Abstract
Transposable elements (TEs) are known to provide DNA for host regulatory functions, but the mechanisms underlying the transformation of TEs into cis-regulatory elements are unclear. In humans two TEs—MER20 and MER39—contribute the enhancer/promoter for decidual prolactin (dPRL), which is dramatically induced during pregnancy. We show that evolution of the strong human dPRL promoter was a multistep process that took millions of years. First, MER39 inserted near MER20 in the primate/rodent ancestor, and then there were two phases of activity enhancement in primates. Through the mapping of causal nucleotide substitutions, we demonstrate that strong promoter activity in apes involves epistasis between transcription factor binding sites (TFBSs) ancestral to MER39 and derived sites. We propose a mode of molecular evolution that describes the process by which MER20/MER39 was transformed into a strong promoter, called “epistatic capture.” Epistatic capture is the stabilization of a TFBS that is ancestral but variable in outgroup lineages, and is fixed in the ingroup because of epistatic interactions with derived TFBSs. Finally, we note that evolution of human promoter activity coincides with the emergence of a unique reproductive character in apes, highly invasive placentation. Because prolactin communicates with immune cells during pregnancy, which regulate fetal invasion into maternal tissues, we speculate that ape dPRL promoter activity evolved in response to increased invasiveness of ape fetal tissue.
Keywords: cis-regulatory evolution, human evolution, human pregnancy
In the pursuit to understand mechanisms underlying the evolution of gene regulation, it has become increasingly clear that transposable elements play a critical role in regulatory evolution by providing novel genetic elements, including new introns, promoters, enhancers, and insulators (1). In addition to individual examples showing that transposable elements (TEs) are associated with host regulatory function, computational work shows that TEs intersect with many promoter regions (2) and are overrepresented in predicted regulatory regions [(3, 4); see review by Feschotte (1)]. Many questions remain unanswered. How frequently are TEs actually co-opted for regulatory function? What is the adaptive significance of these events? Are TEs used by the host genome immediately upon insertion or do they require extensive sequence evolution before acquiring a biological role? What are the molecular changes that transform a transposable element into a cis-regulatory element?
In this paper we address the latter two questions posed above by investigating evolution of the decidual prolactin promoter in primates. In a recent paper we showed that the prolactin gene was independently recruited into uterine expression in primates, mice, and elephants by the co-option of different transposable elements, highlighting the frequency at which TEs can be recruited and their importance in gene regulatory innovation (5). Here we trace the evolutionary history and function of transposable elements in one of these groups—the primates—to understand precisely how uterine prolactin expression evolved via these elements, and why.
Prolactin is one of the most dramatically induced genes in the human endometrial decidua during pregnancy and one of the most abundant secretory products in amniotic fluid. The precise functions of decidual prolactin remain unclear, but in vitro it is known to cause the proliferation of and enhance the cytotoxicity of uterine natural killer (uNK) cells (6–8), immune cells that regulate fetal invasion into maternal tissues (9–12). Prolactin protein concentrations from human decidua are much higher than those from rhesus monkeys, and human decidual tissue from early in pregnancy has a much higher capacity to release prolactin in vitro (13, 14). Because there is a strong correlation between the rate of decidual prolactin (dPRL) gene expression and the rate of dPRL synthesis and secretion (15), it is likely that the gene is regulated differently at the transcriptional level in monkeys and humans.
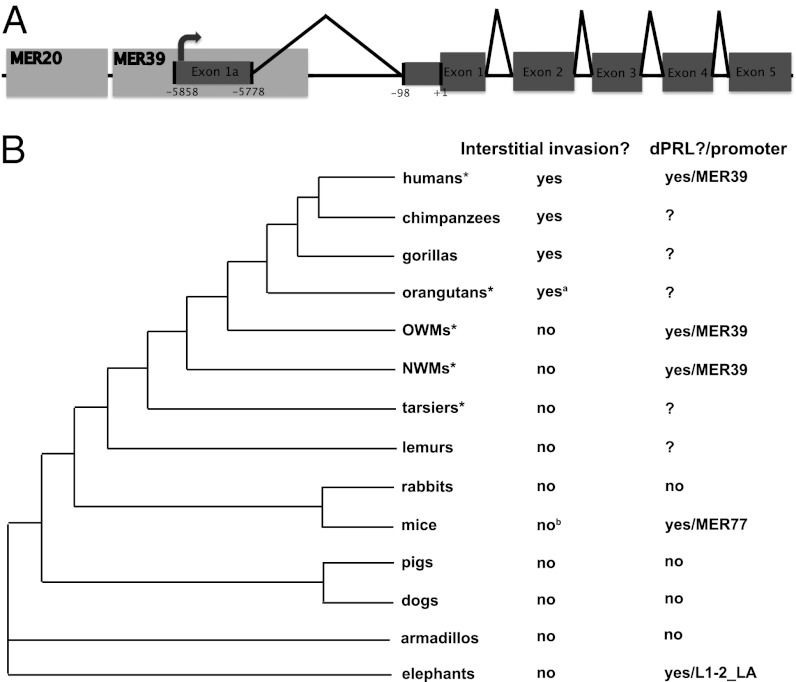
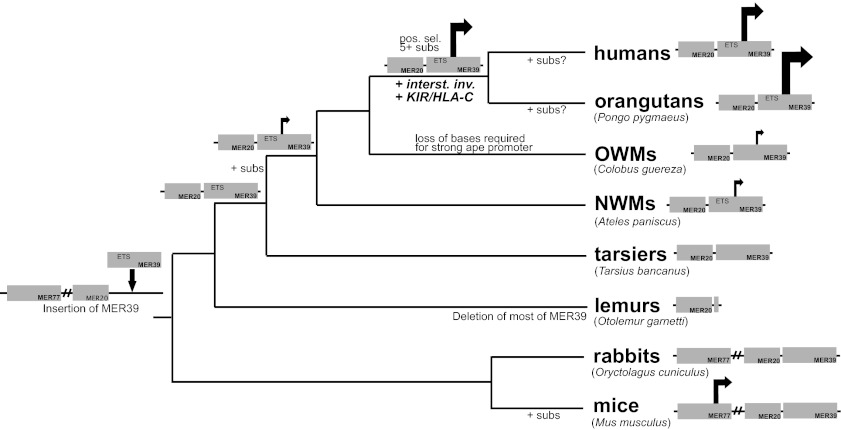
In humans, Old World monkeys (OWMs), and New World monkeys (NWMs), it is known that transcription of dPRL initiates from an alternative promoter than that used in the pituitary, located over 5 kb upstream of coding exon 1 (5). The enhancer/promoter region is derived from two transposable elements (5, 16) (Fig. 1A). A long-terminal repeat called MER39 contains the transcriptional start site (TSS) and is found at the prolactin locus in all primates and rodents (5) (Fig. 1). The DNA transposon MER20, located about 30 bp upstream of MER39, is common to all placental mammals (17) (Fig. 1A). Within this MER20/MER39 region there is a roughly 320-bp sequence shown to be critical for dPRL expression in humans and sufficient to mediate full inducibility by differentiation stimulus (18). Here we examine the evolution and function of the MER20/MER39 region, and show that transformation of MER39 into a strong promoter in humans was a multistep process that took millions of years. We demonstrate that epistatic interactions between ancestral transcription factor binding sites (TFBSs) in MER39 and derived sites are required for strong promoter activity in apes and humans. Finally, we propose a link between the genetic changes in the TE-derived dPRL promoter and a derived reproductive character in apes: interstitial invasion, i.e., highly invasive placentation (Fig. 1B).
