Abstract
Type IV secretion (T4S) systems mediate the transfer of proteins and DNA across the cell envelope of bacteria. These systems play important roles in bacterial pathogenesis and in horizontal transfer of antibiotic resistance. The VirB4 ATPase of the T4S system is essential for both the assembly of the system and substrate transfer. In this article, we present the crystal structure of the C-terminal domain of Thermoanaerobacter pseudethanolicus VirB4. This structure is strikingly similar to that of another T4S ATPase, VirD4, a protein that shares only 12% sequence identity with VirB4. The VirB4 domain purifies as a monomer, but the full-length protein is observed in a monomer-dimer equilibrium, even in the presence of nucleotides and DNAs. We also report the negative stain electron microscopy structure of the core complex of the T4S system of the Escherichia coli pKM101 plasmid, with VirB4 bound. In this structure, VirB4 is also monomeric and bound through its N-terminal domain to the core’s VirB9 protein. Remarkably, VirB4 is observed bound to the side of the complex where it is ideally placed to play its known regulatory role in substrate transfer.
Keywords: macromolecular assembly, structural biology, conjugation
Type IV secretion (T4S) systems are large, multicomponent secretion machines that are found in both Gram-positive and Gram-negative bacteria as well as in some archaea. The T4S system is responsible for horizontal transfer of plasmid DNA between bacteria during conjugation, thereby contributing to the spread of antibiotic resistance genes in healthcare settings (1, 2). T4S systems are also found in pathogens, where they mediate the translocation of effector proteins from pathogen to host cell: for example, in Helicobacter pylori, the causing agent of peptic ulcer disease and related cancers (3). Other important human pathogens that rely on T4S systems are Bordetella pertussis, Legionella pneumophila, Brucella spp., and Bartonella spp. Recently a T4S system in Streptococcus suis was shown to be important for virulence (4).
In Gram-negative bacteria the T4S system span both the inner and outer membranes and are composed of at least 12 components, termed VirB1–VirB11 and VirD4 proteins, after the nomenclature of the Agrobacterium tumefaciens VirB/D T4S system (5). Fourteen copies of three proteins, VirB7, VirB9, VirB10, form the so-called “core” complex, a stable 1.05 MDa complex spanning the two membranes (6). The core complex is composed of two layers, termed I and O layers, linked by thin threads of electron density, each layer inserting in the inner and outer membranes, respectively (Fig. S1A). A more detailed view of the O layer was revealed by the crystal structure of this region, uncovering that the VirB10 component traverses not only the inner but also the outer membrane, forming an α-helical outer membrane pore (7). A pilus is often present and composed of a major and a minor component, VirB2 (8) and VirB5 (9), respectively. VirB5 is thought to act as an adhesin (9). VirB1 and VirB8 are thought to function in assembly and VirB3 and VirB6 are inner-membrane proteins with yet unknown roles (10). Three cytoplasmic ATPases, VirB4, VirB11, and VirD4, energize the system and each of them is essential for substrate transfer. VirD4, also termed “coupling protein,” recruits T4S system substrates to the VirB1–11 secretion channel. Crystal structures of hexameric forms of homologs of both VirD4 and VirB11 have been solved (11–13).
The VirB4 ATPase is the most conserved T4S system component: it is vital for pilus biogenesis (14), substrate transfer, and virulence (15–17). VirB4 proteins are between 600 and 900 residues, often forming two distinct domains (Fig. S1B). The C-terminal domain (CTD) is about 400 residues and contains the ATPase activity. This domain is the most evolutionary conserved part of VirB4 proteins and contains the catalytically essential Walker A and Walker B motifs, characteristic of proteins with RecA-like catalytic domains. A homology model of the VirB4 CTD from A. tumefaciens was proposed (18), based on a structure of the CTD of TrwB, the VirD4 protein of the Escherichia coli R388 plasmid. The less conserved N-terminal domain (NTD) is generally believed to form the interaction with the membrane with one to three predicted transmembrane regions, depending on species. The oligomerization state of VirB4 proteins and their localization within the cell are unclear: monomeric, dimeric, trimeric, and hexameric forms have been reported and VirB4 proteins have been purified successfully from cytoplasmic and membrane fractions (19–25).
Here we present two structures: (i) the crystal structure of the VirB4 CTD from the Gram-positive bacterium Thermoanaerobacter pseudethanolicus (TpsVirB4CTD) and (ii) the negative stain electron microscopy (EM) structure of the VirB4 protein from the T4S system encoded by the E. coli pKM101 plasmid (TraB, referred to hereafter as pkVirB4) bound to the corresponding VirB7/VirB9/VirB10 (referred to thereafter as pkVirB7/pkVirB9/pkVirB10) core complex. TpsVirB4CTD purifies as a monomer. The structure of TpsVirB4CTD demonstrates that VirB4CTD is structurally similar to VirD4CTD. From the structural comparison between TpsVirB4CTD and VirD4CTD, we identify in TpsVirB4 and pkVirB4 a conserved arginine finger at the surface of the protein far from its ATP-binding site, suggesting that both proteins oligomerize to form a functional ATP-binding site. Mass spectrometry experiments on both full-length TpsVirB4 and pkVirB4 indeed reveal dimeric forms in addition to monomeric forms. The EM structure of the core complex bound to pkVirB4 unexpectedly reveal an interaction between a monomeric pkVirB4 molecule and the outer wall of the core complex I layer, the NTD of pkVirB4 and pkVirB9 forming a substantial part of the interface. This side-on interaction provides a rational for the essential regulatory function that VirB4 plays in T4S.
