Abstract
Although reciprocal evolutionary responses between interacting species are a driving force behind the diversity of life, pairwise coevolution between plant competitors has received less attention than other species interactions and has been considered relatively less important in explaining ecological patterns. However, the success of species transported across biogeographic boundaries suggests a stronger role for evolutionary relationships in shaping plant interactions. Alliaria petiolata is a Eurasian species that has invaded North American forest understories, where it competes with native understory species in part by producing compounds that directly and indirectly slow the growth of competing species. Here I show that populations of A. petiolata from areas with a greater density of interspecific competitors invest more in a toxic allelochemical under common conditions. Furthermore, populations of a native competitor from areas with highly toxic invaders are more tolerant to competition from the invader, suggesting coevolutionary dynamics between the species. Field reciprocal transplants confirmed that native populations more tolerant to the invader had higher fitness when the invader was common, but these traits came at a cost when the invader was rare. Exotic species are often detrimentally dominant in their new range due to their evolutionary novelty; however, the development of new coevolutionary relationships may act to integrate exotic species into native communities.
Keywords: allelopathy, glucosinolates, mycorrhizae, Pilea pumila
Reciprocal evolutionary responses between interacting species (i.e., coevolution) are a driving force creating the diversity of life on earth (1–4). Coevolution has been primarily studied in specialized interactions, typically between consumers and hosts [e.g., plant–pollinator (5, 6), plant–herbivore (1, 7), host–parasite (8–10)], where the ecological specificity of interactions allows for partners to exert, and respond to, reciprocal selection on each other. However, for many species the most important biotic interactions are generalized, especially for competitive interactions, and thus each species would potentially face selection from multiple directions (11). Whether, and how, pairwise coevolutionary dynamics can occur in generalized interactions like competition remain open questions (4).
Coevolution between competitors has often been inferred from patterns of character displacement, in which sympatric populations of competing species have reduced niche overlap compared with allopatric ones (12). The majority of character displacement studies infer past coevolutionary processes from extant patterns (12); only a few studies to date have documented ongoing coevolutionary dynamics among competitors (13). Competitor coevolution may be vital to both the development and the maintenance of diversity because theory predicts that long-term coexistence requires that species differ in their niche requirements (14). However, many species appear to coexist despite the lack of obvious niche differences (15). This is especially true for plants, which compete for a limited set of nonsubstitutable resources. For this reason, coevolution between plant competitors has received less attention than that between animals (16), although evidence suggests that some plant populations evolve in response to direct and indirect interactions with other plant species. For example, Turkington and colleagues demonstrated through a series of studies that sympatric populations of two plant species had reduced competitive effects on each other compared with allopatric populations (17, 18). The traits underlying these patterns were not clear, but derive in part from indirect interactions mediated by soil microbes (19). Additionally, several studies of chemically divergent Thymus species have suggested that populations of co-occurring plant species may adapt to the particular Thymus chemotype with which they interact (20–22). Again, this adaptation may be partially mediated by soil properties or communities (20, 23).
Exotic invasions provide an excellent system to study competitor coevolution in action. Exotic invaders can be competitively dominant over native species in their introduced range, leading to ecological and economic costs (24). This dominance results in part from ecological advantages that arise from their lack of evolutionary history with native species (e.g., release from natural enemies or production of novel weapons) (25, 26). However, this dominance likely imposes intense selection pressures on native species, potentially resulting in new coevolutionary relationships that may act to integrate the exotic species into its new community (27). Exotic species can evolve rapidly in their new range (28), and some native species have evolved adaptations to invaders (29), including native plants responding to invasive plant competitors (30–33). However, no study to date has documented reciprocal coevolution between native and exotic plant species (27).
Coevolutionary dynamics are rarely uniform across space or time, but instead vary among populations due to differences in the reciprocity of selection (i.e., coevolutionary “hot” and “cold” spots), selection mosaics, and gene flow and neutral evolutionary processes (3, 34). Therefore, the presence or absence of trait correlations between interacting populations is not sufficient to establish or rule out coevolution (35). Rather, such claims require evidence that (i) the traits of exotic populations differ in response to interactions with natives; (ii) the traits of native populations differ in response to the traits of the invader; and, most importantly, (iii) these trait distributions are adaptive (i.e., confer higher fitness when the two species interact).
Alliaria petiolata is a biennial forb native to Europe that was introduced to North America over 150 y ago. It has aggressively invaded forest understories in the eastern United States, where it can form dense, nearly monospecific stands (36). This dominance is partly attributable to the production of novel allelochemicals (37) that can directly and indirectly interfere with competitor growth by reducing the abundance and diversity of arbuscular mycorrhizal fungi (AMF) (38, 39). These allelochemicals appear to act as “novel weapons” sensu Callaway and Ridenhour (25), because they are more toxic to AMF strains from North America (introduced range) versus Europe (native range) (38). A. petiolata, like all members of the Brassicaceae, does not form mycorrhizal connections (40). Tissue concentrations of sinigrin, one of the major allelochemicals of A. petiolata, have evolved rapidly during this invasion, with higher sinigrin concentrations in populations on the expanding range edge compared with older populations (41). In a related species, sinigrin was selectively favored when plants faced interspecific competition, but was disfavored when those same genotypes competed with conspecifics (42). Variation in sinigrin among A. petiolata individuals and populations partially determines the invader’s impact on mycorrhizal fungal communities and native plant growth (43, 44). To test whether A. petiolata has entered into new coevolutionary relationships with native plant competitors, I first investigated the genetic investment to sinigrin production in A. petiolata roots among populations varying in the density of interspecific competitors. Then I tested the competitive ability of populations of a common native competitor (Pilea pumila) originating from areas with high- or low-sinigrin A. petiolata populations against the invader, as well as the native population’s interactions with soil biota. Finally, I performed a reciprocal transplant experiment in which P. pumila individuals from these six populations were planted into each of six destination sites in plots with ambient densities of A. petiolata or with the invader removed. If chemical competition is driving coevolutionary dynamics between the invader and its native competitors, one would predict that (i) A. petiolata populations from areas with higher densities of interspecific competitors will invest more in sinigrin, (ii) native populations that co-occur with high-sinigrin A. petiolata populations will be more tolerant to competition from the invader, and (iii) native populations that co-occur with high-sinigrin A. petiolata populations will be more fit when planted into invaded sites. Finally, the Geographic Mosaic Theory of Coevolution predicts that the degree of reciprocal selection will vary across landscapes, resulting in coevolutionary “hot” and “cold” spots (3, 34). To explore this possibility, I tested whether the direction or magnitude of selection on putative interaction traits in the native species varied across (i) natural gradients in the density of A. petiolata invasions and/or (ii) plots with or without experimental removal of A. petiolata.