Fig. 1.
Structure and expression of dPRL gene. (A) Human prolactin gene with transposable elements (MER20 and MER39) and the decidual transcriptional start site mapped. Black carets indicate splice junctions. Exons are shaded in dark gray; UTRs are shown as narrow boxes; and coding regions are shown as thicker boxes. (B) Eutherian phylogenetic tree with two characters mapped: interstitial invasion and presence/mechanism of expression of dPRL transcripts. Groups with an asterisk are those from which prolactin promoter reporter constructs were made in this study. aCarter (27) reports that orangutan specimens were not available to determine whether orangutan invasion proceeds through the interstitial route, but Mossman (38), Benirschke (39), and Gruenwald (28) describe fetal invasion in orangutans as interstitial and/or very invasive. bThere is some evidence for interstitial invasion in rats during the last part of pregnancy (40).
Results
Ape MER20/MER39 Region Is a Stronger Promoter than That of Nonapes.
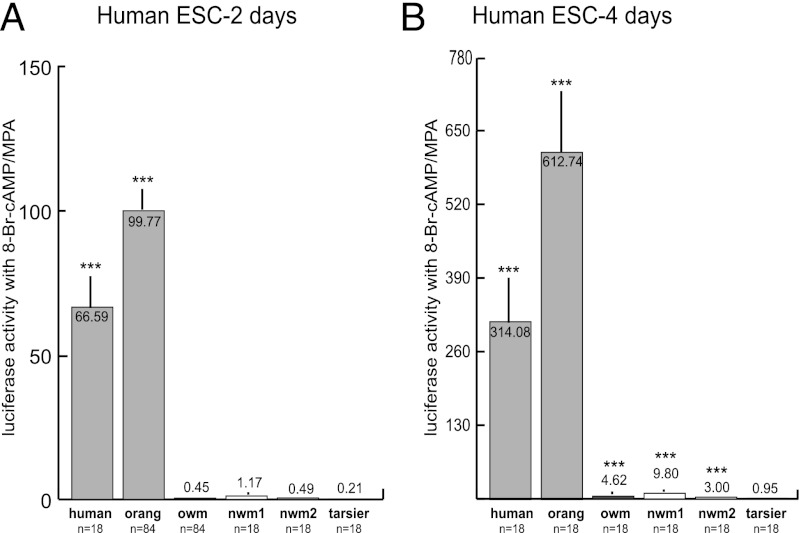
We first tested the MER20/MER39 region from a variety of primates for the ability to activate reporter gene expression in differentiated human endometrial stromal cells. When cells were left for 2 d in the differentiation treatment, only the human and orangutan promoters were significantly more active than the empty reporter vector, on average about 80× more active (Fig. 2A). After 4 d of hormone treatment, the promoters of all primates except the tarsier, the most basal primate tested, were significantly more active than the empty vector, although the ape promoters still drove dramatically stronger expression than those of the nonapes (Fig. 2B). To ensure that the difference between ape and nonape promoter activity was not due to transregulatory factors in human cells, the orangutan and OWM constructs were tested in rabbit endometrial stromal cells. The pattern in rabbit cells was the same as in human cells after 2 d of treatment, with the orangutan construct being ∼50× more active than that of the OWM (OWM construct: 3.31× activity of empty vector; orangutan construct: 150.33× activity of empty vector). Thus, differences between ape and nonape promoter activities are likely caused by differences in cis-regulatory elements.
Fig. 2.
MER20/MER39 promoter activity of various primates in endometrial stromal cells (ESC) treated with medroxyprogesterone acetate (MPA) and 8-bromoadenosine cyclic monophosphate (8-Br-cAMP). Activity in human ESC treated with MPA and 8-Br-cAMP for 2 d (A) and 4 d (B). Apes (human: Homo sapiens; orang: Pongo pygmaeus) are in light gray, Old World monkeys (owm: Colobus guereza) in dark gray, New World monkeys (nwm1: Ateles paniscus; nwm2: Cebus apella) in white, and tarsier (Tarsius bancanus) in black. Values are means ± SEM. ***P < 0.001 (t test) vs. activity of empty vector.
Accelerated Evolution of the dPRL Enhancer/Promoter Occurred in Stem Apes.