Results and Discussion
Structure of TpsVirB4CTD.
Because we failed to crystallize a number of mesophilic VirB4 homologs and fragments, we searched the National Center for Biotechnology Information microbial genome database for VirB4 homologs of thermophilic origin. We identified a VirB4 homolog of 594 residues from the Gram-positive bacterium, T. pseudethanolicus (GeneBank accession no. ABY93733). TpsVirB4 shows ∼30% sequence identity to known Gram-positive VirB4 proteins, such as those found in the broad host-range plasmid pIP501 (26) and Streptococcus aureus plasmid pGO1 (27). The TpsVirB4 gene is located on the chromosome with homologs of VirB11 and VirD4 as neighboring genes. TpsVirB4 shows 20–25% sequence identity to pkVirB4 and other known Gram-negative T4S VirB4 proteins, and is resembling F-like conjugative T4S system VirB4 proteins (28) such as TraC of the E. coli plasmid K12 (29).
A construct of the near full-length TpsVirB4 (residues 10–594), was expressed in E. coli and purified (Fig. S1C). The protein did not crystallize and was therefore subjected to limited proteolysis with trypsin to identify a fragment amenable to crystallographic studies. A mixture of two fragments [residues 190–594 and residues 203–594; also confirmed by mass spectrometry (see Fig. 2A)] was obtained that yielded crystals diffracting to high resolution (Fig. S1C). These fragments encompass the entire CTD of TpsVirB4 (TpsVirB4CTD). The structure of TpsVirB4CTD was solved using single anomalous diffraction phasing to 2.35 Å (Table S1). Subsequently, a complex with bound ADP and Mg2+ was solved to 2.45 Å (Table S1). All crystals contain four molecules in the asymmetric unit.
Fig. 2.
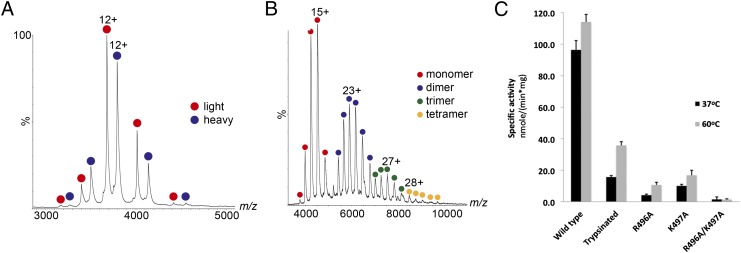
Investigation of the oligomeric state of TpsVirB4 and TpsVirB4CTD. Mass spectrometry analysis of TpsVirB4CTD (A) and TpsVirB4 (B). (A) Mass spectrum showing two dominant products of the TpsVirB4CTD protein after limited proteolysis by trypsin. Using electrospray, multiple charged ions are generated for each of the components present in solution. This result is because of the many different protonation sites being available for each protein. The charge states series corresponding to the lighter and heavier TpsVirB4CTD monomers are indicated in red and blue, respectively. (B) Mass spectrum showing the different oligomerization states of the TpsVirB4 protein. Charge state series corresponding to monomer, dimer, trimer, and tetramer of the protein are indicated in red, blue, green, and yellow, respectively. (C) ATPase activities of TpsVirB4 and its motif D mutants at two different temperatures, 37 °C (black) and 60 °C (gray). From left: wild-type TpsVirB4, TpsVirB4CTD, TpsVirB4R496A, TpsVirB4K497A, and TpsVirB4R496AK497A.
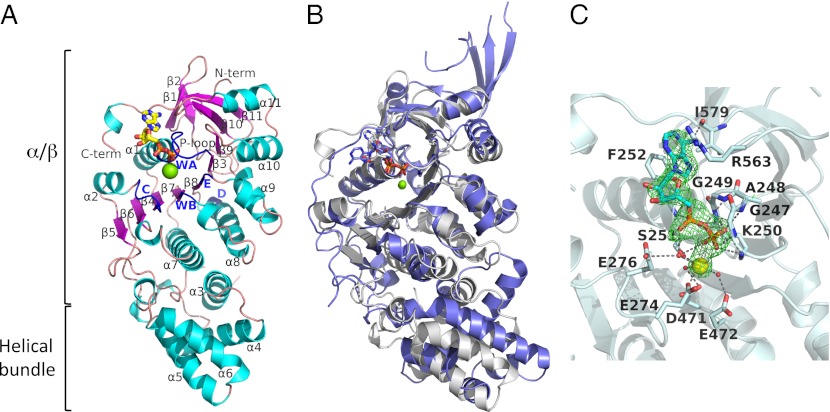
The structure of TpsVirB4CTD can be subdivided into two domains, one RecA-like α/β domain that contains the ATP-binding site and one four-helix bundle domain (Fig. 1A and Fig. S2A). The RecA-like α-/β-domain contains a large parallel β-sheet comprising β-strands β3−β9 sandwiched between helixes α1, α2, and α7−α11, which extends in a sharp turn with four additional antiparallel β-strands, β10, β11, β1, and β2. The four-helix bundle domain is composed of helices α3–α6 (Fig. 1A). A DALI search identified the C-terminal domain of TrwB, the hexameric VirD4 homolog of the T4S system encoded by the E. coli plasmid R388, as the closest structural homolog of TpsVirB4CTD (RMSD in Cα atoms of 3.5 Å) (Fig. 1B and Fig. S2B), despite a shared sequence identity of only 12% over the structurally aligned residues (Fig. S3A).
