Abstract
REPRESSOR OF SILENCING 1 (ROS1) is a DNA demethylation enzyme that was previously identified during a genetic screen for the silencing of both RD29A-LUC and 35S-NPTII transgenes on a T-DNA construct. Here we performed a genetic screen to identify additional mutants in which the 35S-NPTII transgene is silenced. We identified several alleles of ros1 and of the following components of the RNA-directed DNA methylation (RdDM) pathway: NRPD1 (the largest subunit of polymerase IV), RDR2, NRPE1 (the largest subunit of polymerase V), NRPD2, AGO4, and DMS3. Our results show that the silencing of 35S-NPTII in the RdDM pathway mutants is due to the reduced expression of ROS1 in the mutants. We also identified a putative histone acetyltransferase (ROS4) from the genetic screen. The acetyltransferase contains a PHD-finger domain that binds to unmethylated histone H3K4. The mutation in ROS4 led to reduction of H3K18 and H3K23 acetylation levels. We show that the silencing of 35S-NPTII and some transposable element genes was released by the ddm1 mutation but that this also required ROS4. Our study identifies a unique antisilencing factor, and reveals that the RdDM pathway has an antisilencing function due to its role in maintaining ROS1 expression.
Keywords: transcriptional gene silencing, epigenetic, gene silencing, histone modification
DNA cytosine methylation is an important epigenetic modification in plants and animals. The molecular mechanism for RNA-directed DNA methylation (RdDM) has been well-established by both genetic and biochemical studies in Arabidopsis (1–4). Small RNAs (∼24 bp) and long noncoding RNAs produced by a complex transcriptional machinery composed of polymerase (Pol) IV and Pol V, together with other proteins, target specific sequences for de novo DNA methylation (1–4). RdDM mainly occurs in transposons, retrotransposons, rDNA arrays, and centromeric repeat regions, and accounts for about 30% of the DNA methylation in the genome of Arabidopsis (5, 6).
DNA methylation can be removed passively during DNA replication or actively by the DNA glycosylase REPRESSOR OF SILENCING 1 (ROS1) and its homologs DEMETER (DME), DEMETER-LIKE 2 (DML2), and DML3 (1, 2, 7, 8). ROS1 was identified in a genetic screen in which the originally active RD29A-LUC was silenced in ros1 mutants (9). ROS1 is a key DNA demethylation component in vegetative tissues (9), and its transcript level is regulated by MET1 and several RdDM components (10). Screening for second-site suppressors of ros1 mutants by using silenced RD29A-LUC has enabled researchers to identify the main components of the RdDM pathway and has further indicated that RdDM and ROS1 antagonistically regulate DNA methylation (11–14).
ros1 mutations also cause the silencing of another transgene, 35S-NPTII, which is in the same T-DNA construct as RD29A-LUC (9). RD29A-LUC has been used to identify ROS1 and ROS3 genes (9, 15). By screening for second-site suppressors of ros1 using silenced 35S-NPTII, we previously identified genes that are involved in DNA replication and maintenance of transcriptional silencing (7). Our results indicated that RD29A-LUC and 35S-NPTII are silenced and/or maintained by different mechanisms. In the current study, to find additional REPRESSOR OF SILENCING genes, we performed a forward genetic screen by using 35S-NPTII as a selection marker. Unexpectedly, besides three alleles of ros1 and a unique gene (ROS4), the genes we identified are mainly components in the RdDM pathway. We show that the mutations in RdDM components cause the silencing of 35S-NPTII by down-regulating the expression of ROS1. Our results provide genetic evidence for an in vivo role of regulating ROS1 expression by the RdDM pathway. The antisilencing factor ROS4 encodes a putative histone acetyltransferase with a PHD-finger domain and a methyl-CpG–binding domain. ROS4 preferentially binds to unmethylated histone H3K4, and the ros4 mutation results in reduction of the overall H3K23 and H3K18 acetylation levels in plants. We also found that ROS4 is required for the expression of some genes that are normally silenced by DNA methylation.
Results
Isolation of Mutants in Which the Expression of 35S-NPTII is Silenced.