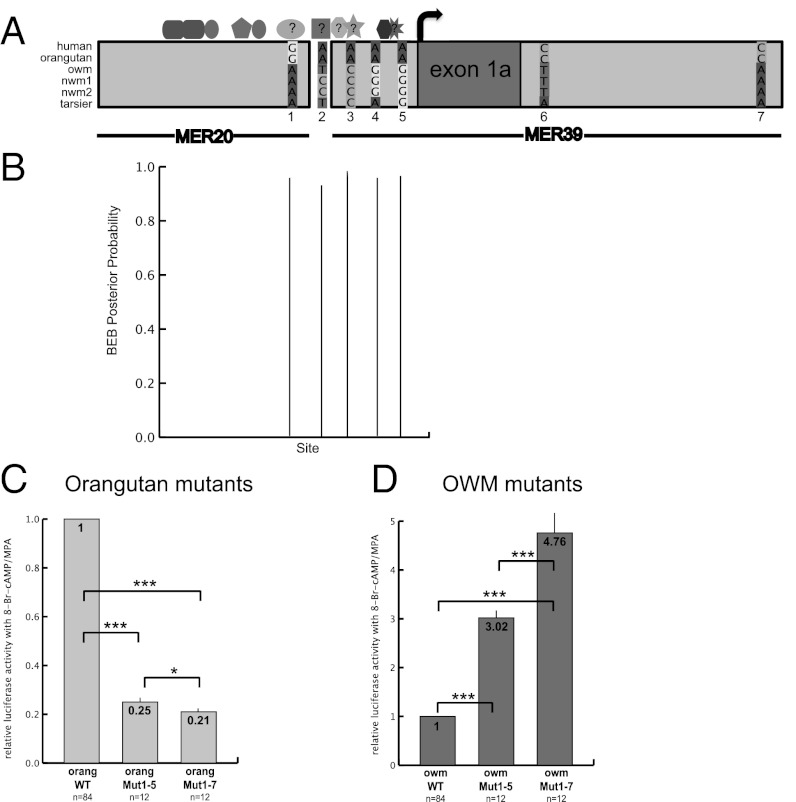
Next we analyzed the evolutionary history of the MER20/MER39 sequence to understand how these functional differences evolved. In a comparison of the promoters tested above and of additional primates with sequence available (Table S1), we noticed a cluster of five nucleotide sites in the region 200 bp upstream of the human TSS, which incurred changes in the ancestral ape lineage and are conserved among the apes we tested (Fig. 3A and Fig. S1). Outside of this cluster are only two other sites, downstream of the TSS in intron 1a, with changes in the stem lineage of apes (Fig. 3A).
Fig. 3.
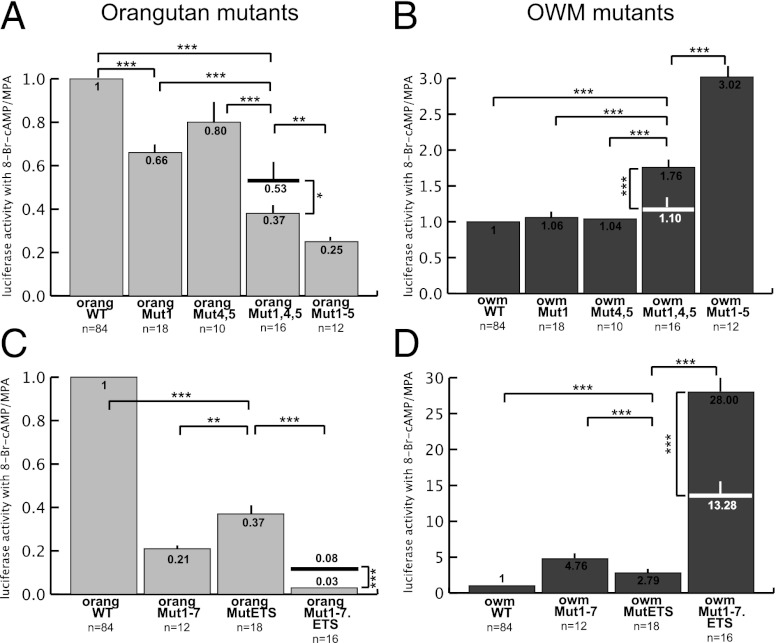
Molecular evolution of the MER20/MER39 sequence and resultant changes in dPRL promoter activity in apes. (A) Map showing location of seven nucleotide changes that occurred in the stem lineage of apes as well as known and hypothesized TFBSs. TFBSs: rounded rectangle, CEBPβ (41); narrow oval, FOX01a (41, 42); pentagon, HOXA11 (17, 42); wide oval, hypothesized site for SP1; square, hypothesized site for YY1; octagon, hypothesized site for progesterone receptor; five-point star, hypothesized site for RUNX1; hexagon, ETS1 (20); eight-point star, hypothesized site for p300. (B) Nucleotide sites that are under positive selection in the stem lineage of apes, identified by the Bayes empirical Bayes method. Only sites with high (>70%) posterior probabilities are shown. Note that sites 1–5 in A, which incurred changes in stem apes, are all under positive selection. (C and D) Activity (relative to wild type) of the mutated orangutan (C) and OWM (D) promoter reporter constructs with sites 1–5 (Mut1–5) and 1–7 (Mut1–7) from A mutated. The orangutan bases were mutated to resemble the OWM bases and vice versa. ***P < 0.001 and *P < 0.1 (t tests) vs. activity of indicated construct.
We were intrigued by the fact that five of the seven changes that occurred in stem apes are clustered in the characterized human enhancer/promoter region. We tested this region for evidence of positive selection using likelihood-based models that allow for rate variation among lineages in a phylogeny (Materials and Methods). We found marginal evidence for an accelerated substitution rate in the stem ape lineage [likelihood ratio test (LRT) P value = 0.063], and the Bayes empirical Bayes method identified the same five sites mentioned above as being under positive selection in the stem apes (Fig. 3B). No other lineages in the phylogeny showed evidence of an accelerated substitution rate in the dPRL enhancer/promoter region. It should be noted, however, that it is unknown if accelerated evolution of a noncoding region is the manner in which adaptive change occurs in regulatory DNA (19).
The orangutan and OWM sequences were analyzed to determine if any of the changes discussed above altered the TFBS profile of the ape promoter. Nucleotide change 1 adds an SP1 binding site to the ape TFBS profile; change 2 adds a YY1 binding site; change 3 adds a RUNX1 binding site; and changes 4 and 5 flank a well-characterized binding site for ETS1 in humans (Fig. 3A and Fig. S1) (20).
Evolution of the MER20/MER39 Sequence Increased dPRL Promoter Activity in Apes.
To determine if any of the substitutions discussed above changed promoter activity in apes, we mutated these sites in the orangutan promoter construct to resemble those of the OWM (referred to as back mutations) and vice versa (referred to as forward mutations). We tested the activity of the mutated constructs in human endometrial stromal cells as above (Fig. 3 C and D). The five back mutations together reduced orangutan promoter activity to 25% of wild type (orangMut1–5 in Fig. 3C); the five forward mutations increased OWM promoter activity by a factor of 3 (owmMut1–5 in Fig. 3D).