Fig. 1.
Structure of TpsVirB4CTD. (A) Ribbon representation of the structure of TpsVirB4CTD, covering residues 205–587. Secondary structure elements, β-strands and α-helixes, are marked β1–β11 and α1–11, respectively. Walker A (WA) and B (WB) motifs and motifs C, D, and E are colored in blue. (B) Superposition of TpsVirB4CTD (white) and TrwBCTD (slate; PDB code 1GKI). ADP molecules and Mg2+ from each structure are shown. (C) Nucleotide-binding site of TpsVirB4CTD. An OMIT Fo-Fc map is shown for the product ADP (green) and the catalytic Mg2+ (yellow) contoured at 3σ. Water molecules are shown as red balls. Oxygen is shown in red, nitrogen in blue, and phosphorus in orange. Residues involved in interactions with the nucleotide are in stick representation with oxygen, nitrogen, and carbon atoms color-coded red, blue, and white, respectively.
Walker A ([A/G]XXXXGK[S/T]) and Walker B (hhDE; h corresponds to any hydrophobic residue) motifs are clearly recognizable and include residues Ala244-Ser251 and residues Val469-Glu472, respectively (Fig. 1C and Fig. S3 A and B). Three additional conserved regions, motifs C, D and E, can be identified, of which motifs C and E line the catalytic site, and motif D is located facing the surface of the protein at the opposite side relative to the active site (Figs. 1A, and Figs. S2A and S3A). Motif C (DX[D/E]X1–3E) is a less conserved motif for VirB4 proteins and includes residues Asp272-Glu276, which line the catalytic Mg2+ site. Motif D (RK) and E ([S/T]Q), containing residues Arg496-Lys497 and Ser507-Gln508, respectively, were previously identified as one motif (21), but because they are clearly separated in space, they are more correctly divided into two motifs (Fig. S3A).
Nucleotide Binding Site.
A structure of TpsVirB4CTD in complex with ADP and Mg2+ was solved to 2.45 Å, showing an ADP molecule bound with full occupancy in two of the four protomers of the asymmetric unit (Fig. 1C and Fig. S3B). The α- and β-phosphates of ADP are recognized similarly as in other RecA-like ATPases with the conserved Walker A lysine, here Lys250, forming an electrostatic interaction with the β-phosphate of ADP (Fig. 1C and Fig. S3B). The α- and β-phosphates also make tight interactions with backbone amides of the P-loop of the Walker A motif, here residues Gly247, Ala248, Gly249, and Lys250. The Mg2+ is coordinated between Ser251 of the Walker A motif, Glu274 of motif C, the β-phosphate of ADP, and three water molecules, which connect the Mg2+ with Asp471 and Glu472 of the Walker B motif. The adenine moiety is stacked between Phe252 and Arg563, and its 6-amino group forms an electrostatic interaction with the carbonyl group of the backbone of Ile579.
Oligomerization State of VirB4 Proteins.
TpsVirB4CTD purifies as a monomer. This finding was confirmed by nanoelectrospray ionization mass spectrometry (nanoESI-MS) analysis of the protein (Fig. 2A). We next examined the oligomerization state of full-length TpsVirB4 by nanoESI-MS and observed mostly monomers and dimers (Fig. 2B). Small populations of trimers and tetramers were also observed. Similar results were obtained when the oligomerization state of the VirB4 homolog encoded by the pKM101 plasmid, TraB (referred to here as pkVirB4), was examined by nanoESI-MS (Fig. S4A). Monomer-dimer equilibria have been observed for a number of ATPases, for example the SecA translocase or the Rep helicase (30, 31). Moreover, ATP-binding has been shown to influence the oligomeric state of ATPases. For example, FliI from the flagellar system purifies as a monomer but hexamerizes readily in the presence of nonhydrolyzable ATP analogs (32; for review on hexameric ATPases, see ref. 33). Thus, we next examined the role of ATP-binding on pkVirB4 protein oligomerization using nanoESI-MS. We observed that addition of MgATPγS, MgAMPPNP, or MgADP-AlF4 to the protein solution did not change the oligomeric state of pkVirB4 (Fig. S4B). The same experiments were also carried out with TpsVirB4 and similar results obtained. Addition of single- and double-stranded DNA to the protein samples did not result in protein-DNA complex formation, nor did it change the oligomerization state of the proteins (Fig. S4C). Thus, both pkVirB4 and TpsVirB4 are found in a monomer-dimer equilibrium under all investigated solution conditions.
Characterization of an Arginine Finger in VirB4 Proteins.
