they both bind to L000195(Clozapine), L001003(Ritanserin) and L001254(Yohimbine). The list of putative off-target sets for the 998 human GPCRs is available at http://cssb.biology.gatech.edu/skolnick/webservice/gpcr/index.html.
Discussion
In this work, we have developed the FINDSITEX method that significantly enhances the performance of our original structure/threading based FINDSITE approach by removing the restriction that the target of interest have a sizable number of solved, threading identified holo structures in the PDB39. For drug target families such as GPCRs, there are only a few solved holo structures in the PDB that have a potential functional relationship to the target protein. On the other hand, there are much biologically characterized binding ligand and drug data55, 63–66 that are not fully exploited for ligand virtual screening. The current approach utilizes remote as well as closely homologous proteins without solved receptor and holo structures but does require experimentally determined ligand binding data that are then employed in structure based virtual screening. FINDSITEX provides the necessary state-of-the-art multiple-template based receptor structures as well as structure-based virtual screening results based on these predicted receptor structures14, 15, 19, 38. FINDSITEX can also predict possible off-targets of a given target.
We demonstrated the application of FINDSITEX to human GPCRs. It is quite likely that the new models using TASSERVMT-lite are better than our old models23 for the intramembrane helical portion of the GPCR’s structures. This could be partly due to the fact that more templates are available24 and due to our new way of utilizing multiple templates through the Variable number of Multiple Templates (VMT) approach of TASSERVMT-lite27. For ligand virtual screening, FINDSITEX provides a top 1% enrichment factor of 22.7, triple that of FINDSITE, when only distantly related proteins with binding data are available (sequence identity < 30% to target). This number reaches 41.4 when closely homologous proteins with binding data exist. This dramatic improvement is mainly due to the dominance of binding data over PDB holo structures for GPCRs. We show that the hybrid structure alignment ranked by an evolutionary score approach works better than BLAST or other sequence/profile based methods as well as the purely structure based method fr-TM-align for functional relationship identification. This could be due to fact that some evolutionarily related sequences are divergent in structure67, and thus do not bind similar ligands. Conversely, the pure structure alignment method, fr-TM-align, is worse than the BLAST because evolutionarily unrelated proteins could have similar structures45. The structure alignment provided by the hybrid approach reduces the false positive alignments of sequence/profile approaches, whereas the evolutionary component of the score reduces the false positives from a purely structure based approach.
FINDSITEX is not limited to the GPCR family. In principle, it is applicable to other drug target families such as ion channels, kinases, proteases, phosphatases, etc. The structure modeling TASSERVMT-lite and ligand-based virtual screening components of EF0.01 are not GPCR specific, although they predict quite accurate structural models and ligand virtual screening results for GPCRs. For human GPCR modeling, the current approach still has poor to moderate accuracy for loop modeling; methods that improve the modeling of the loops will be pursued in the near future. For virtual screening, one missing part of our current work is that it does not provide the actual positions of the potential binding pockets. However, this can be easily remedied by structure-based or geometry-based pocket detection methods68–71. This will be pursued in the very near future.
FINDSITEX is not only an extension of FINDSITE that eliminates the restriction of the existence of sufficient amount of solved PDB holo structures, but it is most importantly a powerful extension of bioactivity databases such as ChEMBL65, GLIDA55, DrugBank63 and PubChem64 for drug discovery. Unlike these databases that provide only sequence based search engines that do not work well for remote targets (targets having no closely homologous proteins in the binding database), for ~2/3 of the remote targets, the structure based FINDSITEX yields an enrichment factor E0.01 > 1 for ligand virtual screening as manifested in the 168 Human benchmark GPCRs.
Supplementary Material
Acknowledgments
The authors thank Dr. Bartosz Ilkowski for managing the cluster on which this work was conducted. We thank the Zhang-lab at University of Michigan for providing us with the GPCR-ITASSER models. This research was supported in part by grants No. GM084222, GM-37408 and GM-48835 of the Division of General Medical Sciences of the National Institutes of Health.