Results
Do Traits of the Exotic A. petiolata Differ in Response to Interaction with Natives?
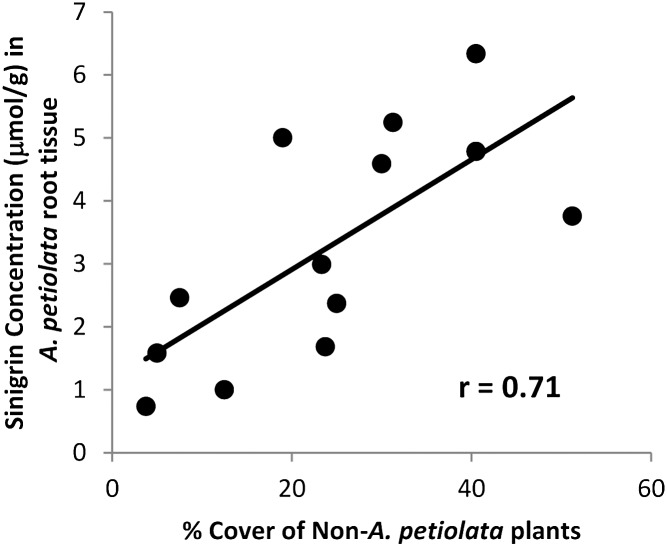
I evaluated root sinigrin concentrations for 13 populations of A. petiolata that came from areas differing in the relative abundance of heterospecific plant species. Replicate individuals from each population were grown in a common environment without any competition to isolate genetic differences in investment to sinigrin. Population mean-sinigrin concentration in the common environment was positively correlated to ground cover of heterospecific plants at the source site, consistent with an evolutionary response in A. petiolata to selection imposed by interspecific competition (r = 0.71, P = 0.007, n = 13) (Fig. 1). The relationship between heterospecific cover and sinigrin concentrations remained significant when controlling for latitude and longitude or soil abiotic conditions (via principal component analysis) using multiple regression.
Fig. 1.
Sinigrin concentration (μmol/g) measured from 5 to 10 individuals from 13 A. petiolata populations grown in a common greenhouse environment, regressed against the percentage of cover of heterospecific (non-A. petiolata) plants at the source site, as determined from 16 to 20 1-m2 plots.
Do the Traits of Native Populations Differ in Response to the Traits of the Invader?
I performed a greenhouse experiment with six populations of a common native understory forb, P. pumila, collected from five sites invaded by A. petiolata and by one uninvaded control. The A. petiolata populations at these sites varied in their sinigrin production and percentage of cover. P. pumila is an annual in the Urticaceae family, one of the most common members of the understory community in mesic deciduous forests, and commonly co-occurs with A. petiolata (R. A. Lankau, personal observation). Each of the six native populations was grown with or without A. petiolata in pots inoculated either with field soil from an uninvaded site containing an intact microbial community or with the same soils after heat sterilization. I then calculated the tolerance to competition, the net response of each population to soil biota, and the ability of each population to maintain AMF colonization in the presence of A. petiolata (see Materials and Methods for calculations).