We noticed that, in the structure-based alignment of the C-terminal domains of TrwB, TpsVirB4, and pkVirB4 (Fig. S3A), motif D is conserved across the VirB4 and VirD4 protein families: this motif contains a conserved Arg residue known in TrwB (Arg375) to act as an arginine finger (34); that is, a residue essential for ATPase activity but yet located in a region of the monomer far away from the ATP-binding site. Arginine fingers are typically contributed to an ATP-binding site in trans by an adjacent subunit within an oligomeric assembly. To provide biochemical evidence for an arginine finger motif in VirB4 proteins, we mutated both residues (Arg and Lys) (Fig. S3A) in motif D of TpsVirB4 and pkVirB4 and compared the ATPase activity of the produced mutants with wild-type enzyme. We produced both single and double mutants of both TpsVirB4 and pkVirB4, resulting in the TpsVirB4R496A, TpsVirB4K497A, TpsVirB4R496AK497A, pkVirB4R730A, pkVirB4K731A and pkVirB4R730AK731A proteins (Fig. S1 C and D). The specific activities were measured for these constructs as well as those of the wild-type TpsVirB4, TpsVirB4CTD, and pkVirB4.
The ATPase activity was measured by monitoring the release of inorganic phosphate (Pi) using an in vitro colorimetric ATPase assay. The activity of both TpsVirB4 and pkVirB4 were affected by the buffer composition as seen previously (20, 23). Both proteins were completely inactive in the presence of 200 mM NaCl but exhibited a relatively low specific activity in a buffer depleted of chloride ions (20, 23).
The specific activity of TpsVirB4 was 96 nmol⋅min-1⋅mg−1 and 114 nmol⋅min-1⋅mg−1 at 37 and 60 °C, respectively. The TpsVirB4CTD protein had a reduced activity. TpsVirB4R496A, TpsVirB4K497A and TpsVirB4R496AK497A all showed a significantly reduced activity compared with the wild-type enzyme, being reduced to 4%, 10%, and 2% of the wild-type activity, respectively (Fig. 2C). The specific activity of pkVirB4 at 37 °C was 16 nmol⋅min-1⋅mg−1 (Fig. S3C). The constructs pkVirB4R730A, pkVirB4K731A, and pkVirB4R730AK731A exhibited an activity of 6%, 5%, and 4% of wild-type pkVirB4 activity, respectively (Fig. S3C).
We therefore conclude that TpsVirB4 or pkVirB4 must oligomerize to hydrolyze ATP. Because dimers are the predominant oligomeric species observed by ESI-MS, VirB4 dimers might be the active form of the enzyme.
EM Structure of the VirB4/core Complex.
We next determined the structure of pkVirB4 (866 residues) bound to the corresponding core complex (pkVirB7-pkVirB9-pkVirB10) by negative stain EM (Fig. 3 and Figs. S1E and S5). This complex was purified using a double pull-down (Fig. S1E), taking advantage of a His-tag at the N terminus of pkVirB4 and a Strep-tag at the C terminus of pkVirB10.
Fig. 3.
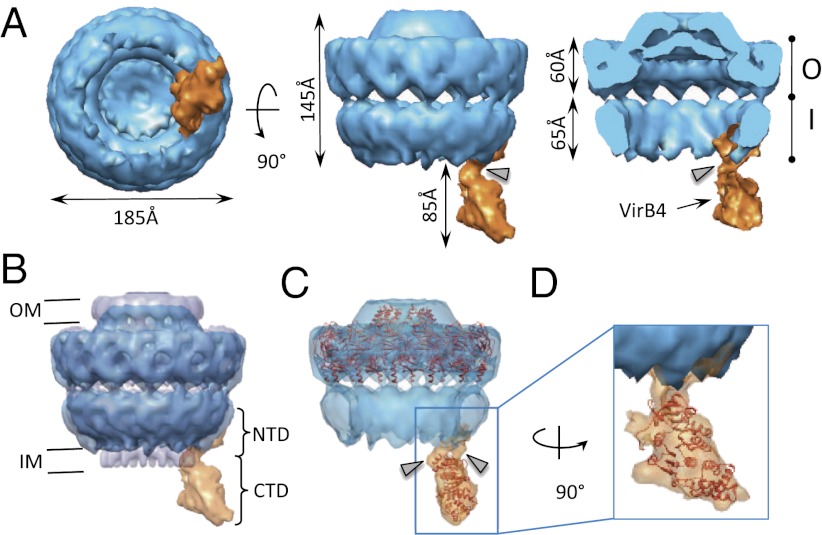
Structure of the pkVirB4 bound to the core complex. (A) Surface representation of the core/pkVirB4 complex. Bottom, side, and section views are shown at Left, Center, and Right, respectively. The core is shown in blue and pkVirB4 is in orange. The O and I layers are indicated as O and I, respectively. This structure was solved to a resolution of 20 Å. (B) Superposition of the negative stain core/pkVirB4 complex structure (in blue) and the cryo-EM structure of the core complex alone. The cryo-EM structure of the core complex is shown in half-transparent gray. (C) Fit of the crystal structure of the O layer (in orange; PDB ID 3JQO) into the negative stain EM structure of core-pkVirB4 complex. (D) TpsVirB4CTD atomic structure (in red ribbon representation) docked into the pkVirB4 electron density of the core-pkVirB4 complex. In A, Right, and in C, the gray arrowhead indicates links between the core and pkVirB4.
Within the negative stain EM dataset (SI Methods), predominantly one momomer of VirB4 was found bound to the core complex (Fig. S5 B–D). There were, however, examples of two or three monomers of pkVirB4 bound to a single core complex; in these cases, binding of these additional pkVirB4 molecules were observed at defined locations illustrated in Fig. S5D. Note that these locations are too far apart (minimum ∼85 Å) to provide a template for binding a higher order oligomeric VirB4 form. Because core complex particles with only one pkVirB4 bound constituted the vast majority of the data, we focused our effort to solve that structure. The core-pkVirB4 complex structure was solved to a resolution of 20 Å, and the pkVirB4 structure derived from images of VirB4 bound to the core complex was refined to a resolution of 17 Å (Fig. 3 and Fig. S5; also see SI Methods).
