Abstract
The limited therapeutic arsenal and the increase in reports of fungal resistance to multiple antifungal agents have made fungal infections a major therapeutic challenge. The polyene antibiotics are the only group of antifungal antibiotics that directly target the plasma membrane via a specific interaction with the main fungal sterol, ergosterol, often resulting in membrane permeabilization. In contrast to other polyene antibiotics that form pores in the membrane, the mode of action of natamycin has remained obscure but is not related to membrane permeabilization. Here, we demonstrate that natamycin inhibits growth of yeasts and fungi via the immediate inhibition of amino acid and glucose transport across the plasma membrane. This is attributable to ergosterol-specific and reversible inhibition of membrane transport proteins. It is proposed that ergosterol-dependent inhibition of membrane proteins is a general mode of action of all the polyene antibiotics, of which some have been shown additionally to permeabilize the plasma membrane. Our results imply that sterol-protein interactions are fundamentally important for protein function even for those proteins that are not known to reside in sterol-rich domains.
In recent years, advances in the treatment of transplant recipients and patients with cancer or AIDS have been accompanied by a dramatic increase in the incidence of life-threatening fungal infections (1). Sepsis, characterized by a whole-body inflammatory state, is the 10th most common cause of death overall, and the number of cases of sepsis caused by fungal organisms has doubled in 20 y (2, 3). Fungal infections are a major therapeutic challenge, because the therapeutic arsenal is limited and the use of drugs is restricted as a result of toxicity or unfavorable pharmacokinetic profiles. Fungal resistance has been turned into a global public health crisis, especially with fungi showing resistance to more than one antifungal agent (4, 5). In contrast to many other antifungal agents, resistance to polyene antibiotics is considered an exceptionally rare event (4, 5).The polyene antibiotics are the only group of antifungal antibiotics that directly target the plasma membrane via a specific interaction with the main fungal sterol, ergosterol (6). Natamycin, a member of the polyene antibiotic family, is widely used in the food industry and in pharmacotherapy for topical treatment. Unlike other polyene antibiotics, the mode of action of natamycin is not based on the ergosterol-dependent permeabilization of the plasma membrane (7). However, the immediate cessation of growth of yeasts by natamycin treatment indicates that there might be an instantaneous effect of natamycin at the level of the plasma membrane (7), which also contains the highest levels of ergosterol (8). Both the rapid action and lack of pore formation lead us to the hypothesis that natamycin may target plasma membrane proteins in an ergosterol-based manner.
Results
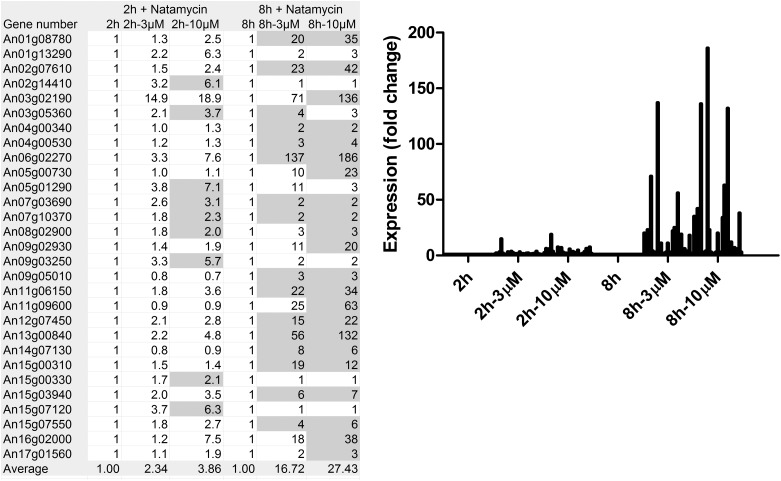
We performed transcriptome analysis on germinating conidia of the fungus Aspergillus niger to identify proteins that may be targeted by natamycin. Germination of these conidia has previously been shown to be blocked by natamycin (9). In the analysis, we focused on the effect of natamycin on transcription of plasma membrane proteins. The most prominent plasma membrane proteins in these germinating spores that were affected in their transcription by natamycin treatment were transport proteins. The majority of sugar and amino acid transporters especially showed a clear tendency of up-regulation in the presence of natamycin. (Table 1). The individual gene regulations of 20 and 31 genes for the sugar and amino acid transporters, respectively, are shown in Table S1. An examination of a subselection of the 29 most strongly up-regulated sugar and amino acid transporters after treatment with natamycin shows that the increase in expression was dose- and time-dependent (Fig. 1). Overall, these results show that conidia react to treatment with natamycin by up-regulating many sugar and amino acid transporters. The expression of other membrane proteins was also affected, resulting in either increased or decreased levels of expression (Table S2). For some membrane proteins, the expression was only slightly affected. These data demonstrate that the binding of natamycin to ergosterol has a strong effect on the expression of a subset of plasma membrane proteins.
