Abstract
The gut microbiota influences numerous aspects of human biology. One facet that has not been thoroughly explored is its impact on the host proteome. We hypothesized that the microbiota may produce certain of its effects through covalent modification of host proteins. We focused on protein lysine ε-acetylation because of its recently discovered roles in regulation of cell metabolism, and the potential for products of microbial fermentation to interact with the lysine acetylation machinery of host cells. Germ-free mice, fed a 15N-labeled diet for two generations, were colonized as adults with a microbiota harvested from conventionally raised mouse donors. Using high-resolution mass spectrometry, we quantified 3,891 liver and proximal colonic proteins, 558 of which contained 1,602 sites of lysine acetylation, 43% not previously described. Multiple proteins from multiple subcellular compartments underwent microbiota-associated increases in their levels of lysine acetylation at one or more residues, in one or both tissues. Acetylated proteins were enriched in functions related to energy production, respiration, and primary metabolism. A number of the acetylation events affect lysine residues at or near the active sites of enzymes, whereas others occur at locations that may affect other facets of protein function. One of these modifications, affecting Lys292 in mouse α-1-antitrypsin, was detected in the corresponding lysine of the human serum protein. Methods described in this report can be applied to other co- or posttranslational modifications, and add quantitation of protein expression and covalent modification to the arsenal of techniques for characterizing the dynamic, important interactions between gut symbionts and their hosts.
Keywords: gut microbiome, quantitative proteomics, spirulina, stable isotopic labeling of mammals
The human intestine is home to a community of tens of trillions of microbes belonging to all three domains of life. The gut microbiota, alone and in concert with the host, produces a wide variety of metabolites in a variety of different dietary contexts, including the products of protein putrefaction (indoles and cresols), polysaccharide fermentation (short-chain fatty acids; SCFA), and lipid metabolism (bile acids and derivatives of phosphatidylcholine) (1–4). These and other, as yet uncharacterized, metabolic products impact numerous aspects of our physiology (5).
Insights concerning the effects of the gut microbiota on host biology have come from comparisons of germ-free (GF) mice with mice that have been exposed to microbes from their environment beginning at birth [“conventionally raised” (CONV-R) animals] or mice that were born GF and at a selected point in their life receive a gut microbiota transplant from CONV-R donors [“conventionalized” (CONV-D) animals] or defined consortia of (sequenced) gut microbes (6–10).
Although MS and NMR spectroscopy have been used to identify metabolites produced by the microbiota or from host and microbial cometabolism, the effect of the microbiota and its metabolic output on the host proteome is less well understood. This reflects the technical challenges involved in performing quantitative proteomics in whole animals. Quantitative MS relies on the use of isotopically labeled internal controls. However, it is not practical to synthesize isotopically labeled standards for each protein or peptide. Therefore, techniques have emerged for metabolic labeling of whole cells or organisms with stable isotopes (11). Stable Isotopic LAbeling of mammalian Cells in culture (SILAC) using labeled amino acids has become a routine approach for characterizing responses to various perturbations at the protein level, as well as at the level of covalent protein modifications (12). Stable Isotopic LAbeling of Mammals (SILAM) is more challenging because isotopically labeled food must be generated in sufficient quantities to feed animals over multiple generations. To date, two approaches have been developed: uniform 15N labeling by feeding a diet containing 15N-labeled blue-green algae as the sole nitrogen source (13), or incorporating select labeled amino acids into a standard mouse diet (14).
Here we explore the general hypothesis that the microbiota impresses its metabolic activities on the host in part through covalent modification of host proteins with various moieties that affect their functions. We tested this hypothesis by using SILAM to characterize changes in lysine ε-acetylation of proteins expressed in the colon and liver upon transplantation of a gut microbiota from a CONV-R mouse into GF recipient animals.