Footnotes
Supporting information available
Detailed description of the TASSERVMT-lite method can be found in Supplementary Materials. This information is available free of charge via the Internet at http://pubs.acs.org/
References
- 1.Skolnick J, Fetrow J, Kolinski A. Structural genomics and its importance for gene function analysis. Nat Biotechnol. 2000;18:283–287. doi: 10.1038/73723. [DOI] [PubMed] [Google Scholar]
- 2.Baker D, Sali A. Protein structure prediction and structural genomics. Science. 2001;294:93–96. doi: 10.1126/science.1065659. [DOI] [PubMed] [Google Scholar]
- 3.Pieper Ursula, Eswar Narayanan, Braberg Hannes, Madhusudhan MS, Davis Fred P, Stuart Ashley C, Mirkovic Nebojsa, Rossi Andrea, Marti-Renom Marc A, Fiser Andras, Webb Ben, Greenblatt Daniel, Huang Conrad C, Ferrin Thomas E, Sali A. MODBASE, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res. 2004;32(Database issue):D217–D222. doi: 10.1093/nar/gkh095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Structural Bioinformatics of Membrane Proteins. Springer; Wien New York: 2010. [Google Scholar]
- 5.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman D-J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acid Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sali AaBTL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 7.Pandit SB, Zhang Y, Skolnick J. TASSER-Lite: An automated tool for protein comparative modeling. Biophysical Journal. 2006;91:4180–4190. doi: 10.1529/biophysj.106.084293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hildebrand A, Remmert M, Biegert A, Söding J. Fast and accurate automatic structure prediction with HHpred. Proteins. 2009;77(Suppl 9):128–32. doi: 10.1002/prot.22499. [DOI] [PubMed] [Google Scholar]
- 9.Jaroszewski L, Rychlewski L, Li W, Godzik A. Comparison of sequence profiles. Strategies for structural predictions using sequence information. Protein Sci. 2000;9:232–241. doi: 10.1110/ps.9.2.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer D. Hybrid fold recognition: combining sequence derived properties with evolutionary information. Pacific Symp. Biocomp; Hawaii. 2000. [PubMed] [Google Scholar]; Altman RB, Dunker AK, Hunter L, Lauderdale K, Klein TE, editors. World Scientific; Hawaii: 2000. pp. 119–130. [Google Scholar]
- 11.Zhou H, Zhou Y. SPARKS 2 and SP3 servers in CASP 6. Proteins(Supplement CASP issue) 2005;(suppl 7):152–156. doi: 10.1002/prot.20732. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y. Progress and challenges in protein structure prediction. Curr Opin Struct Biol. 2008;18(3):342–348. doi: 10.1016/j.sbi.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brylinski M, Skolnick J. FINDSITE: A threading-based method for ligand-binding site prediction and functional annotation. Proc Natl Acad Science. 2008;105:129–134. doi: 10.1073/pnas.0707684105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brylinski M, Skolnick J. Q-Dock: Low-resolution flexible ligand docking with pocket-specific threading restraints. Journal of Computational Chemistry. 2008;29:1574–88. doi: 10.1002/jcc.20917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brylinski M, Skolnick J. Q-Dock(LHM): Low-resolution refinement for ligand comparative modeling. Journal of Computational Chemistry. 2010;31:1093–105. doi: 10.1002/jcc.21395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wass MN, Sternberg MJ. Prediction of ligand binding sites using homologous structures and conservation at CASP8. Proteins. 2009;77(S9):147–151. doi: 10.1002/prot.22513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wass MN, Kelly LA, Sternberg MJ. 3DLigandSite: predicting ligand-binding sites using similar structures. Nucl Acid Res. 2010;38(suppl 2):W469–W473. doi: 10.1093/nar/gkq406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brylinski M, Skolnick J. Comprehensive Structural and Functional Characterization of the Human Kinome by Protein Structure Modeling and Ligand Virtual Screening. J Chem Inf Model. 2010;50(10):1839–1854. doi: 10.1021/ci100235n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee HS, Zhang Y. BSP-SLIM: A blind low-resolution ligand-protein docking approach using predicted protein structures. Proteins. 2011;80:93–110. doi: 10.1002/prot.23165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roy A, Xu D, Poisson J, Zhang Y. A Protocol for Computer-Based Protein Structure and Function Prediction. Journal of Visualized Experiments. 2011:57. doi: 10.3791/3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Filmore D. It’s a GPCR world. Modern Drug Discovery. 2004;2004:24–28. [Google Scholar]
- 22.Zhang Y, Skolnick J. Automated structure prediction of weakly homologous proteins on genomic scale. Proc Natl Acad Sci (USA) 2004;101:7594–7599. doi: 10.1073/pnas.0305695101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, DeVries ME, Skolnick J. Structure modeling of all identified G protein-coupled receptors in the human genome. PLoS Computational Biology. 2006;2:0088–0099. doi: 10.1371/journal.pcbi.0020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yarnitzky T, Levit A, Niv MY. Homology modeling of G-protein-coupled-receptors with X-ray structures on the rise. Curr Opin Drug Discovery & Development. 2010;13:317–325. [PubMed] [Google Scholar]