Table 1.
Effect of natamycin on the differential expression of sugar and amino acid transporters in A. niger conidia
| 2h + natamycin |
8h + natamycin |
|||||
| 2h | 2h, 3μM | 2h, 10μM | 8h | 8h, 3μM | 8h, 10μM | |
| Cumulative values, arbitrary units | ||||||
| Sugar transporters (n = 20) | 3,840 | 5,976 | 7,843 | 3,421 | 14,983 | 20,678 |
| Amino acid transporters (n = 31) | 7,054 | 10,059 | 12,541 | 4,596 | 10,498 | 12,051 |
| Ratio natamycin/control | ||||||
| Sugar transporters | 1 | 1.56 | 2.04 | 1 | 4.38 | 6.04 |
| Amino acid transporters | 1 | 1.43 | 1.78 | 1 | 2.28 | 2.62 |
| All expressed proteins | 1 | 1.23 | 1.34 | 1 | 1.21 | 1.25 |
Fig. 1.
Expression of a selection of sugar and amino acid transporters. The expression of 29 sugar and amino acid transporters that were most strongly up-regulated on treatment with natamycin (>120 arbitrary units after 2 or 8 h) is shown. Together, these dominant transporters make up over 90% of the expression intensity of all sugar and amino acid transporters after natamycin treatment. A detailed description of the genes is provided in Table S1. Expression values are given of conidia during different stages of germination (controls) or in the presence of natamycin at different concentrations. The data are given as fluorescence data from three independent microarray experiments. The gray values are significant below the P = 0.01 level.
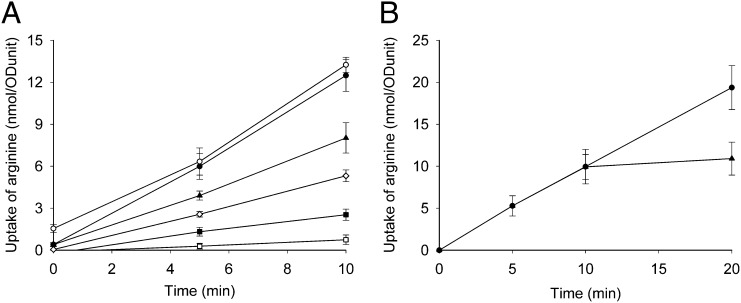
Next, we determined whether the up-regulation of plasma membrane proteins is attributable to a direct effect of natamycin on the functioning of these proteins. For this, we used the yeast Saccharomyces cerevisiae, which is a well-described model system to study functioning of membrane transport proteins (10–12). First, we examined the effects of natamycin on the plasma membrane transporter Can1p. This protein specifically transports arginine into the cell via proton/arginine symport (10). The uptake of arginine was determined in yeast cells that were incubated with different concentrations of natamycin. Fig. 2A shows that natamycin causes a dose-dependent decrease in the uptake of arginine, which was complete at 20 μM natamycin. This can be attributable to either natamycin-dependent inhibition of the transport process or natamycin-induced leakage of arginine. This was examined by allowing the cells to take up arginine for 8 min, after which natamycin was added. Natamycin addition immediately blocked uptake without causing release of the arginine that had already been taken up (Fig. 2B). This indicates that natamycin directly inhibited the arginine import. Next, we tested the effect of natamycin on the growth and the viability of the yeast cells under identical conditions as used during the arginine uptake assay (Fig. S1). These results show that natamycin inhibits arginine uptake at concentrations that inhibit cell growth but without killing the cells. In addition, we could show that natamycin-induced blockade of arginine uptake in yeast cells can be reversed (Fig. S2A). However, relieving the blockade in arginine uptake does not result in immediate regrowth of the yeast culture (Fig. S2B). This suggests that natamycin also affects other plasma membrane proteins that possibly take longer to recuperate after removing the natamycin pressure on the outside of the cells, which is supported by the transcriptome analysis.