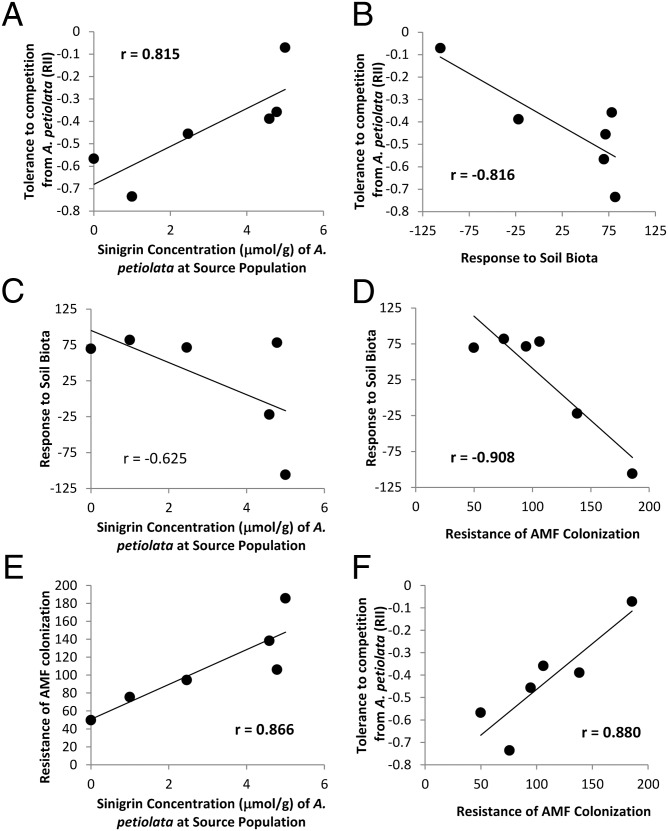
Native populations that co-occur with high-sinigrin populations of A. petiolata were more tolerant to competition from the invader, as evidenced by a significantly positive correlation between a native population’s tolerance to competition from A. petiolata and the sinigrin concentration of the A. petiolata population with which that native population co-occurs in nature (r = 0.815, P = 0.028, n = 6) (Fig. 2). Native populations varied in their response to soil communities in the absence of A. petiolata, with some populations benefiting from soil biota and others growing better in sterilized soil. This response was negatively correlated with their tolerance to competition, such that the native populations that preferred live soils were the worst competitors against A. petiolata (r = −0.816, P = 0.048, n = 6) (Fig. 2). Response to soil communities was not significantly correlated with the sinigrin concentration of the source invader population, although the direction was negative (r = −0.598, P = 0.217, n = 6,) (Fig. 2). Finally, native populations that co-occur with highly toxic A. petiolata were better able to maintain AMF colonization in the presence of the invader (colonization rates were equal or higher in the presence vs. the absence of A. petiolata), whereas those occurring with less toxic A. petiolata or from uninvaded sites lost up to 50% of their AMF colonization (r = 0.866, P = 0.026, n = 6) (Fig. 2). This resistance to loss of AMF colonization may explain the greater tolerance of some populations to competition from the invader, as these two measures were positively correlated (r = 0.881, P = 0.021, n = 6) (Fig. 2). However, resistance may come at a cost bacause less-resistant populations benefited more from growing in live vs. sterilized soil in the absence of A. petiolata (r = −0.908, P = 0.012, n = 6) (Fig. 2). None of the aspects of the P. pumila populations were significantly correlated with seed mass or initial seedling size, suggesting that these results were unlikely to originate from differences in maternal provisioning (SI Text).
Fig. 2.
Pairwise correlations between four aspects of the native P. pumila populations: (i) tolerance to competition from A. petiolata in the greenhouse experiment (A, B, F), (ii) response to soil biota in the absence of A. petiolata in the greenhouse experiment (B–D), (iii) the resistance of AMF colonization in the presence vs. the absence of A. petiolata (D–F), and (iv) the sinigrin concentration of the co-occurring A. petiolata population where native seeds were collected (A, C, E). P values determined by exact permutation tests.
Are Trait Distributions Adaptive When Native and Invasive Species Interact?
I performed a reciprocal transplant experiment with the six populations of P. pumila. Individuals from each of the six source populations were planted back into six invaded destination sites (the five invaded source populations as well as an additional invaded destination) in two types of plots: (i) into control plots with the ambient density of A. petiolata and (ii) into plots where all A. petiolata individuals were weeded out.
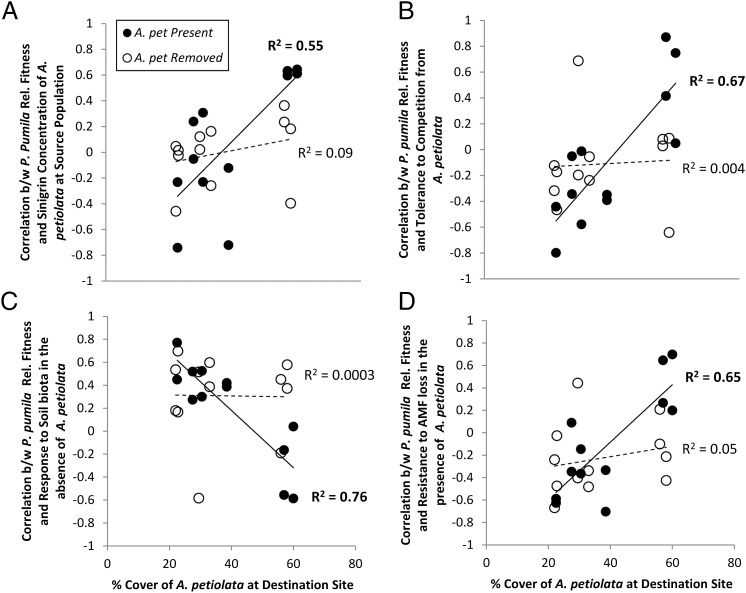
P. pumila populations that historically co-occurred with high-sinigrin A. petiolata tended to have the highest relative fitness in destination sites with a high abundance of the invader. However, this pattern reversed in sites where the invader abundance was low (Fig. 3A). These patterns were not present in plots where the invader was experimentally removed (Fig. 3B), resulting in a significant three-way interaction (F1,128 = 4.78, P = 0.031) (Table S1).
Fig. 3.
Visual representation of the significant three-way interactions in Table 1 between (i) the A. petiolata removal treatment, (ii) the percentage of cover of A. petiolata at destination sites, and (iii) one of four aspects of the P. pumila population. Each point in the graph represents a correlation coefficient, measuring the correlation between the relative fitness of the P. pumila populations and a particular trait or aspect of those populations (analagous to a selection differential). A separate correlation coefficient was calculated for each site, year, and treatment combination: each correlation coefficient was estimated from six underlying data points. For example, in B, each point represents the correlation between the relative fitness of the six native populations in a given site and the tolerance to competition from A. petiolata of each native population as measured in the greenhouse experiment. A positive value implies that populations that were highly tolerant to competition with A. petiolata had the highest relative fitness at that particular site, year, and treatment combination (i.e., would be favored by natural selection). These correlation coefficients were then regressed against the percentage of cover of A. petiolata at a destination site. A shift from negative to positive values across the gradient in percentage of cover of A. petiolata suggests that the selection pressures switch from favoring native populations with low competitive tolerance to those with high tolerance as the intensity of A. petiolata invasion increases. Each panel shows the regression of the correlations between P. pumila relative fitness and a different trait or aspect of those populations against A. petiolata percentage of cover at the destination site. The P. pumila population traits/aspects were the following: (A) the sinigrin concentration of its naturally co-occurring A. petiolata population, (B) the tolerance to competition from A. petiolata, (C) the response to soil biota in the absence of A. petiolata, and (D) the ability of a native population to retain AMF colonization in the presence of A. petiolata. Solid symbols (and solid lines) represent correlation coefficients regressed against A. petiolata cover in the presence of ambient A. petiolata densities, and open symbols (and dotted lines) represent correlation coefficients regressed against (initial) A. petiolata cover in the A. petiolata removal plots. R2 values refer to the variance among correlation coefficients explained by initial A. petiolata cover.