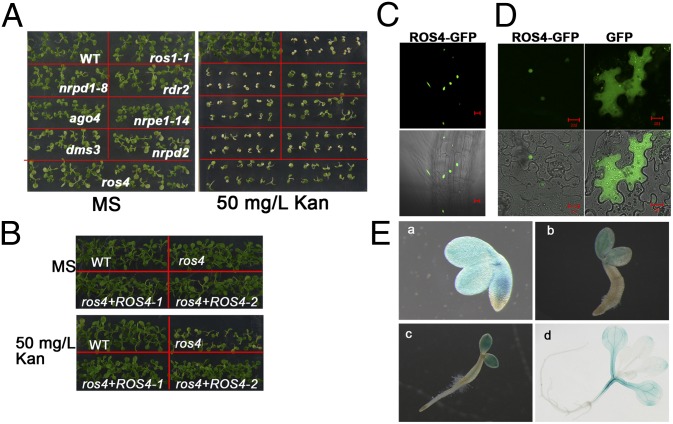
To identify genes that are required for the expression of 35S-NPTII, we performed a forward genetic screen using an ethyl methanesulfonate (EMS)-mutagenized population of C24 transgenic plants carrying a T-DNA locus (used as the wild type). In the T-DNA locus, two transgenes, RD29A-LUC and 35S-NPTII, are actively expressed for many generations without silencing (9). Seedlings carrying the T-DNA are able to grow on Murashige and Skoog (MS) medium containing more than 200 mg/L kanamycin (Kan). To identify mutants with reduced expression of 35S-NPTII, we germinated M2 (germinated seeds of a mutagenized population) seeds on MS medium containing 50 mg/L Kan. After the seedlings had grown for 1 wk, Kan-sensitive plants were transferred to MS medium without Kan and allowed to recover. The putative mutants were confirmed on the Kan medium in the next generation. All of the identified mutants were determined to have lost Kan resistance when seeds were germinated and grown on MS medium supplemented with 50 mg/L Kan (Fig. 1A).
Fig. 1.
Analysis of kanamycin sensitivity of ros4 and other mutants and ROS4 expression. (A) Kan sensitivity of ros4 and RdDM pathway mutants. The seeds of ros1-1, nrpd1-8, rdr2, ago4, nrpe1-14, dms3, nrpd2, and ros4 were germinated on MS medium or MS medium supplemented with 50 mg/L Kan for 7 d. (B) Complementation of the ros4 mutant. A DNA fragment containing the ROS4 promoter (−2499 to −1) driving ROS4 full-length cDNA was transformed into the ros4 mutant. Two independent lines were tested for Kan sensitivity on 50 mg/L Kan MS medium. (C) ROS4-GFP localization in a root of a transgenic line carrying 35S::ROS4-GFP. (Upper) GFP fluorescence image. (Lower) GFP image combined with bright field. (Scale bars, 20 μm.) (D) ROS4-GFP localization in a transient assay using tobacco leaves expressing 35S::ROS4-GFP (Left). GFP was used as a control (Right). (Upper) GFP fluorescence image. (Lower) GFP image combined with bright field. (Scale bars, 20 μm.) (E) The expression analysis of ROS4 promoter::GUS in a transformed line at 12 h (a), 36 h (b), 48 h (c), and 14 d (d) after seed imbibition.
The mutants were crossed with Col-0, and the F2 (second generation from self-fertilized F1) plants that showed the Kan-sensitive phenotype and that carried the NPTII gene (as indicated by PCR) were used for map-based cloning. We identified 15 mutants that include three alleles of ros1, three alleles of nrpd1, four alleles of nrpe1, two alleles of nrpd2, and one allele each of dms3, rdr2, and ago4 (Fig. S1). The nrpd1-8 (P438 mutated to L) and nrpe1-14 (W1360 mutated to a stop codon) mutants were selected as the representative mutants in the RdDM pathway for further characterization. We also isolated a mutant, named ros4. ros4 was created by a G-to-T mutation at position 1351 of AT3G14980, which changes the amino acid E451 to a stop codon. The promoter (−2499 to −1) of ROS4 driving ROS4 full-length cDNA or ROS4 full-length cDNA fused with MYC was able to complement the Kan-sensitive phenotype of ros4 (Fig. 1B, showing only the ROS4 promoter::ROS4 cDNA).
ROS4 Is a Nuclear Protein Expressed in Early Seedling Development.