Fig. 2.
Effect of natamycin on the uptake of arginine by yeast cells. At time 0, 14C-arginine (30 μM) was added to the cells. The uptake of arginine was followed in time and corrected for the amount of cells (ODunit). (A) Yeast cells were incubated for 5 min before the addition of arginine with natamycin at 0 μM (●), 2 μM (▲), 5 μM (◇), 10 μM (■), and 20 μM (□), and DMSO (○) was added as a control. (B) Release of arginine from yeast cells was studied by adding 20 μM natamycin (▲) or no natamycin (●) at 8 min after the addition of arginine, and the uptake of arginine was followed in time. The results shown are the averages of three separately performed experiments with SD.
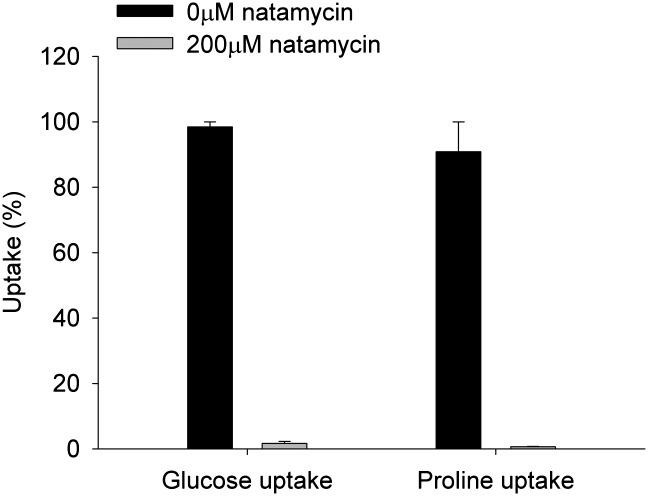
Next, the effects of natamycin on the transport of two other substrates (glucose and proline) were determined. Glucose is transported in yeast via hexose transporters, and of the 20 genes that encode these proteins, 7 are known to encode functional glucose transporters. All of them transport their substrates by passive, energy-independent, facilitated diffusion, with glucose moving down the concentration gradient (12). Proline can be taken up by a specific high-affinity permease Put4p or the general amino acid permease Gap1p, both of which are regulated by nitrogen (10, 13). Other low-affinity systems are also present, such as Agp1p and Gnp1p (13). The uptake of glucose and proline was completely blocked by the addition of natamycin (Fig. 3) at similar antibiotic-to-cell ratios that blocked arginine uptake. These results show that natamycin blocks the uptake of several important nutrients over the plasma membrane of yeast and indicate that natamycin can inhibit different membrane transport proteins.
Fig. 3.
Effect of natamycin on uptake of glucose and proline by yeast cells. Cells were incubated with 0 or 200 μM natamycin, after which the uptake of 14C-glucose or 14C-proline was assayed. The uptake of the different compounds by yeast cells is expressed as the percentage to the uptake of a compound by cells untreated with natamycin after 10 min. Additional details are provided in Experimental Methods. The results shown are the averages of two separately performed experiments with the spread of the data.