The core complex has a diameter of 185 Å and a length of 230 Å from the top of the core complex to the lower tip of pkVirB4 (Fig. 3A). The structure can be divided into three main sections: the O layer of the core complex facing the outer membrane, the I layer of the core complex facing the inner membrane, and the pkVirB4, protruding out of the core complex by about 85 Å (Fig. 3 A and B). The estimation of the molecular mass that could be accommodated within the electron density protruding from the I layer corresponds only to one molecule of pkVirB4, demonstrating that in this core-pkVirB4 complex, only one monomer of pkVirB4 is bound. The reconstruction of the core-pkVirB4 complex is fully consistent with that of the previously determined cryo- EM core complex structure (6) the overall structure of which does not appear to be affected by binding of pkVirB4.
The most striking feature of the core-pkVirB4 complex structure is that pkVirB4 binds to the side of the I layer of the core complex. T4S ATPases were hypothesized to stack underneath the core complex (18). However, the structure presented here suggests that VirB4 instead binds on the side of the T4S system. As expected by the presence of at least one predicted transmembrane segment (23), the electron density corresponding to pkVirB4 stretches through the inner membrane, with pkVirB4 interfacing with the core complex on the periplasmic side and in the membrane. Because the N-terminal part of pkVirB4 was identified to bind to the core complex (see below), we ascribed the parts of pkVirB4 located in the membrane and the periplasm as being formed by the VirB4 NTD (pkVirB4NTD); the part protruding in the cytoplasm and away from the I layer was interpreted as being formed by the CTD (pkVirB4CTD). We estimate pkVirB4NTD to be 65 Å long by 55 Å wide and 45 Å high. Because the structure has been obtained from the images of VirB4 attached to the core complex, the entire reconstruction encompasses the part of the I layer to which VirB4 binds; thus, a portion of the density can be ascribed to VirB9 and VirB10. Part of the density on the periplasmic side, however, protrudes laterally from the I layer and is likely to be the part of pkVirB4NTD that binds to the core complex.
The CTD of VirB4 is linked to the NTD via two connection links (Fig. 3 A and C, gray arrowheads). These links, which are part of the NTD, are shaped like “columns” 7.5 Å long, with a diameter of 15 Å. They are separated by a gap of 15 Å (Fig. 3C). These observations are consistent either with the presence of two transmembrane segments in the sequence of pkVirB4 or just one, the second column being formed by interacting regions within the NTD or between the NTD and the CTD. The CTD itself is large and globular measuring 100 Å down the longest axis and 70 Å at the widest point. The crystal structures of the O layer [PDB ID 3JQO (7)] and that of TpsVirB4CTD (present study) were docked into the negative stain structure using Chimera (35) (Figs. 3 C and D, respectively). Correlation coefficients for these fits were 0.78 and 0.93, respectively, indicating excellent fits for both, and also suggesting that the assignment of the pkVirB4 protruding density to its CTD is correct.
To examine the contribution of the NTD and CTD to the interaction with the core complex, we produced pkVirB4 constructs encompassing either the NTD (residues 1–442; pkVirB4NTD) or the CTD (residues 448–866; pkVirB4CTD). Coexpression of these with the core complex and subsequent purification resulted in the identification of the NTD as interacting with the core (Fig. S6). In addition, to further characterize the binding interface, two N-terminal deletions in pkVirB10 were produced, resulting in pkVirB10Δ60 and pkVirB10Δ130, starting at residue 61 or 131, respectively. The first 60 residues constitute the cytoplasmic and inner membrane regions of the core complex and residues 61–130 constitute a long proline-rich region. The corresponding pkVirB7-pkVirB9-pkVirB10Δ60 and pkVirB7-pkVirB9-pkVirB10Δ130 constructs where coexpressed with pkVirB4 and purified as for wild-type. Both shorter versions of pkVirB10 generate a core complex, which interact with pkVirB4 (Fig. S6). We also attempted to make VirB9-truncated variants of the core complex but none could be stably produced. However, because the pkVirB10Δ130 deletion removes most of the I layer part of pkVirB10 (Fig. 3), then our results suggest that pkVirB4 interacts primarily with pkVirB9, a protein known to reside entirely in the periplasm and decorate the outside of the core complex (7) (also see details in Fig. S6).
It is interesting to note that residues 15–205 of TpsVirB4NTD and residues 260–448 of pkVirB4NTD share significant sequence similarity (34%) and an almost identical predicted secondary structure topology. However, Gram-positive T4S systems do not contain the outer-membrane components of Gram-negative T4S systems, and thus interactions involving Gram-positive VirB4s would then have to be with inner-membrane components VirB3, VirB6, and VirB8. Thus, in addition to interactions between pkVirB4NTD with pkVirB9 observed here, pkVirB4NTD might also interact with the inner-membrane components listed above in the context of a fully assembled T4S system (see ref. 25 and section below).
Mechanistic Implications for VirB4 as a Molecular Motor.