Protein acetylation occurs in two forms: cotranslational α-acetylation at the N termini of most eukaryotic proteins, and posttranslational ε-acetylation of the free amines of lysine residues. Lysine ε-acetylation has been well characterized in the nucleus, where a complex network of histone acetyltransferases (HATs) and deacetylases (HDACs) control the state of acetylation of histone tails to regulate chromatin structure and transcription. Although lysine ε-acetylation of histones has been studied for nearly 50 y, it was not until 2006 that modern MS-based proteomic approaches uncovered the ubiquitous nature of this covalent protein modification (15). Since then, several large-scale proteomic studies have revealed that thousands of proteins in multiple cellular compartments undergo ε-acetylation and that this modification can dynamically regulate the functions of many enzymes (16, 17). In recognition of this fact, HATs and HDACs have been renamed “lysine acetyltransferases” and “lysine deacetylases” (KATs and KDACs) (18). KATs are a diverse group of enzymes comprising several different families, including Gcn5-like N-acetyltransferases, the p300/CBP family, the MYST-family, and others. Most characterized KATs are thought of as nuclear enzymes. However, several important KATs have been shown to shuttle between the nucleus and cytosol in a dynamic and regulated fashion (19). The ubiquity of extranuclear lysine ε-acetylation suggests a more-prominent role for KATs in the cytoplasm than was previously appreciated. KDACs can be grouped into two main families, the zinc-dependent HDACs (KDAC classes I, II, and IV) and the NAD-dependent sirtuins (class III). The subcellular distribution of KDACs is more diverse. For example, HDAC6 is almost exclusively located in the cytosol, and several other class I/II HDACs are also found in this compartment. Sirtuin family deacetylases, Sirt2 and Sirt3, are primarily cytosolic and mitochondrial, respectively. Thus, although lysine ε-acetylation was traditionally considered to be a mechanism for the regulation of chromatin structure, in the past decade several lines of evidence have converged to reveal a far more widespread role for this covalent protein modification outside the nucleus.
There were several reasons why we chose to use lysine ε-acetylation to test the hypothesis that the gut microbiota exerts part of its effect on host cells by causing covalent modification of host metabolic enzymes. As noted above, extranuclear lysine acetylation is emerging as a widespread mechanism for mediating protein function (20) and has been shown to be especially important for regulating metabolic enzymes. Acetate, an end product of microbial fermentation of otherwise indigestible dietary polysaccharides, is the most-abundant SCFA in the intestinal lumen (10–100 mM in nonruminant mammals) and can be transported into cells, where it directly contributes to acetyl-CoA pools (21). Furthermore, butyrate, another SCFA produced by the microbiota, is an inhibitor of class I and II KDACs and when applied to cells results in dramatic elevations in histone acetylation (22, 23). With these considerations in mind, we generated uniformly 15N-labeled GF mice to enable quantitative assessment of lysine ε-acetylation in response to colonization with a mouse gut microbiota.
Results and Discussion
Generation of Gnotobiotic SILAM Mice.
To achieve complete labeling of mice with 15N nitrogen (“heavy” mice) we used extracts of a cyanobacterium, Anthrospira (commonly known as spirulina), that had been grown in the presence of 15N ammonium sulfate. Mouse chow was constructed containing spirulina lyophilisate as the sole protein source (Tables S1 and S2). The 15N- and control 14N-labeled spirulina lyophilisates were from different batches and had been prepared by a commercial vendor on different dates. Therefore, before constructing the labeled mouse diets, we performed macro- and micronutrient analyses of the lyophilisates. We then mixed 15N- or 14N-labeled spirulina with the nonprotein components that constitute a standard mouse chow diet, correcting for minor documented differences in nutrients between the lyophilisates (e.g., lower “neutral detergent fiber” in the 15N spirulina was corrected by adding additional cellulose). The resulting diets were nearly identical in terms of known nutrients (Table S2).
In addition to normalizing the compositions of the two diets, we used a “reciprocal labeling” strategy to control for any slight or undetected differences between the 15N- and 14N-labeled chows (24, 25) (Fig. 1). GF C57BL/6 mice were housed in flexible film gnotobiotic isolators. At the first sign of pregnancy, females were switched to 15N or 14N spirulina chow that had been sterilized by irradiation. The resulting first-generation offspring were weaned onto the appropriate spirulina chow, and upon reaching sexual maturity, were interbred to generate a second generation of pups that were used for proteomics experiments. 15N labeling was continued for two generations because previous reports indicated that a single generation of SILAM labeling results in only 90–95% label incorporation, which considerably complicates quantitative proteomic analyses (26). Although we did not extensively characterize the extent of 15N incorporation into tissues, preliminary results indicated >97% incorporation of label in first-generation (F0) SILAM mice (SI Methods). Moreover, it is important to note that the reciprocal labeling scheme we used also controls for the possibility of incomplete labeling.