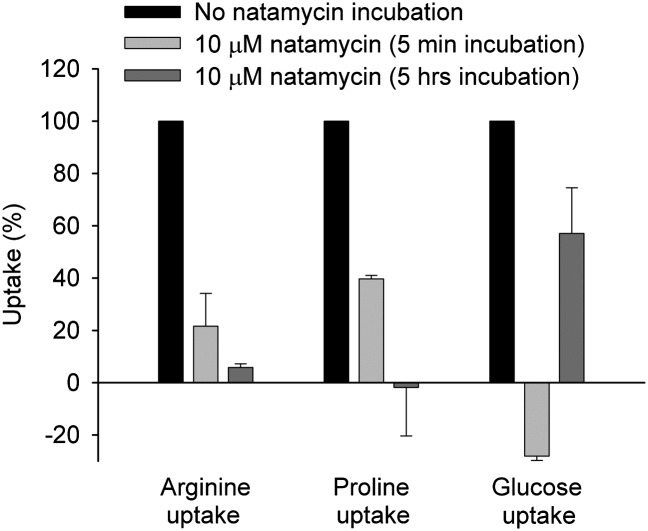
Because the effect of natamycin on transcription of the sugar and amino acid transporters was shown in germinating conidia of A. niger, we adapted the arginine, proline, and glucose uptake assays to these conidia. Fig. 4 shows that natamycin is also able to inhibit the import of all tested substrates in germinating A. niger conidia after only a 5-min incubation. After 5 h of incubating the conidia with natamycin, the inhibition of the uptake of the amino acids was at its maximal level, whereas the conidia seemed to be able to recuperate partially from the blockade in glucose. The twofold general increase in the expression of sugar transporters over that of the amino acid transporters may be the reason for this (Table 1). These results show that natamycin is able to block the uptake of different substrates in baker’s yeast as well as in fungal conidia. Taken together, these data demonstrate that natamycin has a general effect on transport proteins and that the binding of ergosterol by the antifungal has dramatic effects on the expression and functioning of membrane proteins.
Fig. 4.
Effect of natamycin on uptake of arginine, proline, and glucose by A. niger conidia. Conidia were incubated for either 5 min or 5 h with 0 or 10 μM natamycin, after which the uptake of 14C-arginine, 14C-proline, or 14C-glucose was assayed. The uptake of the different compounds by conidia incubated for 0 or 10 min at 30 °C without antibiotic is normalized to 100% and compared with the effect of natamycin on substrate uptake. Negative uptake values occur because a smaller amount of compound was taken up by the spores after incubation with natamycin in comparison to the fast uptake in the absence of the inhibitor. Additional details are provided in Experimental Methods. The results shown are the averages of two separately performed experiments with the spread of the data.
Discussion
Natamycin is the only member of the polyene antibiotic family that does not exert its antifungal action by forming pores and permeabilizing the plasma membrane. It does, however, require binding to the same lipid receptor, ergosterol, as other polyene antibiotics. We have found that on binding ergosterol in the plasma membrane, natamycin is able to inhibit a broad class of essential membrane transport proteins without compromising the barrier function of the membrane.
The differences in substrate specificities, transport mechanisms, and lack of common domains/motifs of the amino acid and glucose transporters make it likely that natamycin inhibits these proteins via a general mechanism (11, 12, 14). What these transporters do have in common is their similarity in length and hydrophobicity profiles (11, 15, 16). Therefore, it is most likely that natamycin affects these proteins through alterations in membrane properties instead of inhibiting each protein involved in a specific way. Moreover, considering the amount of transport proteins that were up-regulated by natamycin treatment in addition to those tested in our assays, we propose that natamycin is able to block virtually all transport processes in the plasma membrane of fungi.
Answering the question of how natamycin changes the membrane properties starts with the premise that the antibiotic action is attributable to binding to ergosterol (7), thereby affecting the functional properties the sterol has on membrane proteins either via ergosterol-rich domains (17) or more directly by interfering with a direct ergosterol-protein interaction. The different oxygen functions of natamycin may play a role in the latter mechanism, as is also suggested for other polyenes (6, 18). A disruption of the equilibrium of free sterols to sphingolipids by natamycin may also be responsible for the inhibition of ergosterol-dependent protein functions because sterols and sphingolipids work together to carry out a wide variety of functions (19).
The unique mechanism revealed in this study for natamycin may also be applicable to other polyene antibiotics that permeabilize the plasma membrane because these antibiotics all exert their action via binding to ergosterol. This is supported by the observation that nystatin affected properties of the arginine and glucose transporters in yeast plasma membrane vesicles (20). Further, the amphotericin B methyl ester inhibited the replication, assembly, and release of HIV-1 by interfering with the ion channel viral protein U (VPU) (18, 21). Microarray studies with S. cerevisiae clearly show an increase of expression of transport proteins on treatment with the polyenes amphotericin B and nystatin (22, 23). However, membrane permeabilization by these polyene antibiotics does mask these effects as a result of the fast collapse of vital ion or substrate gradients.