In addition, the three traits measured on these native populations in the greenhouse experiment (tolerance to competition from A. petiolata, resistance to loss of AMF colonization in the presence of A. petiolata, and response to soil biota in the absence of A. petiolata) were all significantly correlated with relative fitness in the field. Again, the effect of these traits on fitness depended on the abundance of A. petiolata at a site. Native populations that were highly tolerant of competition from A. petiolata in the greenhouse had the highest fitness in sites where A. petiolata was abundant. However, in the site with the lowest A. petiolata abundance, this pattern reversed, suggesting that competitive ability against the invader may come at some cost (Fig. 3B). Similarly, the native populations most able to retain AMF colonization in the presence of A. petiolata in the greenhouse experiment had the highest fitness in heavily invaded sites, but again had lower fitness when the invader was less common. Conversely, native populations that benefited the most from living, uninvaded soil communities in the greenhouse had the highest fitness in sites with low A. petiolata abundance, but the lowest fitness in heavily invaded sites (Fig. 3D). As before, these patterns were not present in plots where A. petiolata was removed, resulting in significant three-way interactions (F1,128 > 5.72, P < 0.02 for all) (Table S1). When A. petiolata was experimentally removed, selection tended to favor native populations that benefited more from live, uninvaded soils (average of all correlations = 0.30 ± 0.24, 95% confidence interval (CI), n = 12) (Fig. 3C) and were less resistant to losses of AMF (average of correlations = −0.23 ± 0.20, 95% CI, n = 12) (Fig. 3D). None of these results changed qualitatively when seed mass or initial seedling size was included in the ANCOVA models. Additionally, neither the sinigrin concentration nor the abundance of the A. petiolata populations correlated significantly with latitude, longitude, or any of 10 soil abiotic metrics, suggesting that the pattern of coevolution was not the result of confounding environmental variables (SI Text).
Discussion
A. petiolata is one of the most aggressive invaders of forest understories, and like many exotic invasive species, appears to gain competitive advantages due to its evolutionary novelty in North America (37). In particular, A. petiolata produces secondary compounds novel to its introduced range that inhibit competitors by interfering with mycorrhizal mutualisms (38). Here, I found that A. petiolata populations that co-occur with a high density of heterospecific competitors invest more in an allelopathic secondary compound, suggesting that this invader has responded evolutionarily to selection imposed by interspecific competition. Additionally, native P. pumila populations that currently co-occur with A. petiolata populations producing high levels of the allelochemical possess some trait(s) that allow for greater competitive ability against the invader (likely involving interactions with mycorrhizal fungi), suggesting that native traits respond evolutionarily to the traits of the invader. Finally, these traits of native populations translated into higher fitness in heavily invaded sites in the field, but were costly in less heavily invaded sites. Together, these results suggest that A. petiolata has entered into a coevolutionary relationship with at least one of its new competitors.
Native populations that co-occur with highly toxic A. petiolata populations and were the most tolerant to competition from the invader had the highest fitness in sites with the most intense invasion. However, these same populations had the lowest fitness when the invader was less common, suggesting a possible cost to these traits. This cost may be related to interactions with soil biota because tolerance to competition from A. petiolata was negatively correlated to a native population’s dependence on soil biota in the absence of the invader. Tolerant native populations tended to have net neutral or negative interactions with an uninvaded soil community, whereas native populations sensitive to competition from the invader responded positively to this same soil community. Populations with neutral or negative responses to native soil biota tended to have the highest fitness in sites with the densest A. petiolata invasion; however, in sites with light invasions, native populations that benefited from soil biota were the most successful. Because A. petiolata is known to have particularly strong impacts on mycorrhizal fungal communities (38, 39, 45), one possible explanation for these patterns is that native populations adapt to the presence of high-sinigrin A. petiolata in part by altering their interactions with mycorrhizal fungi. Native populations with a history of interaction with high-sinigrin A. petiolata, and that were more tolerant to competition from the invader, were better able to maintain AMF colonization in their roots when competing with A. petiolata. In this greenhouse experiment, all plants were grown with the same soil inoculation; therefore, differences in AMF colonization reflect genetic differences among plant populations, rather than differences in the pool of available AMF taxa. The resistance to AMF loss could partially explain the greater competitive tolerance and higher fitness in heavily invaded sites of these populations. However, this trait was also costly: in the absence of A. petiolata, the “resistant” populations gained less benefit from the presence of an intact, uninvaded soil community and had lower relative fitness in the field when A. petiolata was experimentally removed or naturally at low abundance. One potential explanation for the patterns could be that native populations differ in their “choosiness” for AMF partners. AMF species can vary widely in their fitness impact on their plant host (46), and plants may be able to preferentially reward the most beneficial partners (47, 48). When growing with a diverse AMF community in uninvaded soils, native plants may be selected to interact only with the AMF species that optimize their fitness. However, when AMF communities are degraded by A. petiolata, the natives may be selected to make use of whatever AMF strains are available. Some evidence suggests that the AMF taxa most resistant to A. petiolata’s allelochemicals tend to be less beneficial to native plant growth (49). Thus, reduced selectivity could benefit native plants when A. petiolata density is high (and thus AMF abundance/diversity is low), but be detrimental when A. petiolata density is low and AMF abundance/diversity is high.