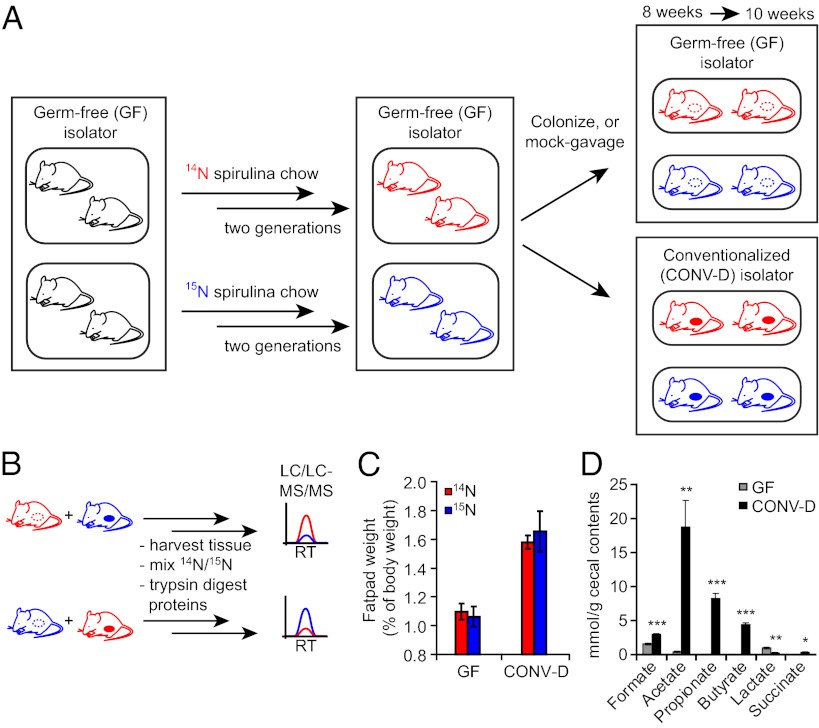
Fig. 1.
Metabolic labeling of GF mice with 15N and colonization with a distal gut microbiota. (A) Schematic illustrating the labeling scheme. GF mice were raised for two generations on a 14N-labeled or a 15N-labeled spirulina chow. At 8 wk of age, a cohort of 15N- or 14N-labeled GF SILAM mice was moved to a separate gnotobiotic isolator and colonized with the cecal microbiota of a conventionally raised mouse [producing conventionalized animals (CONV-D)]. After 2 wk, all mice were killed, and their tissues were harvested. (B) A reciprocal labeling scheme was used to ensure the accuracy and reproducibility of SILAM proteomics. Proteomic comparisons were made using both labeling directions: 15N-CONV-D:14N-GF and 15N-GF:14N-CONV-D. RT, retention time. (C) Epididymal fat-pad weight of GF and CONV-D mice, expressed as a percentage of total body weight. (D) Cecal SCFA concentrations of GF and CONV-D mice, n = 3 mice per group; mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.005 (two-tailed Student t test.)
At 8 wk of age, second-generation “SILAM mice” were either “mock” gavaged with PBS or gavaged with a suspension of freshly harvested cecal contents from a C57BL/6 CONV-R donor. Fourteen days after colonization, mice were killed, and tissues were harvested for protein extraction and analysis (Methods and Fig. S1). Numerous changes in host physiology are known to occur upon conventionalization of GF mice (27). To determine whether these well-documented changes occur in the context of a spirulina-based diet and whether these responses were different in the two diet contexts, we measured changes in two robust biomarkers of colonization: host adiposity and microbial production of SCFA. A ∼50% increase in epididymal fat pad weight occurred 14 d after conventionalization. This increase was not significantly different between mice consuming the 15N- vs. 14N-labeled diets (Fig. 1C). Levels of acetate, butyrate, and propionate also showed significant elevations in the ceca of CONV-D compared with GF mice on a spirulina diet (Fig. 1D).
Changes in the Lysine Acetylome upon Colonization.
We characterized the lysine acetylome in the distal gut, where microbial density and fermentation of polysaccharides to SCFA is highest, and in the liver, which is exposed to the products of gut microbial community metabolism via the portal circulation. Homogenates were subjected to ultracentrifugation to generate two fractions: a soluble fraction that should primarily contain secreted proteins and soluble proteins from the cytosol and lumen of membrane-bound organelles, and a particulate fraction expected to be enriched in integral membrane proteins and proteins bearing membrane anchors. The 15N-labeled and 14N-labeled tissue samples were mixed, either before or after homogenization, digested with trypsin, and analyzed with an Orbitrap Discovery hybrid mass spectrometer using a combination of strong cation exchange fractionation and reverse-phase chromatography (multidimensional protein identification technology, or “MudPIT”; Fig. 1B and SI Methods). To identify sites of lysine ε-acetylation, and to quantify differences in levels of acetylation as a function of colonization status, an additional step was introduced in the analysis pipeline involving immunoprecipitation of tryptic peptides, generated from each of the two fractions from each of the two tissues, with a commercial antibody specific for acetylated lysine residues. Approximately 10% of the peptides in the immunoprecipitated fractions were lysine ε-acetylated, compared with ∼0.2% in the nonenriched fractions, indicating that the immunoprecipitation afforded a ∼50-fold enrichment.