Recently, studies from the laboratory of Burke have shown that deleting one hydroxyl group (at position 35) on the ring structure of such a pore-forming polyene antibiotic, amphotericin B, left this derivative unable to form pores but still able to bind ergosterol and to retain its antifungal activity to a significant extent (24). Gray et al. (24) concluded that amphotericin B primarily kills yeast by simply binding ergosterol and that membrane permeabilization via pore formation represents a second complementary mechanism. Earlier studies from the laboratory of Carreira showed that substituting the hydroxyl group at position 35 of the methyl-ester version of amphotericin B led to a significant but not complete reduction in K+ efflux from model membrane vesicles (25, 26). This finding pointed to the importance of this hydroxyl group for pore formation and led to the conclusion that channel formation is a necessary condition for the antifungal activity of amphotericin B. However, no explanation was given for the residual activity of the compound, and the residual membrane permeabilization that was observed in this study was likely attributable to an aspecific effect caused by the positively charged nature of the methyl-ester version of the Carreira group, as previously suggested (24). Interestingly, the antifungal activity of natamycin is comparable to the activities of both amphotericin derivatives (24, 26). This implies that these amphotericin B derivatives are also able to inhibit a broad class of essential membrane transport proteins by binding to ergosterol, thus explaining their antifungal activities. This is strengthened by the observation that both natamycin and the variant of amphotericin B of the Burke laboratory require ergosterol for interacting with the membrane and executing their antifungal activity (7, 24). Amphotericin B may therefore be a good example of a polyene antibiotic that, via the binding of ergosterol, has a dual mode of action by inhibiting membrane proteins and permeabilizing the plasma membrane. A similar dual mode of action has been observed before in antibacterial lantibiotics that are able to block cell wall synthesis and form pores (27, 28). Likewise, the antibiotics chloramphenicol and tetracycline were shown to act both by inhibiting protein synthesis and by blocking protein translocation into the bacterial membrane (29–31). Because natamycin is the only natural family member known that does not permeabilize the membrane, it is the ideal candidate to study this previously undescribed basic mode of action of the polyene antibiotics further. This unique mode of action by completely blocking the transport function of many (if not all) membrane proteins via binding to a specific lipid receptor in the fungal membrane implies that sterol-protein interactions are fundamentally important for protein function even for those proteins that are not known to reside in sterol-rich membrane domains.
Experimental Methods
A unique RNA extraction method for conidia of A. niger was developed resulting in high-quality intact RNA. Full experimental details are included in SI Experimental Methods. RNA samples from three independent biological replicates were used for hybridization of Affymetrix microarray chips representing 14,509 ORFs of A. niger. Each experiment had conidia pooled from three cDNA labeling experiments. Microarray hybridization and scanning were performed at ServiceXS (Leiden, The Netherlands) according to Affymetrix protocols. The Functional Catalogue (Munich Information Center for Protein Sequences) (32) was used for systematic classification of genes according to their cellular and molecular functions (33).
Transport assays for arginine, glucose, and proline in S. cerevisiae were based on the methods of Robl et al. (10) and Malínská et al. (34). The uptake of arginine was determined in yeast cells that were incubated with different concentrations of natamycin. The substrate uptake assays in fungal conidia were performed in a similar way to the assays with baker’s yeast to allow a comparison of the results. Full experimental details are included in SI Experimental Methods.
Supplementary Material
Acknowledgments
We thank P. Krijgsheld (University of Utrecht) for her contributions to our microarray analyses and A. I. P. M. de Kroon (University of Utrecht) for valuable discussions. This work was supported by the Netherlands Technology Foundation and Applied Science Division of the Netherlands Organization for Scientific Research Grant UBC 6524.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1203375109/-/DCSupplemental.