The covariation between sinigrin concentration among A. petiolata populations and competitive ability among P. pumila populations suggests a pattern of phenotype matching, which is often taken as evidence for ongoing coevolution (3, 50), but must be interpreted with caution (34, 35). This interpretation is strengthened in this study by experimental reciprocal transplants of one of the interacting species, which confirmed that the P. pumila traits that covary with A. petiolata’s sinigrin production were adaptive in heavily invaded sites. Phenotype matching has previously been shown in very specialized systems involving consumer–resource interactions (4). Competitive interactions are rarely so tightly linked, especially in plants, and thus interactions between any two species have been considered too inconsistent to lead to specific evolutionary responses (16). Instead, plants are expected to respond to the average community, rather than to particular species. In this case, it is not clear if A. petiolata is evolving in response to particular competitor species or to the net selection imposed by interspecific competition. However, at least one common native species appears to be evolving specifically in response to A. petiolata. Exotic plant invaders are often extremely abundant and so may be especially likely to create the kind of consistency in selection pressures that can lead to specific evolutionary responses in natives. On the other hand, the exotic species themselves are likely to respond to selection from the most common native species, or to the net effect of the native community, depending on the specificity of competitive interactions.
Because this study (like most studies of adaptation and coevolution) is primarily correlative, it is possible that these patterns arose due to confounding factors rather than direct interactions between these species. Including data on geographic location and soil abiotic conditions did not change any results. Spatial gradients in other biotic interactions, such as herbivory, were not recorded and may play a role if they are (i) positively correlated with the gradients of interest (heterospecific cover and A. petiolata sinigrin concentration) and (ii) tend to select for similar traits (root sinigrin concentration in A. petiolata and tolerance to competition and response to soil biota in P. pumila).
Coevolution likely does not proceed in lockstep across all of the populations of a species, due to differences among populations in the degree of reciprocal selection, the strength and shape of selection, and gene flow among populations (3, 34). Such processes are likely at work in this system as well. For example, because A. petiolata is still increasing its range in North America, there remain populations of P. pumila where A. petiolata has yet to invade, which will be coevolutionary “cold spots” by definition. Additionally, the results of this study suggest that selection on P. pumila by A. petiolata varies with the local density of the invader, likely leading to variation in reciprocal selection. Finally, although the spatial scale from which the six populations studied likely precluded much direct gene flow between them, variation in the presence/absence or density of both of these species can vary across small spatial scales, raising the possibility that gene flow from coevolutionary “cold spots” could interfere with the response to reciprocal selection in “hot spots.”
Many theories of invasive success posit that exotic species gain ecological advantages due to their lack of coevolutionary history with the native community, for example, benefiting from enemy release because native consumers lack the necessary traits to efficiently use the new species (26). This idea has been considered especially important for invasive plants that produce secondary compounds that are novel to native plant, insect, and microbial communities (25). However, novelty cannot last forever, and the high invader abundance created by these evolutionary mismatches may in turn lead to the development of new coevolutionary relationships that, over time, act to integrate exotic species into native communities.
Materials and Methods
A. petiolata Sinigrin Concentration.
Seeds from 13 populations of A. petiolata were collected from 2007 to 2009 (see SI Text for locations). Six of these sites were used for the reciprocal transplant experiment with P. pumila; the other seven were used for a previous field experiment (44). One individual plant from 5 (7 previously studied populations) or 10 (6 current populations) maternal families was grown without competition in a common greenhouse environment for 3 mo, at which point ∼10–20 mg of fine-root material was collected. Glucosinolates were extracted and quantified with HPLC according to established methods (51). Percentage of cover of A. petiolata, all other plants, and bare ground was estimated from 16 to 20 1-m2 plots at each site in 2008 or 2010.
Experimental Tests of Tolerance to Competition and Response to Soil Communities.
P. pumila seeds were collected from six sites in 2009, five sites where A. petiolata seeds were also collected, and one site where A. petiolata has never occurred due to vigilant management (S. Buck, personal communication). Individuals from six of the P. pumila populations were used in a greenhouse competition experiment. Each native plant was grown with an A. petiolata individual or alone in live or sterilized soil. The A. petiolata plants came from two populations, with the representation of each population balanced with respect to P. pumila populations and soil treatments. Each competitive treatment (A. petiolata from population 1, A. petiolata from population 2, or no competitor) by soil (live vs. sterilized) treatment was replicated six times per native population for a total of 216 pots. Plants were grown in 0.5-L pots inoculated with 20 mL of live soil collected from the uninvaded site or an autoclaved sample of that same soil. Soil was autoclaved twice for 2 h each round, with a 24-h cooling period between rounds. Greenhouse temperatures averaged ∼29 °C during the experiment. Day length was unmanipulated (∼16 h/day), but shade cloths were used to reduce light by 60% to better approximate forest understory conditions. After 4 mo of growth, the above- and below-ground biomass was collected, dried, and weighed. A subsample of roots was taken from each plant in the live soil treatment for mycorrhizal colonization. Roots were cleared with hot 10% (wt/vol) KOH for 7 min, bleached with 3% (vol/vol) H2O2 for 30 min, acidified with 1% (vol/vol) HCL for 60 min, and then stained with Direct Blue (Acros Organics). Stained roots were mounted on slides and fungal colonization was scored using the grid-line intersect method (52). At least 50 root intersections were scored per plant, and the presence of arbuscules, vesicles, and hyphae was recorded at each intersection.