All comparisons described below involved analysis of five pairs of SILAM mice (10 animals total): three pairs in which 14N-labeled mice were CONV-D and 15N-labeled mice were GF, and two pairs in which the 14N-labeled mice were GF and the 15N-labeled animals were CONV-D (Table S3). In addition to controlling for possible differences between the diets, this reciprocal labeling scheme also controlled for any possible label-specific biases in our data analysis pipeline. Furthermore, the four groups of mice involved in this study (14N GF, 15N GF, 14N CONV-D, and 15N CONV-D) were from three different litters and were colonized on different days, largely eliminating the possibility that differences observed between GF and CONV-D mice arose from extraneous factors (Table S3). It is important to note that because of the stochastic nature of data-dependant shotgun proteomics, not all proteins or peptides were detected in all five pairs of mice. However, a majority of all proteins and sites of lysine ε-acetylation were detected across multiple pairs of mice (Fig. S2).
The primary focus of this study was the characterization of changes to the lysine acetylome upon conventionalization. When quantifying dynamic posttranslational modifications (PTMs), it is important to discriminate between protein ratios and PTM ratios (28). Thus, we sought to distinguish changes observed for sites of lysine modification from changes in overall protein expression. Because experiments were performed in two different labeling directions (i.e., 15N-CONV-D:14N-GF and 15N-GF:14N-CONV-D) the ratios from the 15N:14N CONV-D:GF experiments were inverted so that all experimental data could be combined. Thus, ratios comparing GF with CONV-D signals will be referred to simply as “SILAM ratios” for the remainder of this report. To calculate these ratios, we developed a custom suite of software tools that separately groups peptide ratios derived from nonacetylated peptides, and acetylated peptides matching each detected site of lysine acetylation. Our data analysis pipeline used ProLuCID and DTASelect for peptide spectral matching and quality filtering, respectively (29, 30). Determination of peptide SILAM ratios was performed using cimage (31) and incorporated stringent requirements for 14N/15N peak coelution and parent-mass accuracy (details in SI Methods). In-house software was then used to group SILAM ratios by protein and site of acetylation.
We quantified a total of 3,891 liver and proximal colonic proteins (SI Results and Discussion, Fig. S3, and Dataset S1); 3,021 in proximal colon and 2,773 in liver, with 1,903 (49%) quantified in both tissues. Of these 3,891 proteins, 558 contained a total of 1,602 sites of lysine acetylation; 387 of the 1,602 sites (24%) were observed in both tissues (Fig. S4). Forty-three percent of the sites were not reported previously (Fig. S4 and Dataset S1), suggesting that existing efforts to characterize ε-lysine acetylation in proteins are not yet approaching comprehensive coverage. Gene ontology (GO) analyses revealed that the acetylated proteins were highly enriched for biological processes involving energy production, respiration, and primary metabolism (32) (Fig. S4). Nuclear-encoded mitochondrial proteins were significantly overrepresented among the acetylated proteins in both tissues, and an enrichment in cofactor-binding enzymes and solute transporters was also observed (Fig. S4).
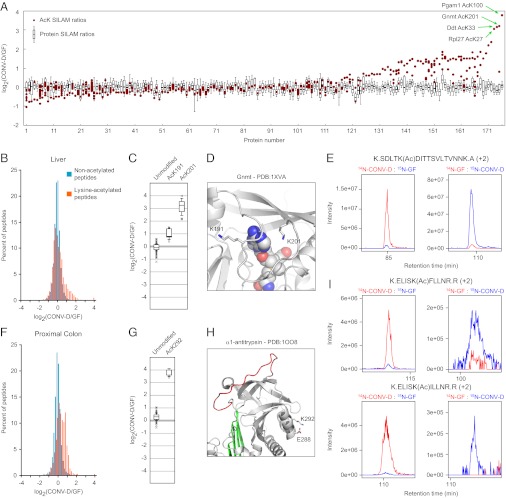
Both colon and liver displayed reproducible increases in lysine ε-acetylation upon conventionalization (Fig. 2 A, B, and F). More dramatic changes were observed in liver, where >10-fold elevations in acetylation of lysine residues were observed on several proteins, in addition to a general increase in acetylation on CONV-D vs. GF proteins from both tissues (Fig. 2 B and F and Dataset S1). An example of a robust and dramatic increase in lysine ε-acetylation involves glycine N-methyl transferase (Gnmt), which plays a central role in regulating concentrations of the cofactor S-adenosylmethionine (S-AdoMet) in liver. K191 and K201 of Gnmt were found to be acetylated in liver. To our knowledge, neither of these sites of acetylation has been reported previously (Dataset S1). Compared with the SILAM ratios for the unmodified peptides, ratios for both acetylation sites were elevated in the livers of CONV-D vs. GF animals: K201 displayed a ∼10-fold elevation, whereas K191 showed a ∼twofold increase in its acetylation upon colonization (Fig. 2 C and E). Both of these residues are located near the cofactor binding site of Gnmt, with the ε-amine of K201 positioned only 5 Å from bound S-AdoMet (Fig. 2D) (33). Interestingly, both K191 and K201 are located on a flexible loop on Gnmt that displays high conformational variability across different crystal structures (34). We speculate that differential acetylation of these residues may represent a mechanism for regulating the activity of this enzyme by controlling the conformation of this loop.