References
- 1.Cuenca-Estrella M, et al. Update on the epidemiology and diagnosis of invasive fungal infection. Int J Antimicrob Agents. 2008;32(Suppl 2):S143–S147. doi: 10.1016/S0924-8579(08)70016-5. [DOI] [PubMed] [Google Scholar]
- 2.Martin GS, Mannino DM, Eaton S, Moss M. The epidemiology of sepsis in the United States from 1979 through 2000. N Engl J Med. 2003;348:1546–1554. doi: 10.1056/NEJMoa022139. [DOI] [PubMed] [Google Scholar]
- 3.Llewelyn MJ, Cohen J. Tracking the microbes in sepsis: Advancements in treatment bring challenges for microbial epidemiology. Clin Infect Dis. 2007;44:1343–1348. doi: 10.1086/515403. [DOI] [PubMed] [Google Scholar]
- 4.Monk BC, Goffeau A. Outwitting multidrug resistance to antifungals. Science. 2008;321:367–369. doi: 10.1126/science.1159746. [DOI] [PubMed] [Google Scholar]
- 5.Kanafani ZA, Perfect JR. Antimicrobial resistance: Resistance to antifungal agents: Mechanisms and clinical impact. Clin Infect Dis. 2008;46:120–128. doi: 10.1086/524071. [DOI] [PubMed] [Google Scholar]
- 6.Bolard J. How do the polyene macrolide antibiotics affect the cellular membrane properties? Biochim Biophys Acta. 1986;864:257–304. doi: 10.1016/0304-4157(86)90002-x. [DOI] [PubMed] [Google Scholar]
- 7.te Welscher YM, et al. Natamycin blocks fungal growth by binding specifically to ergosterol without permeabilizing the membrane. J Biol Chem. 2008;283:6393–6401. doi: 10.1074/jbc.M707821200. [DOI] [PubMed] [Google Scholar]
- 8.Zinser E, Paltauf F, Daum G. Sterol composition of yeast organelle membranes and subcellular distribution of enzymes involved in sterol metabolism. J Bacteriol. 1993;175:2853–2858. doi: 10.1128/jb.175.10.2853-2858.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Leeuwen MR, Van Doorn TM, Golovina EA, Stark J, Dijksterhuis J. Water- and air-distributed conidia differ in sterol content and cytoplasmic microviscosity. Appl Environ Microbiol. 2010;76:366–369. doi: 10.1128/AEM.01632-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robl I, Grassl R, Tanner W, Opekarová M. Construction of phosphatidylethanolamine-less strain of Saccharomyces cerevisiae. Effect on amino acid transport. Yeast. 2001;18:251–260. doi: 10.1002/1097-0061(200102)18:3<251::AID-YEA667>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 11.Regenberg B, Düring-Olsen L, Kielland-Brandt MC, Holmberg S. Substrate specificity and gene expression of the amino-acid permeases in Saccharomyces cerevisiae. Curr Genet. 1999;36:317–328. doi: 10.1007/s002940050506. [DOI] [PubMed] [Google Scholar]
- 12.Ozcan S, Johnston M. Function and regulation of yeast hexose transporters. Microbiol Mol Biol Rev. 1999;63:554–569. doi: 10.1128/mmbr.63.3.554-569.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andréasson C, Neve EP, Ljungdahl PO. Four permeases import proline and the toxic proline analogue azetidine-2-carboxylate into yeast. Yeast. 2004;21:193–199. doi: 10.1002/yea.1052. [DOI] [PubMed] [Google Scholar]
- 14.Chervitz SA, et al. Using the Saccharomyces Genome Database (SGD) for analysis of protein similarities and structure. Nucleic Acids Res. 1999;27:74–78. doi: 10.1093/nar/27.1.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sophianopoulou V, Diallinas G. Amino acid transporters of lower eukaryotes: Regulation, structure and topogenesis. FEMS Microbiol Rev. 1995;16:53–75. doi: 10.1111/j.1574-6976.1995.tb00155.x. [DOI] [PubMed] [Google Scholar]
- 16.Horák J. Yeast nutrient transporters. Biochim Biophys Acta. 1997;1331:41–79. doi: 10.1016/s0304-4157(96)00015-9. [DOI] [PubMed] [Google Scholar]
- 17.Zumbuehl A, Stano P, Heer D, Walde P, Carreira EM. Amphotericin B as a potential probe of the physical state of vesicle membranes. Org Lett. 2004;6:3683–3686. doi: 10.1021/ol0487276. [DOI] [PubMed] [Google Scholar]