In the present study, we describe the crystal structure of a VirB4 protein and also provide a view of a VirB4 protein in a complex with a T4S system core. On their own, the examined VirB4 proteins are in a monomer-dimer equilibrium, where the monomeric form predominates. However, bound to the core machinery, pkVirB4 is monomeric. These two properties of VirB4 proteins are reminiscent of the oligomerization properties of SecA, an ATPase also involved in secretion: SecA, although in a monomer-dimer equilibrium on its own, is monomeric when bound to the SecYEG complex (30, 36). Although others and ourselves have reported hexameric forms of VirB4 proteins (20, 23), we are unable to reproduce these results using nanoESI-MS, a technique considered as the gold standard for oligomerization state studies of proteins and protein complexes (37).
Because VirB4 requires an arginine finger for activity and is monomeric when bound to the secretion machinery, we hypothesize that the required arginine finger is then provided in trans by another component of the machinery. There are precedents for this kind of heterologous intermolecular arginine finger contribution, for example in complexes of small Ras-like GTPases with GTPase-activating proteins (38).
VirB4 proteins may play essential regulatory roles in substrate transfer (39). In the A. tumefaciens system, it does not contact the DNA substrate directly. Instead, its ATPase activity is crucial for substrate hand-over from VirB11 to the inner membrane VirB6/VirB8 channel (39). VirB4 makes a number of interactions, notably with VirB3 (40), a small protein embedded in the inner membrane, and VirB8 (19, 25). Although interaction with VirB10 has been previously described (25, 41), the study presented here suggests that VirB4 interacts more extensively with VirB9 than with VirB10.
The structure of the core-VirB4 complex shows that the VirB4 motor unit is built up on the side of the core complex. Such a location is consistent with the fact that VirB4 does not engage directly with the substrate. Indeed, a positioning of VirB4 underneath the core complex, as was initially expected, would have suggested a substrate gating role as VirB4 would then be obstructing substrate entry inside the core complex: this would have implied VirB4 making contact with the substrate, which it does not. A side position for the VirB4 motor unit is also consistent with VirB4’s regulatory role. In that position, VirB4 would be positioned to relay ATP-driven conformational changes to the entire core complex through its interaction with VirB9. What exactly these conformational changes might be and how they might affect the structure of the core complex remains to be determined. However, a side position for VirB4 would certainly have the potential to remodel the inner membrane complex formed by VirB6, VirB8, and the NTD of VirB10. VirB10 is known to undergo conformational changes upon energization of the system (42), and thus remodeling of the inner-membrane complex by VirB4 might reconfigure VirB10 into an activated state.
Methods
Cloning, Protein Expression, and Purification.
All constructs were cloned, expressed, and purified as described in SI Methods. Primers are listed in Table S2. Core complex bound to pkVirB4 was obtained by coexpression and double pull-down taking advantage of the Strep-tag II at the C terminus of pkVirB10 and a His-tag at the N terminus of pkVirB4.
Limited Proteolysis, Crystallization, Data Collection, and Determination of the Structure of TpsVirB4CTD.
Ni-affinity purified TpsVirB4 was reduced to its C-terminal domain using 1/50 (wt/wt) ratio of trypsin/TpsVirB, and then purified by gel filtration. Crystals were obtained using the hanging-drop method with a reservoir solution containing 30% (wt/vol) PEG3350; 100 mM Bis-Tris, pH 5.75; and 200 mM ammonium sulfate. For the TpsVirB4CTD-ADP complex, the apo form crystal was further soaked in a solution containing 20 mM of ADP-Mg but depleted of 80% of the sulfate ions. Phasing was carried out using the single anomalous diffraction method on data collected on Selenomethonine-substituted crystals as implemented in PHENIX (43). Data collection and refinement statistics are presented in Table S1.
ATPase Activity Assay.
ATPase activities were measured using the ATPase colorimetric assay kit from Innova Biosciences. Assays were carried out at both 37 and 60 °C because of the thermophilic origin of TpsVirB4.
Mass Spectrometry Analysis.
Mass spectrometry experiments were carried out on a Synapt HDMS (Waters) mass spectrometer (44). Sample solutions were injected into the instrument by means of nanoESI. Typical instrumental parameters were as follows: source pressure 6 mbar, capillary voltage 1.20 kV, cone voltage 100 V, trap energy 20 V, transfer energy 10 V, and trap pressure 3.6 × 10−2 mbar.
Electron Microscopy and Image Processing of the Core-pkVirB4 Complex.
Samples were applied on to carbon-coated copper grids (400 mesh, freshly glow-discharged) and stained with 2% Nano-W (methylamine tungstate). Images were taken in a T12 electron microscope (FEI) in a defocus range of 0.7–2 mm and recorded on films at a 42,000 nominal magnification. Films were digitized using a Zeiss scanner with a step size corresponded to 1.67 Å/pixel on the specimen scale. The image analysis was performed using IMAGIC-5 (45). Preprocessing, alignment, and classification of images was performed as previously described (ref. 45 and references therein). Angular orientations of class averages were determined by angular reconstitution. Three-dimensional maps were calculated using the exact-filter back projection algorithm (45) using 330 classes for the core-pkVirB4 complex and 200 classes for the pkVirB4 structure. The resolution for both the core and pkVirB4 was assessed using the Fourier shell correlation at the 0.5 criteria that was 20 Å and 17 Å, respectively. (See SI Methods for details.)