I calculated tolerance to competition for each native population using the relative interaction intensity (RII) (53). RII is calculated as the difference in the mean biomass between a population grown alone vs. grown with a competitor, divided by the sum of the mean biomass alone and with a competitor. Native plants growing with the two A. petiolata populations were pooled. I also calculated the net response to the soil biota in the absence of A. petiolata as the percentage of decline in biomass in sterilized soil, using the following equation:
 |
Only pots without competitors were used to calculate the response to soil biota. I calculated the percentage of AMF colonization as the sum of root intersections containing AMF structures (arbuscules, vesicles, and/or hyphae) divided by the total number of intersections scored. I calculated the ability of a population to resist changes in AMF colonization in the following way:
 |
All populations grew with the same uninvaded soil inoculation. Thus, differences among populations in their ability to retain AMF colonization in the face of A. petiolata reflect genetic differences in the plants, rather than differences in the AMF species/strains present in the soil.
Native Species Reciprocal Transplant Experiment.
Two P. pumila individuals from each of 10 maternal families per population were transplanted as newly germinated seedlings into each of the six invaded sites in May 2010 (SI Text). One individual per family was planted into a 1-m2 plot in which all naturally occurring A. petiolata plants were weeded out, while the other individual was planted in a control plot with no weeding. Each plot received an individual from each of the six populations, for a total of 10 weeded and 10 unweeded plots per site. Plants were surveyed once at midsummer, and all above-ground biomass was collected in September. A new set of seedlings was planted in 2011. With six sites, this yielded a total of 720 plants in each year. Individuals that did not survive to the first census were removed from the analyses to avoid analyzing mortality due to transplant shock. Individuals surviving at the first census but not to the final collection were considered to have 0 fitness. I calculated relative fitness for each population separately for each weeding treatment in each site in each year by first averaging the biomass of all of the replicates of a given population and then dividing that population mean by the grand mean for all populations in that weeding treatment in that year at that site. Calculating relative fitness allows me to compare differences in the performance of populations across sites and treatments while controlling for overall differences in the quality of sites for P. pumila growth (due to differences in canopy openness, soil fertility, etc.).
Statistical Analysis.
Population mean-sinigrin concentration in A. petiolata root tissue was compared with the percentage of cover of heterospecific plants using the Pearson product-moment correlation. Correlations were also calculated between the tolerance to competition, response to soil biota, and resistance of AMF colonization of the P. pumila populations and between these measures and the sinigrin concentration of the A. petiolata population with which the P. pumila population naturally co-occurs. Because these last six correlations are based on only six points (population is the unit of replication), exact permutation tests were used to determine significance. Relative fitness from the field reciprocal transplant experiment was analyzed using ANCOVA, with a model that included year and weeding treatment as fixed categorical factors and the percentage of cover of A. petiolata at the destination site and one of four quantitative aspects of the source population as covariates, with all interactions. Again, source population was the unit of replication; individuals within a population were averaged for each site by treatment by year combination before analysis. I did this, rather than using an individual as the replicate with a random effect of population, so that I could treat population mean trait values as continuous variables. Because fitness was relativized within each site by treatment by year combination, the main effects of these three variables (and their interactions with each other) were constrained to be nonsignificant. However, these variables could show significant interactions with the various aspects of the source populations. The four aspects of the source populations tested were (i) the sinigrin concentration of the A. petiolata population with which the native population originally co-occurred, (ii) the tolerance to competition from A. petiolata measured from the greenhouse experiment, (iii) the net response to soil biota measured from the greenhouse experiment, and (iv) the resistance of AMF colonization to A. petiolata measured from the greenhouse experiment. Including these four aspects of the source populations as covariates tests (i) whether P. pumila populations that co-occur with high sinigrin A. petiolata populations possess traits that lead to high fitness when grown with A. petiolata in the field, (ii) whether P. pumila populations that display high tolerance to A. petiolata in controlled greenhouse conditions have high fitness when grown with A. petiolata in the field, (iii) whether P. pumila populations that have generally positive or negative responses to soil biota in the absence of A. petiolata in the greenhouse experiment had higher or lower fitness when grown with A. petiolata in the field, and (iv) whether P. pumila populations that were best able to maintain AMF colonization in the presence of A. petiolata in the greenhouse experiment had higher or lower fitness when planted with A. petiolata in the field. The interaction of these aspects with the weeding treatment tests whether the fitness consequences of any genetic differences between populations are only evident when the invader is actively present. Finally, the interaction of these aspects with the cover of A. petiolata at the destination sites tests whether the fitness consequences of any genetic differences between populations depend on the intensity of the invasion at a site.
To help visualize the potential higher-order interactions in the ANCOVA models, I also calculated the correlation between P. pumila relative fitness and each of the four population attributes/traits for each site by treatment by year combination. I then plotted those correlations (analogous to selection differentials if you consider the population attributes as traits) against the percentage of cover of A. petiolata at the destination sites separately for the A. petiolata present and removal treatments (percentage of cover of A. petiolata was measured before weeding). The regression line in these plots provides an additional test of whether selection varies significantly with initial A. petiolata percentage of cover. I also calculated the mean and 95% CI for correlations across all sites in the A. petiolata present or removal treatments to test whether selection consistently favors higher (mean > 0) or lower (mean < 0) levels of a trait in the two conditions.
Supplementary Material
Acknowledgments
I thank Rachel Nodurft, Charlene Chalmers, Dalya Abou-El-Seoud, and Diane Mohr for assistance with field and greenhouse experiments. Emily Lankau and Daniel Keymer provided comments on the manuscript. This work was funded by National Science Foundation Division of Environmental Biology Grant 0918450 to R.A.L.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201343109/-/DCSupplemental.