- 18.Waheed AA, et al. Inhibition of HIV-1 replication by amphotericin B methyl ester: Selection for resistant variants. J Biol Chem. 2006;281:28699–28711. doi: 10.1074/jbc.M603609200. [DOI] [PubMed] [Google Scholar]
- 19.Guan XL, et al. Functional interactions between sphingolipids and sterols in biological membranes regulating cell physiology. Mol Biol Cell. 2009;20:2083–2095. doi: 10.1091/mbc.E08-11-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Opekarová M, Tanner W. Nystatin changes the properties of transporters for arginine and sugars. An in vitro study. FEBS Lett. 1994;350:46–50. doi: 10.1016/0014-5793(94)00730-6. [DOI] [PubMed] [Google Scholar]
- 21.Waheed AA, et al. Inhibition of human immunodeficiency virus type 1 assembly and release by the cholesterol-binding compound amphotericin B methyl ester: Evidence for Vpu dependence. J Virol. 2008;82:9776–9781. doi: 10.1128/JVI.00917-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang L, et al. Response of gene expression in Saccharomyces cerevisiae to amphotericin B and nystatin measured by microarrays. J Antimicrob Chemother. 2002;49:905–915. doi: 10.1093/jac/dkf001. [DOI] [PubMed] [Google Scholar]
- 23.Agarwal AK, et al. Genome-wide expression profiling of the response to polyene, pyrimidine, azole, and echinocandin antifungal agents in Saccharomyces cerevisiae. J Biol Chem. 2003;278:34998–35015. doi: 10.1074/jbc.M306291200. [DOI] [PubMed] [Google Scholar]
- 24.Gray KC, et al. Amphotericin primarily kills yeast by simply binding ergosterol. Proc Natl Acad Sci USA. 2012;109:2234–2239. doi: 10.1073/pnas.1117280109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szpilman AM, Cereghetti DM, Wurtz NR, Manthorpe JM, Carreira EM. Synthesis of 35-deoxy amphotericin B methyl ester: A strategy for molecular editing. Angew Chem Int Ed Engl. 2008;47:4335–4338. doi: 10.1002/anie.200800589. [DOI] [PubMed] [Google Scholar]
- 26.Szpilman AM, Manthorpe JM, Carreira EM. Synthesis and biological studies of 35-deoxy amphotericin B methyl ester. Angew Chem Int Ed Engl. 2008;47:4339–4342. doi: 10.1002/anie.200800590. [DOI] [PubMed] [Google Scholar]
- 27.Breukink E, et al. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science. 1999;286:2361–2364. doi: 10.1126/science.286.5448.2361. [DOI] [PubMed] [Google Scholar]
- 28.Hasper HE, et al. An alternative bactericidal mechanism of action for lantibiotic peptides that target lipid II. Science. 2006;313:1636–1637. doi: 10.1126/science.1129818. [DOI] [PubMed] [Google Scholar]
- 29.van Stelten J, Silva F, Belin D, Silhavy TJ. Effects of antibiotics and a proto-oncogene homolog on destruction of protein translocator SecY. Science. 2009;325:753–756. doi: 10.1126/science.1172221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pioletti M, et al. Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J. 2001;20:1829–1839. doi: 10.1093/emboj/20.8.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlünzen F, et al. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature. 2001;413:814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- 32.Ruepp A, et al. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 2004;32:5539–5545. doi: 10.1093/nar/gkh894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pel HJ, et al. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol. 2007;25:221–231. doi: 10.1038/nbt1282. [DOI] [PubMed] [Google Scholar]
- 34.Malínská K, Malínský J, Opekarová M, Tanner W. Visualization of protein compartmentation within the plasma membrane of living yeast cells. Mol Biol Cell. 2003;14:4427–4436. doi: 10.1091/mbc.E03-04-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.