Fig. 2.
Changes to the lysine acetylome in the liver (A–E) and proximal colon (F–I) of gnotobiotic mice induced by colonization with a gut microbiota. (A) Protein ratios (box plots) and SILAM ratios for individual sites of lysine ε-acetylation (red circles) are shown on the same graph. The box plots represent the distribution of nonacetylated peptide ratios for a given protein, and red circles indicate the average ratios for acetylated peptides corresponding to a particular site of acetylation. Only proteins harboring acetylation marks observed in at least three pairs of mice are shown. On the x axis proteins are ordered by the difference between the median protein ratio and the average SILAM ratio of the most-altered site of acetylation. Complete data can be found in Datasets S1 and S3. (B and F) Histograms showing the distribution of SILAM ratios of nonacetylated peptides (teal) and of lysine-acetylated peptides (orange) in liver (B) and colon (F). (C–E) Sites of lysine ε-acetylation of Gnmt in the liver. (G–I) Sites of lysine acetylation of A1AT in the colon. (C and G) Peptide ratio distributions for unmodified peptides and acetylated peptides matching the indicated lysine residues. (D and H) Crystal structures showing lysine residues whose ε-acetylation is increased upon conventionalization. (E and I) MS1 chromatographs documenting the marked microbiota-induced acetylation of lysine 201 on Gnmt (E) and of lysine 292 on two different isoforms of A1AT (I). A description of the box plots used in C and G can be found in SI Methods.
Colonization also dramatically affected acetylation of lysines in several other metabolic enzymes expressed in the liver, including the glycolytic enzyme phosphoglycerate mutase (Pgam1) and the melanin-biosynthetic enzyme dopachrome decarboxylase (Ddt) (Fig. S5). In both cases, the acetylated residues were also positioned near their active sites (Fig. S5). Notably, several proteins displayed multiple sites of acetylation that were modestly (two- to fourfold) but reproducibly elevated upon colonization, indicating that lysine ε-acetylation in response to microbial colonization may involve colocalization of the substrate proteins with a particular acetyltransferase, deacetylase, or acetyl-CoA pools (Fig. S5). Strikingly, no sites of lysine ε-acetylation were detected that were reproducibly reduced by more than twofold in liver or colon upon conventionalization.
In the proximal colon, the most dramatically elevated site of lysine ε-acetylation was found in α-1-antitrypsin (A1AT). A single acetylated lysine, K292, was detected, and it showed a dramatic (>10-fold) elevation upon conventionalization (Fig. 2 G–I). A1AT is also expressed in the liver and secreted into circulation. Accordingly, we also detected A1AT in liver tissue with the same dynamic acetylation mark at K292 (Fig. S6). A1AT, a member of the serpin family of serine protease inhibitors, undergoes a marked conformational rearrangement upon binding and covalently inactivating its target proteases, suggesting that residues far from the reactive central loop (“C-loop”) may contribute to its activity. Many mutations in A1AT outside the reactive C-loop lead to destabilization of the protein and multimerization, causing a collection of disorders known as serpinopathies (35). Despite the fact that K292 is not localized to a region with known functional significance, this residue also displays considerable variability across multiple crystal structures and may participate in a salt bridge with Glu288, suggesting a potentially important role in protein stabilization that would be blocked by lysine ε-acetylation (Fig. 2H).
Humans have a single A1AT gene, whereas mice have a cluster of highly similar genes encoding A1AT isoforms (36). The acetylated residue, K292, is conserved in most but not all mouse isoforms, and is also conserved in the human ortholog (Fig. S6). Because the protein sequences of the mouse isoforms are so similar, it was not possible to determine precisely which of the isoforms were present. Peptides from at least two different A1AT isoforms were detected containing the highly responsive K292 site [K.ELISK(Ac)FLLNR.R and K.ELISK(Ac)ILLNR.R; Fig. 2I and Fig. S6]. Thus, this robustly regulated lysine ε-acetylation event was observed in at least two different protein isoforms in both proximal colon and liver.