References
- 1.Ehrlich P, Raven P. Butterflies and plants: A study in coevolution. Evolution. 1964;18:586–608. [Google Scholar]
- 2.Janzen DH. When is it coevolution? Evolution. 1980;34:611–612. doi: 10.1111/j.1558-5646.1980.tb04849.x. [DOI] [PubMed] [Google Scholar]
- 3.Thompson JN. The Geographic Mosaic of Coevolution. Chicago: University of Chicago Press; 2005. [Google Scholar]
- 4.Thompson JN. The coevolving web of life. Am Nat. 2009;173:125–140. doi: 10.1086/595752. [DOI] [PubMed] [Google Scholar]
- 5.Thompson JN, Cunningham BM. Geographic structure and dynamics of coevolutionary selection. Nature. 2002;417:735–738. doi: 10.1038/nature00810. [DOI] [PubMed] [Google Scholar]
- 6.Bascompte J, Jordano P, Olesen JM. Asymmetric coevolutionary networks facilitate biodiversity maintenance. Science. 2006;312:431–433. doi: 10.1126/science.1123412. [DOI] [PubMed] [Google Scholar]
- 7.Zangerl AR, Berenbaum MR. Increase in toxicity of an invasive weed after reassociation with its coevolved herbivore. Proc Natl Acad Sci USA. 2005;102:15529–15532. doi: 10.1073/pnas.0507805102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burdon JJ, Thrall PH. Coevolution of plants and their pathogens in natural habitats. Science. 2009;324:755–756. doi: 10.1126/science.1171663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gómez P, Buckling A. Bacteria-phage antagonistic coevolution in soil. Science. 2011;332:106–109. doi: 10.1126/science.1198767. [DOI] [PubMed] [Google Scholar]
- 10.Decaestecker E, et al. Host-parasite ‘Red Queen’ dynamics archived in pond sediment. Nature. 2007;450:870–873. doi: 10.1038/nature06291. [DOI] [PubMed] [Google Scholar]
- 11.Iwao K, Rausher MD. Evolution of plant resistance to multiple herbivores: Quantifying diffuse coevolution. Am Nat. 1997;149:316–335. [Google Scholar]
- 12.Dayan T, Simberloff D. Ecological and community-wide character displacement: The next generation. Ecol Lett. 2005;8:875–894. [Google Scholar]
- 13.Grant PR, Grant BR. Evolution of character displacement in Darwin’s finches. Science. 2006;313:224–226. doi: 10.1126/science.1128374. [DOI] [PubMed] [Google Scholar]
- 14.Chesson P. Mechanisms of maintenance of species diversity. Annu Rev Ecol Syst. 2000;31:343–366. [Google Scholar]
- 15.Hubbell SP. The Unified Neutral Theory of Biodiversity and Biogeography. Princeton, NJ: Princeton University Press; 2001. [Google Scholar]
- 16.Thorpe AS, Aschehoug ET, Atwater DZ, Callaway RM. Interactions among plants and evolution. J Ecol. 2011;99:729–740. [Google Scholar]
- 17.Aarssen LW, Turkington R. Biotic specialization between neighboring genotypes in Lolium perenne and Trifolium repens from a permanent pasture. J Ecol. 1985;73:605–614. [Google Scholar]
- 18.Turkington R. The growth, distribution and neighbor relationships of Trifolium repens in a permanent pasture. 5. The coevolution of competitors. J Ecol. 1989;77:717–733. [Google Scholar]
- 19.Chanway CP, Holl FB, Turkington R. Effect of Rhizobium leguminosarum biovar trifolii genotype on specificity between Trifolium repens and Lolium perenne. J Ecol. 1989;77:1150–1160. [Google Scholar]
- 20.Ehlers BK, Thompson J. Do co-occurring plant species adapt to one another? The response of Bromus erectus to the presence of different Thymus vulgaris chemotypes. Oecologia. 2004;141:511–518. doi: 10.1007/s00442-004-1663-7. [DOI] [PubMed] [Google Scholar]
- 21.Grondahl E, Ehlers BK. Local adaptation to biotic factors: Reciprocal transplants of four species associated with aromatic Thymus pulegioides and T. serpyllum. J Ecol. 2008;96:981–992. [Google Scholar]
- 22.Jensen CG, Ehlers BK. Genetic variation for sensitivity to a thyme monoterpene in associated plant species. Oecologia. 2010;162:1017–1025. doi: 10.1007/s00442-009-1501-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ehlers BK. Soil microorganisms alleviate the allelochemical effects of a thyme monoterpene on the performance of an associated grass species. PLoS ONE. 2011;6:e26321. doi: 10.1371/journal.pone.0026321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pimentel D, Zuniga R, Morrison D. Update on the environmental and economic costs associated with alien-invasive species in the United States. Ecol Econ. 2005;52:273–288. [Google Scholar]
- 25.Callaway RM, Ridenour WM. Novel weapons: A biochemically based hypothesis for invasive success and the evolution of increased competitive ability. Front Ecol Environ. 2004;2:436–443. [Google Scholar]
- 26.Hallett SG. Dislocation from coevolved relationships: A unifying theory for plant invasion and naturalization? Weed Sci. 2006;54:282–290. [Google Scholar]
- 27.Leger EA, Espeland EK. Coevolution between native and invasive plant competitors: Implications for invasive species management. Evolutionary Applications. 2010;3:169–178. doi: 10.1111/j.1752-4571.2009.00105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prentis PJ, Wilson JRU, Dormontt EE, Richardson DM, Lowe AJ. Adaptive evolution in invasive species. Trends Plant Sci. 2008;13:288–294. doi: 10.1016/j.tplants.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 29.Strauss SY, Lau JA, Carroll SP. Evolutionary responses of natives to introduced species: What do introductions tell us about natural communities? Ecol Lett. 2006;9:357–374. doi: 10.1111/j.1461-0248.2005.00874.x. [DOI] [PubMed] [Google Scholar]