Supplementary Material
Acknowledgments
We thank the staff of beamlines I02 and I04 at Diamond Light Source. This work was funded by Grant 082227 from the Wellcome Trust (to G.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4AG5 and 4AG6 for ADP-Mg2+-bound and selenomethionine–incorporated TpsVirB4, respectively). EM maps have been deposited to the Electron Microscopy Data Bank under the entry codes EMD-2136 and EMD-2137 for the 20-Å map and the 17-Å map, respectively.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201428109/-/DCSupplemental.
References
- 1.Grohmann E, Muth G, Espinosa M. Conjugative plasmid transfer in gram-positive bacteria. Microbiol Mol Biol Rev. 2003;67:277–301. doi: 10.1128/MMBR.67.2.277-301.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wallden K, Rivera-Calzada A, Waksman G. Type IV secretion systems: Versatility and diversity in function. Cell Microbiol. 2010;12:1203–1212. doi: 10.1111/j.1462-5822.2010.01499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kwok T, et al. Helicobacter exploits integrin for type IV secretion and kinase activation. Nature. 2007;449:862–866. doi: 10.1038/nature06187. [DOI] [PubMed] [Google Scholar]
- 4.Zhao Y, et al. Role of a type IV-like secretion system of Streptococcus suis 2 in the development of streptococcal toxic shock syndrome. J Infect Dis. 2011;204:274–281. doi: 10.1093/infdis/jir261. [DOI] [PubMed] [Google Scholar]
- 5.Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu Rev Microbiol. 2005;59:451–485. doi: 10.1146/annurev.micro.58.030603.123630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fronzes R, et al. Structure of a type IV secretion system core complex. Science. 2009;323:266–268. doi: 10.1126/science.1166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandran V, et al. Structure of the outer membrane complex of a type IV secretion system. Nature. 2009;462:1011–1015. doi: 10.1038/nature08588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eisenbrandt R, et al. Conjugative pili of IncP plasmids, and the Ti plasmid T pilus are composed of cyclic subunits. J Biol Chem. 1999;274:22548–22555. doi: 10.1074/jbc.274.32.22548. [DOI] [PubMed] [Google Scholar]
- 9.Aly KA, Baron C. The VirB5 protein localizes to the T-pilus tips in Agrobacterium tumefaciens. Microbiology. 2007;153:3766–3775. doi: 10.1099/mic.0.2007/010462-0. [DOI] [PubMed] [Google Scholar]
- 10.Fronzes R, Christie PJ, Waksman G. The structural biology of type IV secretion systems. Nat Rev Microbiol. 2009;7:703–714. doi: 10.1038/nrmicro2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeo HJ, Savvides SN, Herr AB, Lanka E, Waksman G. Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV secretion system. Mol Cell. 2000;6:1461–1472. doi: 10.1016/s1097-2765(00)00142-8. [DOI] [PubMed] [Google Scholar]
- 12.Gomis-Rüth FX, et al. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature. 2001;409:637–641. doi: 10.1038/35054586. [DOI] [PubMed] [Google Scholar]
- 13.Hare S, Bayliss R, Baron C, Waksman G. A large domain swap in the VirB11 ATPase of Brucella suis leaves the hexameric assembly intact. J Mol Biol. 2006;360:56–66. doi: 10.1016/j.jmb.2006.04.060. [DOI] [PubMed] [Google Scholar]
- 14.Fullner KJ, Lara JC, Nester EW. Pilus assembly by Agrobacterium T-DNA transfer genes. Science. 1996;273:1107–1109. doi: 10.1126/science.273.5278.1107. [DOI] [PubMed] [Google Scholar]
- 15.Watarai M, Makino S, Shirahata T. An essential virulence protein of Brucella abortus, VirB4, requires an intact nucleoside-triphosphate-binding domain. Microbiology. 2002;148:1439–1446. doi: 10.1099/00221287-148-5-1439. [DOI] [PubMed] [Google Scholar]
- 16.Berger BR, Christie PJ. The Agrobacterium tumefaciens virB4 gene product is an essential virulence protein requiring an intact nucleoside triphosphate-binding domain. J Bacteriol. 1993;175:1723–1734. doi: 10.1128/jb.175.6.1723-1734.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fullner KJ, Stephens KM, Nester EW. An essential virulence protein of Agrobacterium tumefaciens, VirB4, requires an intact mononucleotide binding domain to function in transfer of T-DNA. Mol Gen Genet. 1994;245:704–715. doi: 10.1007/BF00297277. [DOI] [PubMed] [Google Scholar]
- 18.Middleton R, Sjölander K, Krishnamurthy N, Foley J, Zambryski P. Predicted hexameric structure of the Agrobacterium VirB4 C terminus suggests VirB4 acts as a docking site during type IV secretion. Proc Natl Acad Sci USA. 2005;102:1685–1690. doi: 10.1073/pnas.0409399102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan Q, et al. Identification of the VirB4-VirB8-VirB5-VirB2 pilus assembly sequence of type IV secretion systems. J Biol Chem. 2005;280:26349–26359. doi: 10.1074/jbc.M502347200. [DOI] [PubMed] [Google Scholar]