To determine whether K298, the orthologous residue in human A1AT to K292 in the mouse protein, was acetylated, we immunoprecipitated acetylated tryptic peptides derived from human serum and analyzed them by MS. Although we could not measure the dynamics of lysine ε-acetylation in human serum, we were able to ascertain that K298 is also acetylated, indicating that acetylation may represent a conserved mechanism for regulating the function of this important protease inhibitor in mice and humans (Fig. S6).
The majority of ε-acetylated lysine residues detected were found in nuclear encoded mitochondrial proteins (60.2% and 57.8% of the acetylated residues were distributed among 220 and 139 proteins in liver and colon, respectively; Dataset S2). The gut microbiota is known to influence energy homeostasis by regulating mitochondrial metabolism through alterations in the rate of free fatty acid oxidation, and by providing a substrate for production of acetyl-CoA (6, 7). Nonetheless, it is interesting to note that those proteins displaying the most dramatic alterations were cytosolic (Pgam1, Gnmt, Ddt) or secreted (A1AT). The dynamic nature of lysine ε-acetylation of cytosolic proteins can likely be explained by regulation of one or more of the cytosolic KATs or KDACs—either at the transcript level or at the level of enzyme activity. These regulated lysine ε-acetylation events seem to be quite specific, because other lysine ε-acetylation events on other proteins or even on the same protein (in the case of Gnmt and Pgam1) did not display elevations of similar magnitude. However, the dynamic acetylation of the lysine residue in A1AT is surprising because it is a secreted protein that is not exposed to cytosolic or nuclear enzymes. Very little is known about KATs or KDACs localized to the secretory pathway. Interestingly, two endoplasmic reticulum/Golgi-localized acetyltransferases, ATase1 and ATase2, have recently been characterized and shown to play an important role in the acetylation, secretion, and stabilization of β-secretase 1 (37–39). It is possible that ATase1 or ATase2 or other as yet unknown KATs or KDACs participate in the secretory pathway and display altered activity in response to colonization. Future work to understand the scale and dynamics of lysine ε-acetylation of serum proteins may yield important insights.
Prospectus.
We have shown that using quantitative proteomics to study gnotobiotic mice is a powerful approach to uncover ways in which the host and microbiota interact to affect the host epiproteome. In this study we focused on lysine ε-acetylation, but quantitative proteomics can be used to study many other posttranslational covalent modifications (40, 41). Serine/threonine phosphorylation is the most well characterized, largely owing to the development of facile chromatographic techniques for phosphopeptide enrichment. However, other modifications such as ubiquitination, methylation, O-GlcNAcylation, and oxidation likely play unexpected roles in regulating facets of host–microbe interactions and should be thoroughly explored. Gnotobiotic SILAM mice colonized with defined collections of cultured mouse or human gut symbionts, or with mouse or human gut microbiota from donors representing various physiologic or disease states, provide an opportunity to explore these ideas further under highly controlled experimental conditions in multiple tissues and, perhaps in the future, in purified cellular populations. The field of quantitative epiproteomics in gnotobiotic animals is currently in its infancy but holds great promise for enhancing our understanding of how various microbial communities impact the biology of their hosts.
Methods
15N and 14N Spirulina Diets.
Mouse chow containing 15N or 14N-labeled spirulina lyophilisate as the sole nitrogen source (13, 42, 43) was generated by mixing spirulina lyophilisate (Cambridge Isotope Laboratories) with protein-free diet (Harlan Teklad). Additional information regarding the composition of these diets can be found in Tables S1 and S2. Diets were extruded into pellets, vacuum-sealed, and sterilized by irradiation before being introduced into gnotobiotic isolators.
Labeling and Colonization of GF Mice.
All studies involving mice used protocols approved by Washington University’s Institutional Animal Care and Use Committee. GF C57BL/6 mice were reared in flexible film gnotobiotic isolators (Class Biological Clean). At the first sign of pregnancy, female mice were switched to one or the other spirulina diets. Pups were suckled by mothers consuming the spirulina diet, then weaned onto the spirulina diet and subsequently maintained on it for the remainder of their lives. These first-generation SILAM mice were interbred to generate a second generation of SILAM mice used for the experiments described here (Fig. S1). At 8 wk of age, second-generation SILAM mice were colonized with a single oral gavage composed of a 200-μL suspension of freshly harvested cecal contents (diluted 1:10 in PBS) obtained from a CONV-R C57BL/6 donor. The resulting CONV-D mice were maintained on spirulina diets ad libitum for 14 d before killing.