- 30.Goergen EM, Leger EA, Espeland EK. Native perennial grasses show evolutionary response to Bromus tectorum (cheatgrass) invasion. PLoS ONE. 2011;6:e18145. doi: 10.1371/journal.pone.0018145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mealor BA, Hild AL. Potential selection in native grass populations by exotic invasion. Mol Ecol. 2006;15:2291–2300. doi: 10.1111/j.1365-294X.2006.02931.x. [DOI] [PubMed] [Google Scholar]
- 32.Mealor BA, Hild AL. Post-invasion evolution of native plant populations: A test of biological resilience. Oikos. 2007;116:1493–1500. [Google Scholar]
- 33.Callaway RM, Ridenour WM, Laboski T, Weir T, Vivanco JM. Natural selection for resistance to the allelopathic effects of invasive plants. J Ecol. 2005;93:576–583. [Google Scholar]
- 34.Gomulkiewicz R, et al. Dos and don’ts of testing the geographic mosaic theory of coevolution. Heredity (Edinb) 2007;98:249–258. doi: 10.1038/sj.hdy.6800949. [DOI] [PubMed] [Google Scholar]
- 35.Nuismer SL, Gomulkiewicz R, Ridenhour BJ. When is correlation coevolution? Am Nat. 2010;175:525–537. doi: 10.1086/651591. [DOI] [PubMed] [Google Scholar]
- 36.Rodgers VL, Stinson KA, Finzi AC. Ready or not, garlic mustard is moving in: Alliaria petiolata as a member of eastern North American forests. Bioscience. 2008;58:426–436. [Google Scholar]
- 37.Barto EK, Powell JR, Cipollini D. How novel are the chemical weapons of garlic mustard in North American forest understories? Biol Invasions. 2010;12:3465–3471. [Google Scholar]
- 38.Callaway RM, et al. Novel weapons: Invasive plant suppresses fungal mutualists in America but not in its native Europe. Ecology. 2008;89:1043–1055. doi: 10.1890/07-0370.1. [DOI] [PubMed] [Google Scholar]
- 39.Stinson KA, et al. Invasive plant suppresses the growth of native tree seedlings by disrupting belowground mutualisms. PLoS Biol. 2006;4:e140. doi: 10.1371/journal.pbio.0040140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schreiner RP, Koide RT. Mustards, mustard oils and mycorrhizas. New Phytol. 1993;123:107–113. [Google Scholar]
- 41.Lankau RA, Nuzzo V, Spyreas G, Davis AS. Evolutionary limits ameliorate the negative impact of an invasive plant. Proc Natl Acad Sci USA. 2009;106:15362–15367. doi: 10.1073/pnas.0905446106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lankau RA, Strauss SY. Mutual feedbacks maintain both genetic and species diversity in a plant community. Science. 2007;317:1561–1563. doi: 10.1126/science.1147455. [DOI] [PubMed] [Google Scholar]
- 43.Lankau RA. Intraspecific variation in allelochemistry determines an invasive species’ impact on soil microbial communities. Oecologia. 2011;165:453–463. doi: 10.1007/s00442-010-1736-8. [DOI] [PubMed] [Google Scholar]
- 44.Lankau RA. Interpopulation variation in allelopathic traits informs restoration of invaded landscapes. Evolutionary Applications. 2012;5:270–282. doi: 10.1111/j.1752-4571.2011.00218.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barto EK, et al. Differences in arbuscular mycorrhizal fungal communities associated with sugar maple seedlings in and outside of invaded garlic mustard forest patches. Biol Invasions. 2011;13:2755–2762. [Google Scholar]
- 46.Klironomos JN. Variation in plant response to native and exotic arbuscular mycorrhizal fungi. Ecology. 2003;84:2292–2301. [Google Scholar]
- 47.Bever JD, Richardson SC, Lawrence BM, Holmes J, Watson M. Preferential allocation to beneficial symbiont with spatial structure maintains mycorrhizal mutualism. Ecol Lett. 2009;12:13–21. doi: 10.1111/j.1461-0248.2008.01254.x. [DOI] [PubMed] [Google Scholar]
- 48.Kiers ET, et al. Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science. 2011;333:880–882. doi: 10.1126/science.1208473. [DOI] [PubMed] [Google Scholar]
- 49.Lankau RA. Intraspecific variation in allelochemistry determines an invasive species’ impact on soil microbial communities. Oecologia. 2011;165:453–463. doi: 10.1007/s00442-010-1736-8. [DOI] [PubMed] [Google Scholar]
- 50.Berenbaum MR, Zangerl AR. Chemical phenotype matching between a plant and its insect herbivore. Proc Natl Acad Sci USA. 1998;95:13743–13748. doi: 10.1073/pnas.95.23.13743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lankau RA, Kliebenstein DJ. Competition, herbivory, and genetics interact to determine the accumulation and fitness consequences of a defensive metabolite. J Ecol. 2009;97:78–88. [Google Scholar]
- 52.McGonigle TP, Miller MH, Evans DG, Fairchild GL, Swan JA. A new method which gives an objective measure of colonization of roots by vesicular arbuscular mycorrhizal fungi. New Phytol. 1990;115:495–501. doi: 10.1111/j.1469-8137.1990.tb00476.x. [DOI] [PubMed] [Google Scholar]
- 53.Weigelt A, Jolliffe P. Indices of plant competition. J Ecol. 2003;91:707–720. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.