For determination of subcellular localization, ROS4 cDNA was amplified and fused with the N terminus of GFP and stably expressed in Arabidopsis or transiently expressed in tobacco epidermal cells under a 35S promoter. The 35S::ROS4-GFP transgene was able to complement the Kan-sensitive phenotype of ros4. Confocal microscopy showed that ROS4-GFP was localized in the nucleus when stably expressed in the root cells of transgenic Arabidopsis plants (Fig. 1C) or when transiently expressed in tobacco epidermal cells (Fig. 1D, Left). As controls, GFP alone was localized mainly in the cytoplasm (Fig. 1D, Right).
To determine the expression pattern of ROS4, we used transgenic plants carrying the ROS4 promoter (−2499 to −1)::GUS (β-glucuronidase) fusion. GUS staining indicated that ROS4 was expressed in cotyledons and hypocotyls during early seedling development, but the expression became less in true leaves and roots (Fig. 1E).
ROS4 Encodes a Putative Histone Acetyltransferase with a PHD Domain That Preferentially Binds to Unmethylated Histone H3K4.
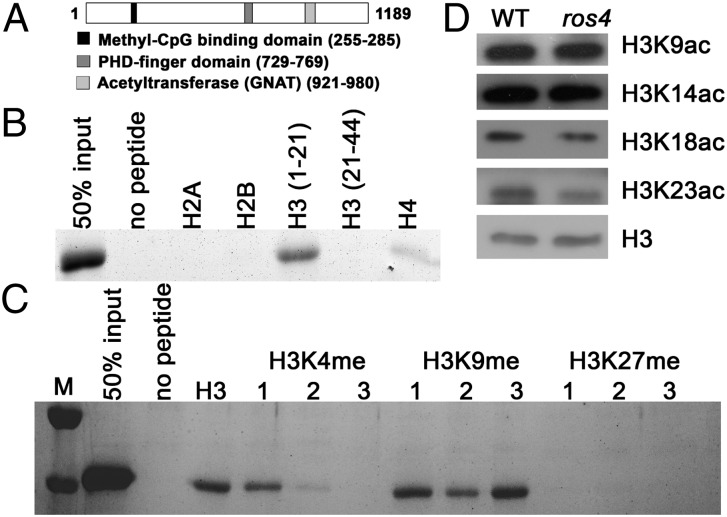
ROS4 encodes a predicted polypeptide with 1,189 amino acids that has a high similarity to acyl-CoA N-acetyltransferases (GNAT family). There is a putative PHD-finger domain around residues 729–769, a putative methyl-CpG–binding domain around amino acids 255–285, and an acyl-CoA N-acyltransferase domain around amino acids 921–980 (Fig. 2A). Previous studies have shown that PHD domain-containing proteins preferentially bind to H3K4me2/3 in Arabidopsis (16) and animals (17) or to H3K4me0 in animals (18, 19). Because many studies have shown that the PHD domain is responsible for histone binding (20), we expressed the PHD domain (amino acids 723–769) of ROS4 in Escherichia coli and conducted a peptide pull-down assay with different combinations of histones. The PHD fragment bound strongly to the H3 N-terminal tail (amino acids 1–21) without any modifications and bound very weakly to H4, but did not bind to H3 (amino acids 21–44), H2A, or H2B (Fig. 2B). The binding between the PHD domain of ROS4 and H3 (amino acids 1–21) was substantially reduced by H3K4me2/3 and somewhat reduced by H3K4me1, but was not reduced substantially by H3K9 methylation. The fragment also did not bind to H3K27me1/2/3 (H3, amino acids 21–44) (Fig. 2C). These results indicate that, in contrast to the PHD fingers of AtING and Alfin-like proteins that were previously characterized in Arabidopsis (16) and bind to H3K4me2/3, the PHD finger of ROS4 preferentially binds to unmethylated H3K4 (H3K4me0).
Fig. 2.
PHD domain of ROS4 binds to H3K4me0, and the mutation in ROS4 leads to reduction of acetylation of H3K18 and H3K23. (A) Structural diagram of the ROS4 protein. (B) In vitro assays of the binding of the recombinant PHD domain to unmodified histone peptides. H3 was divided into two fragments, H3 (1–21) and H3 (21–44). Note that the PHD domain also weakly binds to H4. (C) Effect of histone methylation on the binding of the PHD domain to the H3 tail. At least three independent experiments were done with similar results. (D) H3K9ac, H3K14ac, H3K18ac, and H3K23ac levels in the wild type and ros4. Total histone proteins were extracted from 5- to 7-d-old seedlings, and different antibodies were used for Western blotting; H3 was used as a loading control. M, size markers.