Sample Preparation, MS, and Data Analysis.
Proteins were extracted from frozen tissue and subjected to overnight trypsin digestion. The resulting peptides were either analyzed directly or subjected to immunoprecipitation with a commercial anti-acetyl lysine antibody from ImmuneChem (#ICP0388). Peptides were chromatographed via multidimensional chromatography (“MudPIT”) directly into an Orbitrap Discovery hybrid mass spectrometer (Thermo Scientific). Our data analysis pipeline used ProLuCID for peptide identification and cimage for quantitation of 14N/15N ratios (29, 31). Subsequent data analysis steps used custom software for grouping and interpretation of the data. Refer to SI Methods for additional details.
Supplementary Material
Acknowledgments
We thank Barbara Mickelson (Harlan Teklad) for expert guidance and for preparataion of the spirulina diets; and David O’Donnell, Maria Karlsson, and Sabrina Wagoner for gnotobiotic husbandry and technical assistance. This work was supported by National Institutes of Health Grants DK30292 and DK70977 and by grants from the Crohns and Colitis Foundation of America. G.M.S. was supported by a postdoctoral fellowship from The Helen Hay Whitney Foundation.
Footnotes
The authors declare no conflict of interest.
Data deposition: The raw MS data reported in this paper were deposited in the Proteome Commons database, https://proteomecommons.org, and are accessible via the following hashes: ClxC6eoabIQRspEyyyV47kWSQneOKi0nZGMBz6ZptdlitxuoeGIgYtoZCofTjwtsuJQFqF0i4kOzh3diPjJ6iit7uzoAAAAAAADVPw== and vr/a4z8SnZyISjRNqf27hOgvuRsdKOohHTuasbp+Ob2fNMqkirkM1TaGdWenNi0Yil48Pxpbn4BGGNPdofnoj6GdTi4AAAAAAACtwA==.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1208669109/-/DCSupplemental.
References
- 1.Heinzmann SS, et al. Stability and robustness of human metabolic phenotypes in response to sequential food challenges. J Proteome Res. 2012;11:643–655. doi: 10.1021/pr2005764. [DOI] [PubMed] [Google Scholar]
- 2.Wikoff WR, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci USA. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holmes E, et al. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature. 2008;453:396–400. doi: 10.1038/nature06882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Z, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holmes E, Li JV, Athanasiou T, Ashrafian H, Nicholson JK. Understanding the role of gut microbiome-host metabolic signal disruption in health and disease. Trends Microbiol. 2011;19:349–359. doi: 10.1016/j.tim.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 6.Bäckhed F, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crawford PA, et al. Regulation of myocardial ketone body metabolism by the gut microbiota during nutrient deprivation. Proc Natl Acad Sci USA. 2009;106:11276–11281. doi: 10.1073/pnas.0902366106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reinhardt C, et al. Tissue factor and PAR1 promote microbiota-induced intestinal vascular remodelling. Nature. 2012;483:627–631. doi: 10.1038/nature10893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gouw JW, Krijgsveld J, Heck AJ. Quantitative proteomics by metabolic labeling of model organisms. Mol Cell Proteomics. 2010;9:11–24. doi: 10.1074/mcp.R900001-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ong SE, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 13.Wu CC, MacCoss MJ, Howell KE, Matthews DE, Yates JR., 3rd Metabolic labeling of mammalian organisms with stable isotopes for quantitative proteomic analysis. Anal Chem. 2004;76:4951–4959. doi: 10.1021/ac049208j. [DOI] [PubMed] [Google Scholar]
- 14.Krüger M, et al. SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell. 2008;134:353–364. doi: 10.1016/j.cell.2008.05.033. [DOI] [PubMed] [Google Scholar]
- 15.Kim SC, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 16.Choudhary C, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 17.Zhao S, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allis CD, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 19.Sadoul K, Wang J, Diagouraga B, Khochbin S. The tale of protein lysine acetylation in the cytoplasm. J Biomed Biotechnol. 2011;2011:970382. doi: 10.1155/2011/970382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim GW, Yang XJ. Comprehensive lysine acetylomes emerging from bacteria to humans. Trends Biochem Sci. 2011;36:211–220. doi: 10.1016/j.tibs.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 21.Wellen KE, et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blackwell L, Norris J, Suto CM, Janzen WP. The use of diversity profiling to characterize chemical modulators of the histone deacetylases. Life Sci. 2008;82:1050–1058. doi: 10.1016/j.lfs.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 23.Davie JR. Inhibition of histone deacetylase activity by butyrate. J Nutr. 2003;133(7 Suppl):2485S–2493S. doi: 10.1093/jn/133.7.2485S. [DOI] [PubMed] [Google Scholar]