These five substitutions only partially explain the difference between orangutan and OWM promoter activity: the orangutan wild-type promoter is over 200× more active than the OWM wild-type promoter (Fig. 2). To more completely reproduce this expression pattern, we next added the two sites outside the cluster of five (changes 6 and 7 in Fig. 3A) that incurred changes in the stem apes. The addition of these mutations to the orangutan construct reduced activity from 25% to 21% of orangutan wild type (orangMut1–7 in Fig. 3C); the addition to the OWM construct increased activity from 3× to ∼5× the activity of OWM wild type (owmMut1–7 in Fig. 3D).
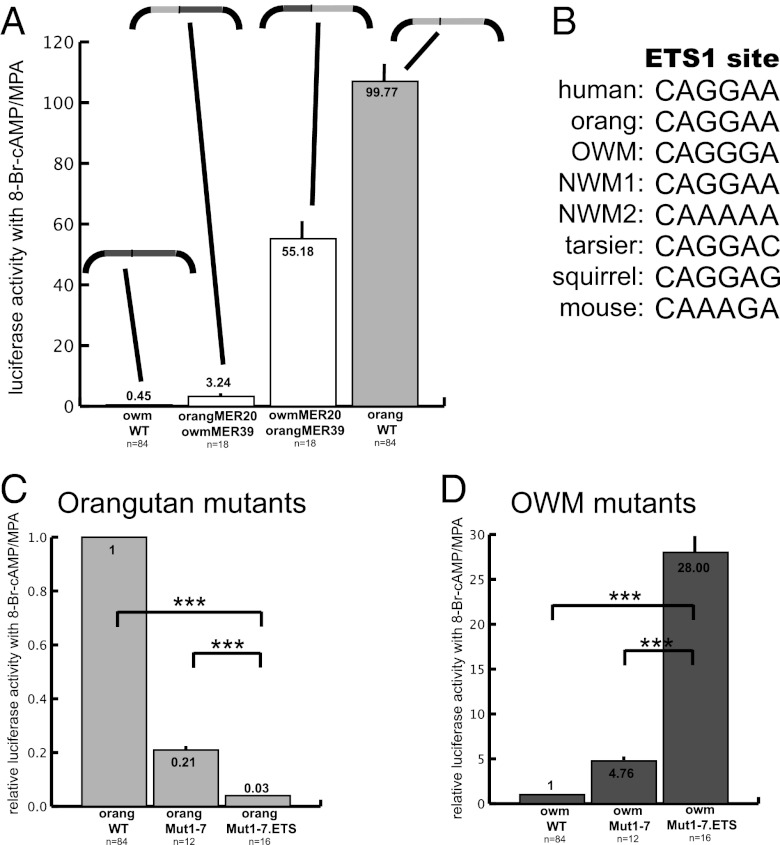
Including the seven ape-derived sites tested above, the orangutan and OWM sequences have 39 differences between them (36 SNPs and three insertions/deletions; Fig. S1). To aid in the decision of which of these sites to test next, we swapped the MER20 and MER39 elements between the orangutan and OWM constructs (Fig. S2), and tested them as above. The OWM MER20/orangutan MER39 chimeric promoter was ∼55× more active than the empty vector, whereas the orangutan MER20/OWM MER39 promoter was only 3× more active (Fig. 4A). Because the orangutan MER39 directs stronger promoter activity, we chose to look in MER39 for additional substitutions that could also be involved in the difference between orangutan and OWM promoter activities.
Fig. 4.
Contribution of MER20 vs. MER39 to activity of ape dPRL promoter. (A) Activity of chimeric MER20–MER39 promoters from orangutan and OWM. Structure of each promoter–reporter construct tested is indicated above activity level: light gray indicates region is derived from orangutan and dark gray from OWM; black represents the reporter construct. (B) Core ETS1 sequence in apes, and orthologous positions in other primates and rodents. (C and D) Activity of mutated orangutan (C) and OWM (D) promoters (Mut1–7) with additional ETS1 site mutated (Mut1–7.ETS). Note that the ETS1 site and sites 3–7 (Fig. 3A) are in MER39. ***P < 0.001 (t test) vs. activity of indicated construct. Significant differences shown in Fig. 3 are not repeated.
We next tested an ETS1 site located within MER39, with dramatic results. This site was chosen because previous work showed it to be essential for dPRL expression in humans (20). The ETS1 binding site (CAGGAA) is located 3 bp downstream of site 4 and 11 bp upstream of site 5 (Fig. 3A and Fig. S1). The core ETS1 site is conserved in all apes but variably present in nonapes (and absent from the OWM species we tested; Fig. 4B and Discussion). In fact, it is likely that the ETS1 site was present upon insertion of MER39 at the PRL locus because the core ETS1 binding site is present in a consensus of MER39 sequences from the human genome (Fig. S3A). The back mutation on the orangMut1–7 construct (A to G mutation in position 5 of the core site) reduced promoter activity from 21% to 3% of the wild type (orangMut1–7.ETS in Fig. 4C). The forward ETS1 mutation on the owmMut1–7 construct increased activity from 5× to 28× the activity of wild type (owmMut1–7.ETS in Fig. 4D).
Epistasis Between Ancestral and Evolved Sites Contributes to Activity of the Ape dPRL Promoter.
To estimate the contributions of sites 1–7 and the ETS1 site to ape dPRL promoter activity, we tested some of the sites alone and/or in smaller groups (Fig. 5). Mutation of site 1 alone did significantly change OWM promoter activity but significantly reduced orangutan activity to 66% of wild type (Fig. 5 A and B). Mutation of sites 4 and 5 together did not significantly change activity of the OWM or orangutan promoters (Fig. 5 A and B). The combination of mutations 1, 4, and 5 significantly increased OWM promoter activity to 1.76× wild type (Fig. 5B), and reduced orangutan to 37% of wild type, whereas the expected activity of both mutations combined, without epistasis on the multiplicative scale, would be 53% (Fig. 5A). Thus, the impact of mutation 1 and mutations 4 and 5 are clearly dependent on each other, because there is not a significant change from OWM wild type when the mutations are alone, but there is when they are together, and the combination in orangutan causes a significantly greater reduction in activity than the addition of the two sets alone. These data also show that mutations 2 and/or 3 are involved in the difference between orangutan and OWM promoter activity, because the activities of Mut1,4,5 and Mut1–5 are significantly different for both the orangutan and OWM (Fig. 5 A and B).