Because ROS4 is a putative acetyltransferase that binds to H3K4me0, we explored whether H3 acetylation is affected by ros4 mutation by performing Western blot analysis with different H3 acetylation antibodies. Because ROS4 is mainly expressed early in seedling development, we extracted proteins from 5- to 7-d-old seedlings. Western blot analysis indicated that, relative to the wild type, H3K23ac was clearly reduced, H3K18ac was weakly reduced, and H3K9ac and H3K14ac were not changed in ros4 (Fig. 2D).
RdDM Pathway Mutations Lead to Silencing of 35S-NPTII Due to a Lack of Expression of ROS1.
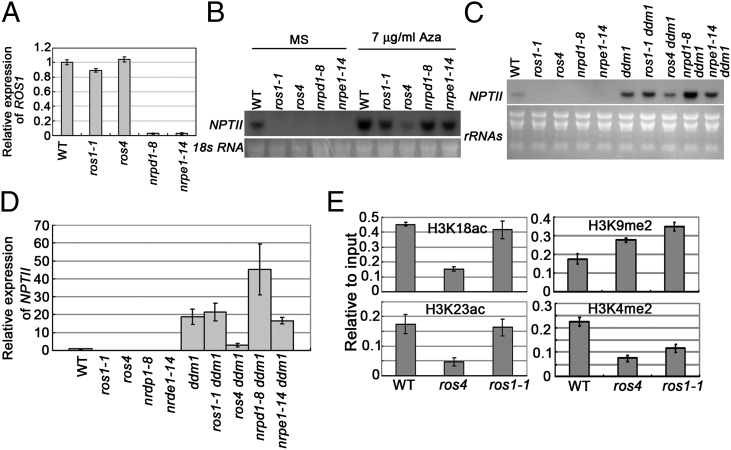
Previous studies showed that the expression of ROS1 is regulated by the RdDM pathway (10). We confirmed that the expression of ROS1 was greatly reduced by nrpd1 and nrpe1 mutations (Fig. 3A). In ros4, ROS1 expression was not changed (Fig. 3A). Given that the RdDM pathway is known to silence the expression of target genes through siRNA-guided DNA methylation, mutations in RdDM pathway components should release rather than lead to transcriptional gene silencing (TGS) if 35S-NPTII is directly regulated by the RdDM pathway. We suspected that the TGS of 35S-NPTII in RdDM mutants might be due to down-regulation of ROS1. To test this hypothesis, we expressed ROS1 cDNA under the constitutive 35S promoter in ros1, nrpd1, nrpe1, and ros4 mutants and found that the transgenic plants became resistant to Kan in ros1, nrpd1, and nrpe1 (Fig. S2) but not in ros4 (24 independent transgenic lines were tested). However, ROS1 cDNA driven by its native promoter was able to complement the ros1-1 mutant but was unable to recover the Kan-resistant phenotype in nrpd1 (63 lines tested) or nrpe1 (24 lines tested). These results indicate that NRPD1 and NRPE1, but not ROS4, regulate the expression of ROS1 and that the silencing of 35S-NPTII in nrpd1 and nrpe1 mutants is the result of the down-regulation of ROS1.
Fig. 3.
ROS4 suppresses DNA methylation-mediated transcriptional gene silencing of 35S-NPTII. (A) Quantitative RT-PCR analysis of ROS1 expression in different mutants. Three independent experiments (each with three biological replicates) were done with similar results. (B) NPTII expression as indicated by Northern analysis in ros4 and other mutants treated or not treated with 7 μg/mL 5-aza-cytidine. 18S RNA was used as the loading control. (C) The effect of the ros4 mutation on ddm1-promoted NPTII expression as determined by comparing single and double mutants. (D) Quantitative RT-PCR assays of NTPII transcripts in different single and double mutants. Three independent experiments (each with three biological replicates) were done with similar results. (E) ChIP assay of H3K18ac, H3K23ac, H3K9me2, and K3H4me2 in the wild type, ros4, and ros1-1. Seven-day-old seedlings were used for ChIP assay with antibodies against H3K18ac, H3K23ac, H3K9me2, or H3K4me2. Three independent experiments (each with three biological replicates) were done with similar results. In A, D and E, error bars refer to standard error (n = 3).