- 24.Huttlin EL, Hegeman AD, Sussman MR. Metabolic labeling approaches for the relative quantification of proteins. In: Julian PW, editor. Comprehensive Analytical Chemistry. Vol 52. Amsterdam: Elsevier; 2008. pp. 479–530. [Google Scholar]
- 25.Huttlin EL, et al. Discovery and validation of colonic tumor-associated proteins via metabolic labeling and stable isotopic dilution. Proc Natl Acad Sci USA. 2009;106:17235–17240. doi: 10.1073/pnas.0909282106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McClatchy DB, Dong MQ, Wu CC, Venable JD, Yates JR., 3rd 15N metabolic labeling of mammalian tissue with slow protein turnover. J Proteome Res. 2007;6:2005–2010. doi: 10.1021/pr060599n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin Immunol. 2007;19:59–69. doi: 10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 28.Wu R, et al. Correct interpretation of comprehensive phosphorylation dynamics requires normalization by protein expression changes. Mol Cell Proteomics. 2011;10:M111.009654. doi: 10.1074/mcp.M111.009654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu T, et al. ProLuCID, a fast and sensitive tandem mass spectra-based protein identification program. Mol Cell Proteomics. 2006;5:S174. [Google Scholar]
- 30.Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: Tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weerapana E, et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 33.Fu Z, et al. Crystal structure of glycine N-methyltransferase from rat liver. Biochemistry. 1996;35:11985–11993. doi: 10.1021/bi961068n. [DOI] [PubMed] [Google Scholar]
- 34.Pakhomova S, Luka Z, Grohmann S, Wagner C, Newcomer ME. Glycine N-methyltransferases: A comparison of the crystal structures and kinetic properties of recombinant human, mouse and rat enzymes. Proteins. 2004;57:331–337. doi: 10.1002/prot.20209. [DOI] [PubMed] [Google Scholar]
- 35.Ekeowa UI, et al. alpha1-Antitrypsin deficiency, chronic obstructive pulmonary disease and the serpinopathies. Clin Sci (Lond) 2009;116:837–850. doi: 10.1042/CS20080484. [DOI] [PubMed] [Google Scholar]
- 36.Forsyth S, Horvath A, Coughlin P. A review and comparison of the murine alpha1-antitrypsin and alpha1-antichymotrypsin multigene clusters with the human clade A serpins. Genomics. 2003;81:336–345. doi: 10.1016/s0888-7543(02)00041-1. [DOI] [PubMed] [Google Scholar]
- 37.Ko MH, Puglielli L. Two endoplasmic reticulum (ER)/ER Golgi intermediate compartment-based lysine acetyltransferases post-translationally regulate BACE1 levels. J Biol Chem. 2009;284:2482–2492. doi: 10.1074/jbc.M804901200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jonas MC, Pehar M, Puglielli L. AT-1 is the ER membrane acetyl-CoA transporter and is essential for cell viability. J Cell Sci. 2010;123:3378–3388. doi: 10.1242/jcs.068841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Costantini C, Ko MH, Jonas MC, Puglielli L. A reversible form of lysine acetylation in the ER and Golgi lumen controls the molecular stabilization of BACE1. Biochem J. 2007;407:383–395. doi: 10.1042/BJ20070040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Witze ES, Old WM, Resing KA, Ahn NG. Mapping protein post-translational modifications with mass spectrometry. Nat Methods. 2007;4:798–806. doi: 10.1038/nmeth1100. [DOI] [PubMed] [Google Scholar]
- 41.Macek B, Mann M, Olsen JV. Global and site-specific quantitative phosphoproteomics: Principles and applications. Annu Rev Pharmacol Toxicol. 2009;49:199–221. doi: 10.1146/annurev.pharmtox.011008.145606. [DOI] [PubMed] [Google Scholar]
- 42.Frank E, et al. Stable isotope metabolic labeling with a novel N-enriched bacteria diet for improved proteomic analyses of mouse models for psychopathologies. PLoS ONE. 2009;4:e7821. doi: 10.1371/journal.pone.0007821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Price JC, Guan S, Burlingame A, Prusiner SB, Ghaemmaghami S. Analysis of proteome dynamics in the mouse brain. Proc Natl Acad Sci USA. 2010;107:14508–14513. doi: 10.1073/pnas.1006551107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.