Fig. 5.
Epistasis between sites in the dPRL enhancer/promoter of orangutan (A and C) and OWM (B and D). Comparison of effects of mutation 1; mutations 4 and 5; mutations 1, 4, and 5; and mutations 1–5 (A and B). Comparison of effects of ETS1 mutation, mutations 1–7, and mutations 1–7 plus the ETS1 mutation (C and D). ***P < 0.001; **P < 0.01; and *P < 0.1 (t tests) vs. activity of indicated construct. Significant differences shown in Figs. 3 and 4 are not repeated. The expected activity of mutations combined, without epistasis on the multiplicative scale, is indicated on the columns showing actual activity of combined mutations. Note that mutations are partly dependent on each other, because the expected effect of individual mutations combined is significantly different from the actual effect of all mutations together.
We see a similar pattern of epistasis with the ETS1 site. Mutation of the ETS1 site alone in the OWM increased activity of the promoter 2.8× wild type; mutation of sites 1–7 increased activity 4.8×; and the two sets together increased activity 28× (Fig. 5D). There is an epistatic relationship between these two sets of mutations, because you would expect a 13.3-fold increase on the multiplicative scale if they acted independently of each other. In the orangutan, mutation of the ETS1 site alone reduced activity to 37% of wild type; mutation of sites 1–7 reduced activity to 21%; and the two sets together reduced activity to 3% of wild type (Fig. 5C). If the two sets of mutations were multiplicative, as expected without interaction, promoter activity would have been 8%, which is significantly higher than what we found.
From these data we conclude that at least five sites are involved in the difference between orangutan and OWM promoter activity: site 1, site 2 and/or 3, site 4 and/or 5, site 6 and/or 7 (all derived sites in apes), and the ETS1 site (an ancestral site in apes). These sites contribute in an epistatic way, because the product of fold changes of individual sites/small groups does not equal the fold change of the combination. The deviations from expectation are significant, and range from 1.4- to 2.7-fold. Finally, it is apparent that more sites that were not tested here are involved. The most parsimonious explanation for the remaining difference in promoter activities is that additional bases required for strong orangutan activity were present in the ancestor of OWMs and apes but changed along the OWM lineage (or the lineage of the OWM species we tested). A non-mutually exclusive possibility is that additional mutations occurred on both the human and orangutan lineages that independently increased activity of the promoter in both groups (Fig. 6).
Fig. 6.
Summary of the origin and evolution of the MER20/MER39-derived dPRL enhancer/promoter. TEs MER20, MER39, and MER77 are indicated. Note that MER77 is present at the prolactin locus in all species on this tree, even if not depicted. An arrow indicates that a region has promoter activity; size of arrow indicates strength of promoter. Note that strength of mouse promoter has not been tested but location has been determined; rabbits do not express decidual prolactin, so their MER20/MER39 region is assumed here to lack a promoter (5). Also note that the presence of decidual prolactin transcripts has not been tested in lemurs. Key events are indicated, such as insertion or deletion of TEs, positive selection (pos. sel.), substitutions that changed activity of the promoter (+ subs), evolution of interstitial invasion (+ interst. inv.), and interaction between killer immunoglobulin-like receptor and major histocompatability complex class I antigen HLA-C (+ KIR/HLA-C).
Discussion
In this study we examine the evolutionary history and function of the transposable element-derived dPRL promoter in primates to understand mechanisms of cis-regulatory evolution by transposable elements. Our previous work on decidual prolactin expression and the work reported here shows that the MER20/MER39 region was not a promoter upon insertion of MER39 (Fig. 6); it evolved to be a weak promoter in monkeys and then a strong promoter in apes (Fig. 6). Most of our work here elucidates the mechanisms underlying the evolution from a weak promoter in monkeys to a strong promoter in apes. By mutagenesis experiments we show that at least four of seven ape-derived substitutions contributed to increased promoter activity in the apes. We show that an ETS1 site that was previously determined to be critical for decidual prolactin expression in humans is a key player promoting strong ape dPRL promoter activity (20). We also demonstrate positive/synergistic epistatic interactions between many of the individual sites, with epistatic effects ranging from 1.4- to 2.7-fold of the expectation (Fig. 5).
The ETS1 site seems to be the most critical of the sites tested for activity of the ape dPRL promoter, because the forward and back mutations alone caused large changes in activity of the promoter (Fig. 5). As mentioned above, the core ETS1 site is conserved in apes but variable in nonapes: one NWM we tested (Ateles paniscus) has the core site, whereas the other (Cebus apella) does not; also, the OWM we tested, Colobus guereza, does not have an intact site, but rhesus macaques do (Fig. 4B). Interestingly, Ateles paniscus has stronger promoter activity than the other nonapes we tested that lack the ETS1 site (Fig. 2). Nevertheless, the ETS1 site must interact with other sites to direct strong activity at this promoter, because Ateles paniscus still has very weak promoter activity relative to the apes (Fig. 2), and addition of the ETS1 site in Colobus guereza increases activity of the promoter less than threefold, compared with the almost 30-fold increase we saw with the combination of all mutations (Fig. 5). Thus, it appears that stabilization of the ETS1 site, a key player at the ape enhancer/promoter, required substitutions to nearby interacting sites.
We call the mode of molecular evolution described above “epistatic capture.” Epistatic capture is the process by which a TFBS, like the ETS1 site in MER39, comes under increased purifying selection due to epistatic interactions with derived TFBSs. Thus, a TFBS that is present in a TE upon insertion but variable up to a certain point in a phylogeny becomes stabilized as a result of new interactions with derived sites. Epistasis has previously been observed in other cis-regulatory regions—e.g., in an enhancer of the shavenbaby gene in Drosophila—where many subtle-effect substitutions with epistatic effects were involved in its loss of function in one species (21). The loss of enhancer function in that study was shown to be involved in morphological evolution of Drosophila larvae, although the study did not explore how epistasis evolved or how it affected the evolutionary process (21). Another example of epistasis in evolution is found in glucocorticoid receptor (GR) evolution (22, 23). Two different patterns have been observed: an “epistatic ratchet” (23) and what we call here epistatic capture (22). In the epistatic ratchet, amino acids that acted as epistatic modifiers necessary for evolution to the derived GR state are conserved in ancestral GRs but variable in those with the derived structure/function, making it difficult to reevolve the protein to its ancestral state (23). In addition, some amino acids show a pattern of variation that is identical to that observed here, where residues become less variable because of epistasis with derived amino acids (22).