ROS4 Is Required for the Expression of 35S-NPTII Even When DNA Methylation Is Inhibited.
We next determined whether the TGS of 35S-NPTII is regulated in a similar manner by ROS1 and ROS4. When 7 μg/mL of 5-aza-2′-deoxycytidine (Aza), a DNA methylation enzyme inhibitor, was combined with 50 mg/L Kan in the medium, Kan resistance was recovered in all four mutants (Fig. S3), indicating that the silencing of 35S-NPTII is caused by DNA methylation. Northern blot analyses did not detect NPTII expression in any of the Kan-sensitive mutants, but detected NPTII expression in the wild type (Fig. 3 B and C). Addition of 5-aza-cytidine increased the expression of NPTII in the wild type and in all mutants (Fig. 3B). Interestingly, expression of NPTII induced by 5-aza-cytidine treatment was much lower in ros4 than in the wild type, ros1, nrpd1, or nrpe1 (Fig. 3B). This result suggests that ROS4 but not ROS1 is required for the expression of NPTII that is released from silencing by 5-aza-cytidine. Furthermore, we used ddm1, a DNA hypomethylation mutant (in C24 ecotype, identified in another screen) (21), to compare the effects of mutations in ROS1 or ROS4 on NPTII expression by Northern blot and quantitative (q)RT-PCR analyses. NPTII transcripts were more abundant in ddm1 than in the wild type, indicating that the TGS of 35S-NPTII could be released by the mutation in DDM1 (Fig. 3 C and D). The expression of NPTII in ddm1 was not affected by the ros1 mutation but was greatly reduced by the ros4 mutation, indicating that the ros4 mutation inhibits ddm1-promoted expression of NPTII. We also found that the expression of NPTII in ddm1 was enhanced by nrpd1 but not by nrpe1 (Fig. 3 C and D), suggesting that NRPD1 and NRPE1 play different roles in regulating the DNA methylation and/or chromatin structure of the 35S promoter.
Next, we used the chromatin immunoprecipitation (ChIP) assay and different antibodies to analyze the histone H3 modifications in the 35S promoter. We included ros1-1 as a control. Previous studies indicated that H3K9me was increased and H3K4me was reduced in the ros1 mutant (22, 23). As shown in Fig. 3E, both H3K18ac and H3K23ac levels were decreased in ros4 relative to the wild type, but ros1-1 did not show a clear change in the levels of H3K18ac and H3K23ac. Both ros4 and ros1-1 had a higher level of H3K9 dimethylation and a lower level of H3K4 dimethylation than the wild type. These results further support ROS4 as important for histone H3K18 and H3K23 acetylation in vivo and suggest that ROS1 may function either downstream of ROS4 or independently of ROS4 in the silencing of 35S-NPTII. Our results also suggest that ROS4 is critical for the expression of the 35S-NPTII transgene that is normally silenced by DNA methylation, even when the DNA methylation is inhibited by 5-aza-cytidine treatment or by the ddm1 mutation.
Effect of ROS4 and RdDM Factors on the Expression of RD29A-LUC or Endogenous RD29A.
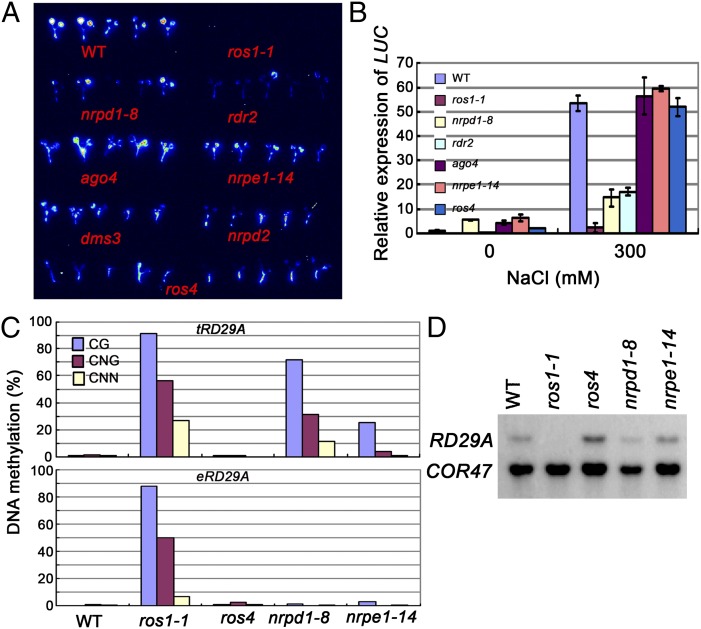
Because ROS1 is required for the expression of RD29A-LUC as well as 35S-NPTII, we tested whether RD29A-LUC is silenced by ros4 or the RdDM mutations. As shown previously (9) and confirmed here, ros1-1 emitted undetectable luminescence (Fig. 4A). nrpd1-8 and rdr2 emitted less luminescence than nrpe1-14, ago4, dms3, and nrpd2, and the latter four emitted similar luminescence as the wild type. ros4 mutation did not change the luminescence level. The expression of RD29A-LUC as indicated by luminescence was confirmed by qRT-PCR (Fig. 4B).