- 25.Peng J, Xu J. A multiple-template approach to protein threading. Proteins. 2011;79(6):1930–39. doi: 10.1002/prot.23016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng J. A multi-template combination algorithm for protein comparative modeling. BMC Struc Biol. 2008;8:18. doi: 10.1186/1472-6807-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou H, Skolnick J. Template-based protein structure modeling using TASSERVMT. Proteins. 2011;80(2):352–361. doi: 10.1002/prot.23183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J, Zhang Y. GPCRRD: G protein-coupled receptor spatial restraint database for 3D structure modeling and function annotation. Bioinformatics. 2010;26:3004–05. doi: 10.1093/bioinformatics/btq563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y. Template-based modeling and free modeling by I-TASSER in CASP7. Proteins. 2007;69(Suppl 8):108–117. doi: 10.1002/prot.21702. [DOI] [PubMed] [Google Scholar]
- 30.Barth P, Wallner B, Baker D. Prediction of membrane protein structures with complex topologies using limited constraints. PNAS. 2009;106(5):1409–1414. doi: 10.1073/pnas.0808323106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nikiforovich G, Taylor C, Marshall G, Baranski T. Modeling the possible conformations of the extracellular loops in G-protein-coupled receptors. Proteins. 2010;78(2):271–285. doi: 10.1002/prot.22537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mehler E, Hassen S, Kortagere S, Weinstein H. Ab initio computational modeling of loops in G-protein-coupled recepotrs: Lessons from the crystal structures of rhodopsin. Proteins. 2006;64(3):673–690. doi: 10.1002/prot.21022. [DOI] [PubMed] [Google Scholar]
- 33.Niv M, Skrabanek L, Filizola M, Weinstein H. Modeling activated states of GPCRs: The rhodopsin template. J Comput Aided Mol Des. 2006;20:437–448. doi: 10.1007/s10822-006-9061-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Foquet N, M’Kadmi C, Perahia D, Gagne D, Berge G, Marie J, Baneres J, Galleyrand J, Hehrentz J, Martinez J. Activation of the ghrelin receptor is described by a privileged collective motion: A model for constitutive and agonist-induced activation of sub-class A G-protein-coupled receptor (GPCR) J Mol Biol. 2010;395(4):769–784. doi: 10.1016/j.jmb.2009.09.051. [DOI] [PubMed] [Google Scholar]
- 35.Abrol R, Bray J, Goddard W., III Bihelix: Towards de novo structure prediction of an ensemble of G-protein coupled receptor conformations. Proteins. 2012;80(2):505–518. doi: 10.1002/prot.23216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kimura S, Tebben A, Langley D. Exapnding GPCR homology model binding sites via a balloon potential: A molecular dynamics refinement approach. Prteoins. 2008;71(4):1919–1929. doi: 10.1002/prot.21906. [DOI] [PubMed] [Google Scholar]
- 37.Capra J, Laskowski R, Thornton J, Singh M, Funkhouser T. Predicting Protein Ligand Binding Sites by Combining Evolutionary Sequence Conservation and 3D Structure. Plos Comput Biol. 2009;5(12) doi: 10.1371/journal.pcbi.1000585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Radestock S, Weil T, Renner S. Homology Model-Based Virtual Screening for GPCR Ligands Using Docking and Target-Biased Scoring. J Chem Inf Model. 2008;48(5):1104–1117. doi: 10.1021/ci8000265. [DOI] [PubMed] [Google Scholar]
- 39.Bernstein FC, Koetzle TF, Williams GJB, Meyer EF, Jr, MDB, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. The Protein Data Bank: A Computer-based Archival File for Macromolecular Structures. J Mol Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 40.Evers A, Klabunde T. Structure-based drug discovery using GPCR homology modeling: successful virtual screening for antagonists of the Alpha1A adrenergic receptor. J Med Chem. 2005;48:1088–1097. doi: 10.1021/jm0491804. [DOI] [PubMed] [Google Scholar]
- 41.Pandit S, Skolnick J. Fr-TM-align: a new protein structural alignment method based on fragment alignments and the TM-score. BMC Bioinformatics. 2008;(9):531. doi: 10.1186/1471-2105-9-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henikoff S, Henikoff JG. Amino Acid Substitution Matrices from Protein Blocks. PNAS. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Altschul SF, Gish W, Miller W, Myers E, Lipman D. Basic local alignment tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucl Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, Hubner I, Arakaki A, Shakhnovich E, Skolnick J. On the origin and highly likely completeness of single-domain protein structures. Proc Natl Acad Sci (USA) 2006;103:2605–2610. doi: 10.1073/pnas.0509379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brylinski M, Skolnick J. Comparison of structure-based and threading-based approaches to protein functional annotation. Proteins. 2010;78(1):118–34. doi: 10.1002/prot.22566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Camerino D, Tricarico D, Desaphy J. Ion channel pharmacology. Neurotherapeutics. 2007;4(2):184–98. doi: 10.1016/j.nurt.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 48.Manning G, Whyte D, Martinez R, Hunter T, Sudarssanam S. The protein kinase complement of the human genome. Science. 2002;298:5600. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 49.Barrett AJ, Rawlings N, Woessner J. The Handbook of Proteolytic Enzymes. 2. Academic Press; 2003. [Google Scholar]