Epistatic capture, as we define it, is not required to occur within a TE but it may be particularly applicable to TEs. Upon insertion at the prolactin locus, MER20 and MER39 provided many near-perfect or perfect TFBSs that would later allow for strong activity of the promoter. As mentioned previously, the ETS1 site was likely present upon insertion of MER39 at the PRL locus. Also, the hypothesized SP1 binding site in which site 1 is located has a core GGCAG motif; other than the A-to-G change that occurred in stem apes (Fig. 3A), this core site is present in the consensus MER20 sequence, suggesting that a close TFBS was present upon insertion of the TE (Fig. S3B). The spatial arrangement of these TFBSs probably also facilitated evolution of the promoter, because many of them are located in close proximity to each other and the factors that (may) bind to them are known to interact in other contexts (e.g., ETS1 with RUNX1 and SP1) (24, 25). It is possible that epistatic capture is a common mechanism by which TEs are converted into functional regulatory elements (26), because TEs are often full of perfect and near-perfect binding sites (1).
Though we have answered the question of how the ape decidual prolactin promoter became so active, the question of why high PRL expression evolved in apes remains. The marginal evidence of positive selection on the dPRL promoter region in the stem lineage of apes suggests that there may have been an evolutionary force driving the increase in promoter activity. We note here that increased invasiveness of the ape placenta coincided with the increase in promoter activity. As shown in Figs. 1 and 6, fetal invasion in humans and other apes is extremely invasive (referred to as interstitial invasion). Fetal placental cells invade the ape uterus via endovascular and interstitial routes, in contrast to most other mammals, including monkeys, in which migration of fetal cells mainly occurs in arteries (27). Interstitial invasion allows the ape fetus to remodel maternal vasculature more extensively than monkeys and other mammals (27, 28). As a consequence of these differences, placentation in the apes is deep and potentially dangerous. In humans, for example, pregnancies that occur in the fallopian tubes or on uterine scar tissue can lead to hemorrhaging and death. Though it is unclear why this aggressive form of placentation evolved, many hypotheses have been put forth, including selection for increased nutrient transfer to the fetus and a larger brain (12, 29).
In normal human pregnancies fetal invasion is restrained by the endometrial decidua, which consists of differentiated endometrial stromal cells, uNK cells, and an extensive network of vascular tissue. Decidual stromal cells produce many products that inhibit the activity of invasive fetal proteins (30, 31), and they produce molecules that signal to uNK cells, which are known to regulate placental invasion in humans among other functions (9–12). Prolactin is one such product of decidual cells: it has been shown to increase proliferation of uNK cells and increase their cytotoxicity in vitro (6–8). It is therefore plausible that as interstitial invasion evolved in the stem lineage of apes, one maternal response was to up-regulate expression of prolactin. This hypothesis remains speculative without further evidence. It is interesting to note, however, that the receptor–ligand interaction that allows uNKs to inhibit placental cells also evolved in the stem lineage of apes. HLA-C molecules on placental cells, and the lineage of KIR genes expressed by uNKs that recognize HLA-C, are only present in the great apes and not in monkeys or more basal primates (12, 27). Thus, one maternal response to increased aggressiveness of the ape placenta may have been the inhibition of placental cells by uNK cells. A second response may have been to up-regulate prolactin expression to enhance the uNK response.
Materials and Methods
Reporter Constructs.
Genomic DNA was obtained from the following species: Homo sapiens (human), Pongo pygmaeus (orangutan), Colobus guereza (mantled guereza, an OWM), Cebus apella (tufted capuchin, a NWM), Ateles paniscus (red-faced spider monkey, a NWM), and Tarsius bancanus (western tarsier). Primers were designed to span MER20 and MER39 for each species, using the annotated genome for that species or that of a closely related species (Table S2). PCR products were cloned into the pGL4.70 vector (Promega). PGL4.23 (Promega) was used as the internal control.
Cells.
Immortalized human endometrial stromal cells (ATCC CRL-4003) were used for most of the experiments and maintained in hormone-free media with 10% charcoal-stripped FBS. Primary rabbit endometrial stromal cells were harvested for some of the transient transfection experiments using the protocol from Bigsby (32), and maintained in the same media as the human cells.
Transient Transfection Experiments.
Cells were plated at a density of 6.0 × 104 cells/well in 24-well plates in media with 10% charcoal-stripped FBS. A total of 250 ng of the promoter reporter constructs were cotransfected with 25 ng of the control vector using Lipofectamine LTX reagent (Invitrogen). Six hours later, media was changed to one containing 2% charcoal-stripped FBS. At 12 h later, the media was changed to one containing 10−6 M medroxyprogesterone acetate (MPA, a stable synthetic progestin) and 0.5 mM 8-bromoadenosine cyclic monophosphate (8-Br-cAMP, a stable cAMP analog). At 48 or 96 h after treatment, a luciferase assay was performed using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. Cells harvested at 96 h had a media change at 48 h. Renilla luciferase activity was normalized to firefly luciferase activity. Transfections were performed with four or six replicates, and experiments were performed at least twice, usually three or more times. Values are means ± SEM.
Molecular Evolution of dPRL Enhancer/Promoter.
MER20 and MER39 sequences were retrieved from Ensembl (http://www.ensembl.org/). Sequences were aligned using ClustalW2 (33) and adjusted by eye. Consensus MER20 and MER39 sequences were retrieved from Repbase (34).
The prolactin promoter region (chr6: 22,303,070–22,303,383 on human GRCh37 assembly) was tested for evidence of positive selection using likelihood-based models implemented in an updated version of the software EvoNC (35), which allows for rate variation among lineages in the phylogeny (O. Fedrigo, Duke University, Durham, NC; software available at http://www.duke.edu/∼ofedrigo/Olivier_Fedrigo/HyPhyScripts.html). This test identifies lineage-specific accelerated substitution rates in noncoding sequences, relative to a nearby neutrally evolving sequence. We used the synonymous substitution rate in the prolactin coding region as the neutral proxy for the promoter region, and we included all primates and rodents with available and alignable promoter and coding sequences in the analysis (Table S1). A LRT was performed comparing the null hypothesis (no positive selection allowed in the stem apes) to the alternative (positive selection allowed). A χ2 test with 1 degree of freedom was used to determine the significance of the LRT. The Bayes empirical Bayes method was used to identify sites under positive selection.