Fig. 4.
ROS4 does not regulate the expression of RD29A-LUC or of the endogenous RD29A gene. (A) Luminescence images of different mutants. Ten-day-old seedlings were treated with 300 mM NaCl for 3.5 h before luminescence images were obtained. (B) Quantitative RT-PCR analyses of LUC transcripts in different mutants. Three independent experiments (each with three biological replicates) were done with similar results. Error bars refer to standard error (n = 3). (C) DNA methylation analyses of the transgene RD29A (tRD29A) and the endogenous RD29A (eRD29A) promoter by bisulfite sequencing in the wild type, ros1-1, ros4, nrpd1-8, and nrpe1-14. (D) The expression of the endogenous RD29A gene in the wild type, ros1-1, ros4, nrpd1-8, and nrpe1-14 under 300 mM NaCl treatment. The stress-inducible gene COR47 was used as a control for effective stress treatments.
We measured the DNA methylation by bisulfite sequencing. In both the wild type and ros4, no DNA methylation could be detected in the RD29A-LUC transgene (tRD29A) promoter (Fig. 4C). However, the DNA methylation was increased to a higher level in nrpd1-8 and nrpe1-14 than in the wild type. DNA methylation was higher in nrpd1-8 than in nrpe1-14, and was lower in both nrpd1-8 and nrpe1-14 than in ros1-1. These results indicate that NRPD1 and NRPE1 have different effects on RD29A-LUC and suggest that RD29A-LUC may also be regulated by an RdDM-independent pathway. The increased DNA methylation in nrpd1-8 and nrpe1-14 is probably due to down-regulation of ROS1, which leads to severe DNA hypermethylation, and part of this may be independent of the RdDM pathway and not suppressed by the nrpd1 and nrpe1 mutations.
DNA methylation of the endogenous RD29A (eRD29A) promoter was not detected in nrpd1-8, nrpe1-14, or ros4 but was detected at a high level in ros1 (Fig. 4C) (9). The expression of RD29A was consistently silenced in ros1-1 but not in ros4, nrpd1-8, or nrpe1-14 (Fig. 4D).
ROS4 Is Important for ddm1-Promoted Expression of some Transposable Element Genes.