- 20.Arechaga I, et al. ATPase activity and oligomeric state of TrwK, the VirB4 homologue of the plasmid R388 type IV secretion system. J Bacteriol. 2008;190:5472–5479. doi: 10.1128/JB.00321-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rabel C, Grahn AM, Lurz R, Lanka E. The VirB4 family of proposed traffic nucleoside triphosphatases: Common motifs in plasmid RP4 TrbE are essential for conjugation and phage adsorption. J Bacteriol. 2003;185:1045–1058. doi: 10.1128/JB.185.3.1045-1058.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dang TA, Zhou XR, Graf B, Christie PJ. Dimerization of the Agrobacterium tumefaciens VirB4 ATPase and the effect of ATP-binding cassette mutations on the assembly and function of the T-DNA transporter. Mol Microbiol. 1999;32:1239–1253. doi: 10.1046/j.1365-2958.1999.01436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Durand E, Oomen C, Waksman G. Biochemical dissection of the ATPase TraB, the VirB4 homologue of the Escherichia coli pKM101 conjugation machinery. J Bacteriol. 2010;192:2315–2323. doi: 10.1128/JB.01384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Durand E, Waksman G, Receveur-Brechot V. Structural insights into the membrane-extracted dimeric form of the ATPase TraB from the Escherichia coli pKM101 conjugation system. BMC Struct Biol. 2011;11:4. doi: 10.1186/1472-6807-11-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Draper O, Middleton R, Doucleff M, Zambryski PC. Topology of the VirB4 C terminus in the Agrobacterium tumefaciens VirB/D4 type IV secretion system. J Biol Chem. 2006;281:37628–37635. doi: 10.1074/jbc.M606403200. [DOI] [PubMed] [Google Scholar]
- 26.Abajy MY, et al. A type IV-secretion-like system is required for conjugative DNA transport of broad-host-range plasmid pIP501 in gram-positive bacteria. J Bacteriol. 2007;189:2487–2496. doi: 10.1128/JB.01491-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caryl JA, O’Neill AJ. Complete nucleotide sequence of pGO1, the prototype conjugative plasmid from the Staphylococci. Plasmid. 2009;62:35–38. doi: 10.1016/j.plasmid.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 28.Lawley TD, Klimke WA, Gubbins MJ, Frost LS. F factor conjugation is a true type IV secretion system. FEMS Microbiol Lett. 2003;224:1–15. doi: 10.1016/S0378-1097(03)00430-0. [DOI] [PubMed] [Google Scholar]
- 29.Schandel KA, Muller MM, Webster RE. Localization of TraC, a protein involved in assembly of the F conjugative pilus. J Bacteriol. 1992;174:3800–3806. doi: 10.1128/jb.174.11.3800-3806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woodbury RL, Hardy SJ, Randall LL. Complex behavior in solution of homodimeric SecA. Protein Sci. 2002;11:875–882. doi: 10.1110/ps.4090102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korolev S, Hsieh J, Gauss GH, Lohman TM, Waksman G. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90:635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- 32.Kazetani K, Minamino T, Miyata T, Kato T, Namba K. ATP-induced FliI hexamerization facilitates bacterial flagellar protein export. Biochem Biophys Res Commun. 2009;388:323–327. doi: 10.1016/j.bbrc.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 33.Tucker PA, Sallai L. The AAA+ superfamily—A myriad of motions. Curr Opin Struct Biol. 2007;17:641–652. doi: 10.1016/j.sbi.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 34.de Paz HD, et al. Functional dissection of the conjugative coupling protein TrwB. J Bacteriol. 2010;192:2655–2669. doi: 10.1128/JB.01692-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 36.Zimmer J, Nam Y, Rapoport TA. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature. 2008;455:936–943. doi: 10.1038/nature07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heck AJ. Native mass spectrometry: A bridge between interactomics and structural biology. Nat Methods. 2008;5:927–933. doi: 10.1038/nmeth.1265. [DOI] [PubMed] [Google Scholar]
- 38.Bourne HR. G proteins. The arginine finger strikes again. Nature. 1997;389:673–674. doi: 10.1038/39470. [DOI] [PubMed] [Google Scholar]
- 39.Cascales E, Christie PJ. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science. 2004;304:1170–1173. doi: 10.1126/science.1095211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mossey P, Hudacek A, Das A. Agrobacterium tumefaciens type IV secretion protein VirB3 is an inner membrane protein and requires VirB4, VirB7, and VirB8 for stabilization. J Bacteriol. 2010;192:2830–2838. doi: 10.1128/JB.01331-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Terradot L, et al. Biochemical characterization of protein complexes from the Helicobacter pylori protein interaction map: strategies for complex formation and evidence for novel interactions within type IV secretion systems. Mol Cell Proteomics. 2004;3:809–819. doi: 10.1074/mcp.M400048-MCP200. [DOI] [PubMed] [Google Scholar]
- 42.Cascales E, Christie PJ. Agrobacterium VirB10, an ATP energy sensor required for type IV secretion. Proc Natl Acad Sci USA. 2004;101:17228–17233. doi: 10.1073/pnas.0405843101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pringle SD, et al. An investigation of the mobility separation of some peptide and protein ions using a new hybrid quadrupole/travelling wave IMS/oa-ToF instrument. Int J Mass Spectrom. 2007;261(1):1–12. [Google Scholar]
- 45.van Heel M, et al. Single-particle electron cryo-microscopy: Towards atomic resolution. Q Rev Biophys. 2000;33:307–369. doi: 10.1017/s0033583500003644. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.