Identification of TFBSs in the dPRL Enhancer/Promoter.
Potential TFBS were identified using the MATCH tool (http://www.biobase-international.com/gene-regulation), which uses a library of TRANSFAC binding-site matrices. Only TFBS matches with >90% identity to the core binding site motif are reported.
Mutagenesis of dPRL Enhancer/Promoter.
Mutations of the promoter reporter constructs were made using the QuikChange Lightning Kit (Stratagene). Mutated reporter constructs were transfected into human endometrial stromal cells and tested for luciferase activity as above. Activities of the mutant constructs are presented as fold changes relative to wild type.
MER20/MER39 Swaps.
Chimeric primate MER20/MER39 promoters were made using an overlap extension PCR technique described in Wurch et al. (36). Fig. S2 summarizes the technique. Primers were designed according to the recommendations in Wurch et al. (36).
Statistics.
A two-sample t test was performed to determine if interactions exist between activities of different mutations. The expected activity of combined mutations was determined by multiplying mean fold changes of individual mutations together. The default expectation without interaction is multiplicative, because fold changes are defined as multiplicative quantities and thus are expected to accumulate multiplicatively if there is no interaction (epistasis). Hence the multiplicative model of nonepistatic effects is a consequence of the scale-type measures expressed as fold changes (37).
To determine the SD and SEM for the expected activity of combined mutations, we estimated the propagation of error from individual experiments onto the product using the following equations: SD 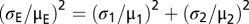 and SEM
and SEM 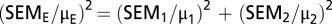 .
.
These equations are approximations based on error propagation theory and assume that there is no correlation between measurements for each set of mutations. For a comparison of the expected and actual activity of combined mutations, a two-sample t test was performed. The number of cases for the expected activity of combined mutations was estimated as follows: 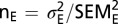 .
.
Supplementary Material
Acknowledgments
We thank Dr. Birgit Gellersen for advice on the transfection experiments and Dr. Derek Wildman for DNA samples. This study was supported by John Templeton Foundation Grant 12793.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118566109/-/DCSupplemental.
References
- 1.Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jordan IK, Rogozin IB, Glazko GV, Koonin EV. Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet. 2003;19:68–72. doi: 10.1016/s0168-9525(02)00006-9. [DOI] [PubMed] [Google Scholar]
- 3.Gentles AJ, et al. Evolutionary dynamics of transposable elements in the short-tailed opossum Monodelphis domestica. Genome Res. 2007;17:992–1004. doi: 10.1101/gr.6070707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lynch VJ, Leclerc RD, May G, Wagner GP. Transposon-mediated rewiring of gene regulatory networks contributed to the evolution of pregnancy in mammals. Nat Genet. 2011;43:1154–1159. doi: 10.1038/ng.917. [DOI] [PubMed] [Google Scholar]
- 5.Emera D, et al. Convergent evolution of endometrial prolactin expression in primates, mice, and elephants through the independent recruitment of transposable elements. Mol Biol Evol. 2012;29:239–247. doi: 10.1093/molbev/msr189. [DOI] [PubMed] [Google Scholar]
- 6.Gubbay O, Critchley HOD, Bowen JM, King A, Jabbour HN. Prolactin induces ERK phosphorylation in epithelial and CD56(+) natural killer cells of the human endometrium. J Clin Endocrinol Metab. 2002;87:2329–2335. doi: 10.1210/jcem.87.5.8515. [DOI] [PubMed] [Google Scholar]
- 7.Sun R, Li AL, Wei HM, Tian ZG. Expression of prolactin receptor and response to prolactin stimulation of human NK cell lines. Cell Res. 2004;14:67–73. doi: 10.1038/sj.cr.7290204. [DOI] [PubMed] [Google Scholar]
- 8.Chen YZ, Zhuang YL, Chen XY, Huang LL. Effect of human endometrial stromal cell-derived conditioned medium on uterine natural killer (uNK) cells’ proliferation and cytotoxicity. Am J Reprod Immunol. 2011;65:589–596. doi: 10.1111/j.1600-0897.2010.00955.x. [DOI] [PubMed] [Google Scholar]
- 9.Acar N, Ustunel I, Demir R. Uterine natural killer (uNK) cells and their missions during pregnancy: a review. Acta Histochem. 2011;113:82–91. doi: 10.1016/j.acthis.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 10.von Rango U, et al. Apoptosis of extravillous trophoblast cells limits the trophoblast invasion in uterine but not in tubal pregnancy during first trimester. Placenta. 2003;24:929–940. doi: 10.1016/s0143-4004(03)00168-1. [DOI] [PubMed] [Google Scholar]
- 11.von Rango U, Classen-Linke I, Kertschanska S, Kemp B, Beier HM. Effects of trophoblast invasion on the distribution of leukocytes in uterine and tubal implantation sites. Fertil Steril. 2001;76:116–124. doi: 10.1016/s0015-0282(01)01859-3. [DOI] [PubMed] [Google Scholar]
- 12.Moffett A, Loke C. Immunology of placentation in eutherian mammals. Nat Rev Immunol. 2006;6:584–594. doi: 10.1038/nri1897. [DOI] [PubMed] [Google Scholar]
- 13.Maslar IA, Kaplan BM, Luciano AA, Riddick DH. Prolactin production by the endometrium of early human pregnancy. J Clin Endocrinol Metab. 1980;51:78–83. doi: 10.1210/jcem-51-1-78. [DOI] [PubMed] [Google Scholar]
- 14.Maslar IA, Lazur JJ, Norman RL, Spies HG. Amniotic fluid and decidual prolactin during pregnancy in rhesus macaques. Biol Reprod. 1988;38:1067–1076. doi: 10.1095/biolreprod38.5.1067. [DOI] [PubMed] [Google Scholar]
- 15.Wu WX, Brooks J, Glasier AF, McNeilly AS. The relationship between decidualization and prolactin mRNA and production at different stages of human pregnancy. J Mol Endocrinol. 1995;14:255–261. doi: 10.1677/jme.0.0140255. [DOI] [PubMed] [Google Scholar]