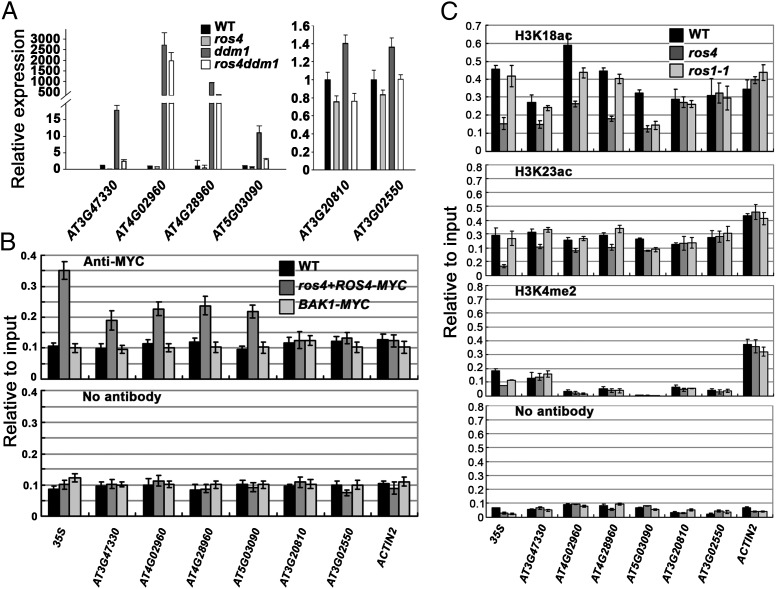
Because the ros4 mutation led to reduction of the ddm1-promoted expression of 35S-NPTII, we hypothesized that the expression of ROS4-targeted endogenous genes would be lower in the ros4 ddm1 double mutant than in the ddm1 single mutant. We compared the expression profiles among the wild type, ddm1, ros4 ddm1, and ros4 by a microarray assay. We chose genes whose expression was reduced in ros4, was increased in ddm1, and was lower in ddm1 ros4 than in ddm1. We selected six genes from our preliminary microarray data and checked their expression by qRT-PCR (Fig. 5A). Among them, the expression of three transposable element genes, AT3G47330, AT4G02960, and AT4G28960, was highly induced by the ddm1 mutation, but the expression was attenuated when the ddm1 mutation was combined with the ros4 mutation. Because the expression of each is already silenced in the wild type, further reduced expression by ros4 could not be detected. The expression of AT5G03090, AT3G20810, and AT3G02550 was consistent with the microarray data, that is, these genes were expressed at higher levels in ddm1 than in ddm1 ros4 and at lower levels in ros4 than in the wild type. Our results suggest that ROS4 is critical for the expression of some endogenous genes that are normally silenced by DNA methylation, even when the DNA methylation is inhibited by the ddm1 mutation.
Fig. 5.
ROS4 is important for the ddm1-promoted expression of endogenous genes. (A) The expression of six selected genes as determined by qRT-PCR in the wild type, ros4, ddm1, and ros4 ddm1. At least three independent experiments (each with three biological replicates) were done with similar results. (B) ROS4-Myc binds to the 35S promoter and the promoters of four endogenous genes as indicated by ChIP assay. The ChIP assay was performed on 7-d-old seedlings of the ros4 mutant complemented by the ROS4 promoter::ROS4-Myc, the wild type, and a transgenic plant expressing BAK1-Myc. At least three independent experiments (each with three biological replicates) were done with similar results. (C) H3K18ac, H3K23ac, and H3K4me2 levels in the wild type, ros4, and ros1-1. The ChIP assay used 7-d-old seedlings and H3K18ac, H3K23ac, or H3K4me3 antibodies. No antibody was used as a negative control. In A–C, error bars refer to standard error (n = 3).
ROS4 Targets 35S-NPTII and Three Transposable Element Genes.
To determine whether ROS4 directly targets 35S-NPTII and the other six genes, we used a ChIP assay. We transformed the ros4 mutant with the ROS4 promoter::ROS4-Myc and obtained transgenic plants in which the Kan-sensitive phenotype of ros4 was complemented. Because ROS4 can be detected in early seedling development, we used 5- to 7-d-old seedlings for our assays. ChIP assays using Myc antibody indicated that ROS4-Myc, but not BAK1-Myc (BRASSINOSTEROID INSENSITIVE 1-ASSOCIATED KINASE 1, as an unrelated control protein), was enriched in the 35S promoter, AT3G47330, AT4G02960, AT4G28960, and AT5G03090, but not in AT3G20810, AT3G02550, or ACTIN2 (Fig. 5B), suggesting that the latter three genes are not targeted by ROS4. Although the expression of AT3G20810 and AT3G02550 was increased by ddm1 and repressed when ddm1 was combined with ros4, it is unlikely that ROS4 directly targets these two genes.
We then used antibodies for H3K18ac and H3K23ac in the ChIP assay, and found that H3K18ac and H3K23ac in the promoter regions of AT3G47330, AT4G02960, AT4G28960, and AT5G03090 were apparently reduced in ros4 relative to the wild type (Fig. 5C). H3K18ac and H3K23ac in the promoter regions of AT3G20810, AT3G02550, and ACTIN2 were not changed by the ros4 mutation. The mutation in ROS1 only resulted in the reduction of H3K18ac and H3K23ac in the promoter of AT5G03090, but not in other promoter regions. Consistent with the results in Fig. 3E, H3K18ac and H3K23ac of the 35S promoter were greatly reduced in ros4 relative to the wild type, but this was not the case in ros1-1 (Fig. 5C). H3K4me2 levels were very low in the 35S promoter and the five selected genes; as a control, the ACTIN2 promoter had a high level of H3K4me2. These results indicate that besides affecting the 35S promoter, ROS4 directly affects histone acetylation of some endogenous genes.