- 50.Barford D. Molecular mechanisms of the protein serine/threonine phosphatases. Trends Biochem Sci. 1996;21(11):407–12. doi: 10.1016/s0968-0004(96)10060-8. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Z. Protein tyrosine phosphatases: structure and function, substrate specificity, and inhibitor development. Annu Rev Pharmacol Toxicol. 2002;42(1):209–34. doi: 10.1146/annurev.pharmtox.42.083001.144616. [DOI] [PubMed] [Google Scholar]
- 52.Tanimoto TT. An elementary mathematical theory of classification and prediction. IBM Interanl Report. 1958 Nov;1958 [Google Scholar]
- 53.Zhang Y, Skolnick J. A scoring function for the automated assessment of protein structure template quality. Proteins. 2004;57:702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- 54.Willett P. Chemical similariry searching. J Chem Inf Model. 1998;38:983–996. [Google Scholar]
- 55.Okuno Y, Tamon A, Yabuuchi H, Niijima S, Minowa Y, Tonomura K, Kunimoto R, Feng C. GLIDA: GPCR—ligand database for chemical genomics drug discovery—database and tools update. Nucl Acid Res. 2007;36:D907–D912. doi: 10.1093/nar/gkm948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou H, Skolnick J. Ab initio protein structure prediction using chunk-TASSER. Biophys J. 2007;93:1510–1518. doi: 10.1529/biophysj.107.109959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones TD. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 58.Irwin JJ, Shoichet BK. ZINC - A Free Database of Commercially Available Compounds for Virtual Screening. J Chem Inf Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Needleman S, Wunsch C. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 60.Nordstroem K, Almen M, Edstam M, Fredriksson R, Schioeth H. Independent HHsearch, Needleman-Wunsch-Based, and Motif Ananylysses Reveal the Over Hierarchy for Most of the G Protein-Coupled Recepltor Families. Mol Biol Evol. 2011;28(9):2471–2480. doi: 10.1093/molbev/msr061. [DOI] [PubMed] [Google Scholar]
- 61.Kendall MG. A new measure of rank correlation. Biometrika. 1938;30(Part 1–2):81–89. [Google Scholar]
- 62.Brylinski M, Skolnick J. Cross-Reactivity Virtual Profiling of the Human kinome by X-ReactKIN: A chemical Systems Biology Approach. Molecular Pharmaceutics. 2010;7(6):2324–33. doi: 10.1021/mp1002976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wishart D, Knox C, Guo A, Shrivastava S, Hassanali M, Stothard P, Chang Z, Woolsey J. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucl Acid Res. 2006;34(Database):D668–72. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Y, Xiao J, Suzek T, Zhang J, Wang J, Zhou Z, Han L, Karapetyan K, Dracheva S, Shoemaker B, Bolton E, Gindulyte A, Bryant S. PubChem’s BioAssay Database. Nucl Acid Res. 2012;40(1):D400–12. doi: 10.1093/nar/gkr1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gaulton A, Bellis L, Bento A, Chambers J, Davies M, Hersey A, Light Y, McGlinchey S, Michalovich D, Al-Lazikani B, Overington J. ChEMBL: a large-scale bioactivity database for drug discovery. Nucl Acid Res. 2012;40(D1):D1100–07. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucl Acid Res. 2004;32:D277–80. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kosloff M, Kolodny R. Sequence-similar, structure-dissimilar protein pairs in the PDB. Proteins. 2008;71(2):891–902. doi: 10.1002/prot.21770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang B, Schroeder M. LIGSITEcsc: predicting protein binding sites using the Connolly surface and degree of conservation. BMC Structural Biology. 2006;6:19. doi: 10.1186/1472-6807-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tan K, Varadarajan R, Madhusudhan M. DEPTH: a web server to compute depth and predict small-molecule binding cavities in proteins. Nucl Acid Res. 2011;39(Web Server):W242–8. doi: 10.1093/nar/gkr356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Z, Li Y, Lin B, Schroeder M, Huang B. Identification of cavities on protein surface using multiple computational approaches for drug binding site prediction. Bioinformatics. 2011;27(15):2083–88. doi: 10.1093/bioinformatics/btr331. [DOI] [PubMed] [Google Scholar]
- 71.Ngan C, Hall D, Zerbe B, Grove L, Kozakov D, Vajda S. FTSite: High accuracy detection of ligand binding sites on unbound protein structures. Bioinformatics. 2012;28(2):286–87. doi: 10.1093/bioinformatics/btr651. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.