- 16.Gerlo S, Davis JRE, Mager DL, Kooijman R. Prolactin in man: A tale of two promoters. Bioessays. 2006;28:1051–1055. doi: 10.1002/bies.20468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lynch VJ, et al. Adaptive changes in the transcription factor HoxA-11 are essential for the evolution of pregnancy in mammals. Proc Natl Acad Sci USA. 2008;105:14928–14933. doi: 10.1073/pnas.0802355105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Telgmann R, Maronde E, Taskén K, Gellersen B. Activated protein kinase A is required for differentiation-dependent transcription of the decidual prolactin gene in human endometrial stromal cells. Endocrinology. 1997;138:929–937. doi: 10.1210/endo.138.3.5004. [DOI] [PubMed] [Google Scholar]
- 19.Hahn MW. Detecting natural selection on cis-regulatory DNA. Genetica. 2007;129:7–18. doi: 10.1007/s10709-006-0029-y. [DOI] [PubMed] [Google Scholar]
- 20.Brar AK, Kessler CA, Handwerger S. An Ets motif in the proximal decidual prolactin promoter is essential for basal gene expression. J Mol Endocrinol. 2002;29:99–112. doi: 10.1677/jme.0.0290099. [DOI] [PubMed] [Google Scholar]
- 21.Frankel N, et al. Morphological evolution caused by many subtle-effect substitutions in regulatory DNA. Nature. 2011;474:598–603. doi: 10.1038/nature10200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ortlund EA, Bridgham JT, Redinbo MR, Thornton JW. Crystal structure of an ancient protein: Evolution by conformational epistasis. Science. 2007;317:1544–1548. doi: 10.1126/science.1142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bridgham JT, Ortlund EA, Thornton JW. An epistatic ratchet constrains the direction of glucocorticoid receptor evolution. Nature. 2009;461:515–519. doi: 10.1038/nature08249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arman M, et al. The human CD6 gene is transcriptionally regulated by RUNX and Ets transcription factors in T cells. Mol Immunol. 2009;46:2226–2235. doi: 10.1016/j.molimm.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 25.Santiago FS, Khachigian LM. Ets-1 stimulates platelet-derived growth factor A-chain gene transcription and vascular smooth muscle cell growth via cooperative interactions with Sp1. Circ Res. 2004;95:479–487. doi: 10.1161/01.RES.0000141135.36279.67. [DOI] [PubMed] [Google Scholar]
- 26.Emera D, Wagner GP. Transposable element recruitments in the mammalian placenta: Impacts and mechanisms. Brief Funct Genomics. 2012 doi: 10.1093/bfgp/els013. in press. [DOI] [PubMed] [Google Scholar]
- 27.Carter AM. Comparative studies of placentation and immunology in non-human primates suggest a scenario for the evolution of deep trophoblast invasion and an explanation for human pregnancy disorders. Reproduction. 2011;141:391–396. doi: 10.1530/REP-10-0530. [DOI] [PubMed] [Google Scholar]
- 28.Gruenwald P. Expansion of placental site and maternal blood supply of primate placentas. Anat Rec. 1972;173:189–203. doi: 10.1002/ar.1091730208. [DOI] [PubMed] [Google Scholar]
- 29.Haig D. Genetic conflicts in human pregnancy. Q Rev Biol. 1993;68:495–532. doi: 10.1086/418300. [DOI] [PubMed] [Google Scholar]
- 30.Esadeg S, He H, Pijnenborg R, Van Leuven F, Croy BA. Alpha-2 macroglobulin controls trophoblast positioning in mouse implantation sites. Placenta. 2003;24:912–921. doi: 10.1016/s0143-4004(03)00148-6. [DOI] [PubMed] [Google Scholar]
- 31.Floridon C, et al. Does plasminogen activator inhibitor-1 (PAI-1) control trophoblast invasion? A study of fetal and maternal tissue in intrauterine, tubal and molar pregnancies. Placenta. 2000;21:754–762. doi: 10.1053/plac.2000.0573. [DOI] [PubMed] [Google Scholar]
- 32.Bigsby RM. Progesterone induces a secretory protein in cultured rabbit endometrial stromal cells. J Steroid Biochem. 1986;25:937–942. doi: 10.1016/0022-4731(86)90326-2. [DOI] [PubMed] [Google Scholar]
- 33.Larkin MA, et al. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 34.Jurka J, et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 35.Wong WSW, Nielsen R. Detecting selection in noncoding regions of nucleotide sequences. Genetics. 2004;167:949–958. doi: 10.1534/genetics.102.010959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wurch T, Lestienne F, Pauwels PJ. A modified overlap extension PCR method to create chimeric genes in the absence of restriction enzymes. Biotechnol Tech. 1998;12:653–657. [Google Scholar]
- 37.Houle D, Pélabon C, Wagner GP, Hansen TF. Measurement and meaning in biology. Q Rev Biol. 2011;86:3–34. doi: 10.1086/658408. [DOI] [PubMed] [Google Scholar]
- 38.Mossman H. Vertebrate Fetal Membranes: Comparative Ontogeny and Morphology, Evolution, Phylogenetic Significance, Basic Functions, Research Opportunities. New Brunswick, NJ: Rutgers Univ Press; 1987. [Google Scholar]
- 39.Benirschke K. 2011. Comparative Placentation. Available at http://placentation.ucsd.edu/homefs.html.
- 40.Vercruysse L, Caluwaerts S, Luyten C, Pijnenborg R. Interstitial trophoblast invasion in the decidua and mesometrial triangle during the last third of pregnancy in the rat. Placenta. 2006;27:22–33. doi: 10.1016/j.placenta.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 41.Christian M, et al. Cyclic AMP-induced forkhead transcription factor, FKHR, cooperates with CCAAT/enhancer-binding protein beta in differentiating human endometrial stromal cells. J Biol Chem. 2002;277:20825–20832. doi: 10.1074/jbc.M201018200. [DOI] [PubMed] [Google Scholar]
- 42.Lynch VJ, Brayer K, Gellersen B, Wagner GP. HoxA-11 and FOXO1A cooperate to regulate decidual prolactin expression: Towards inferring the core transcriptional regulators of decidual genes. PLoS ONE. 2009;4:e6845. doi: 10.1371/journal.pone.0006845. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.