Discussion
DNA methylation and histone modifications are crucial for regulating chromatin structure and gene transcription (3). DNA methylation tends to increase heterochromatin status and silence genes. Active DNA demethylation is an important mechanism for maintaining gene activity in both plants and animals (24). In this study, we characterized a PHD finger-containing protein that binds to H3K4me0 and suppresses gene silencing caused by DNA methylation. We showed that the expression of 35S-NPTII can be enhanced by 5-aza-cytidine treatment and by the ddm1 mutation in the wild-type C24, indicating that the 35S promoter is already silenced in C24 wild-type plants by DNA methylation, although this silencing is not complete because there is still considerable expression of the transgene. Both ROS1 and ROS4 are needed to maintain the expression of 35S-NPTII in the wild type (Fig. S4). ROS1 and ROS4 may function in the same pathway. Because ROS4 does not control the RD29A-LUC transgene and ROS4-dependent H3K18 and H3K23 acetylation are not affected by ROS1, ROS4 may regulate the activity or targeting of ROS1 to a subset of loci subjected to active DNA demethylation. The histone acetylation activity of ROS4 may be important for creating a chromatin environment necessary for ROS1 to function. However, ROS4 may have additional functions in promoting the expression of some genes. This is indicated by our observation that when 5-aza-cytidine treatment or the ddm1 mutation led to up-regulation of the expression of NPTII and some endogenous genes, the expression was reduced by the ros4 mutation but not by the ros1 mutation. It is also possible that ROS4 and ROS1 may prevent the silencing of 35S-NPTII and other genes via different mechanisms. During the preparation of this manuscript, Qian et al. (25) reported that ROS4/IDM1 recognizes methyl-CG, unmethylated H3K4, and unmethylated H3R2 marks on a subset of target loci of active DNA demethylation, and generates acetylated H3K18 and H3K23 marks, which are important for the DNA demethylation and attenuation of silencing of the recognized loci.
The T-DNA locus used for the genetic screening contains 35S-NPTII and RD29A-LUC. Previous studies indicated that the expression of these two genes is regulated by different epigenetic mechanisms (7). In this study, we found that both 35S-NPTII and RD29A-LUC are not regulated by one simple mechanism. We used Kan as a selection marker to identify the genes that positively regulate the expression of NPTII. Strikingly, most of the identified genes are components in the RdDM pathway. Our results revealed that the down-regulation of ROS1 by mutations in these RdDM components was the cause of 35S-NPTII silencing (10).
It seems that NRPD1 and NRPE1 play similar roles in controlling 35S-NPTII due to their regulation of ROS1, because both mutations led to the silencing of 35S-NPTII. In the ddm1 mutant background, however, nrpd1 but not nrpe1 appeared to be important in negatively regulating 35S-NPTII: NPTII transcripts were more abundant in the nrpd1-8 ddm1 double mutant than in the ddm1 single mutant or the nrpe1-14 ddm1 double mutant. It is possible that in addition to indirectly controlling 35S-NPTII via ROS1, Pol IV may also directly regulate this locus by generating silencing RNAs. The differing roles of Pol IV and Pol V are further suggested by our observation that nrpd1-8 and nrpe1-14 exhibited different levels of RD29A-LUC expression and different levels of transgene RD29A promoter DNA methylation. Given that Pol IV is required for siRNA biogenesis and Pol V transcripts are needed for the siRNAs to cause DNA methylation at RdDM target loci, it is possible that other transcripts from RNA polymerases such as Pol II might substitute for Pol V transcripts in the RdDM pathway (26).
Histone hyperacetylation is a major characteristic of actively transcribed genes, whereas histone hypoacetylation is a hallmark of transcriptionally inactive genes (27). In Arabidopsis, there are four histone acetyltransferase (HAT) families: GNAT (Gcn5-related N-acetyltransferase), MYST (MOZ, Ybt2, Sas2, and Tip60-like), p300/CBP, and TAFII250. Arabidopsis also has three histone deacetyltransferase (HDAC) families: RPD3/HDA1, HD2, and SIR2 (27). Previous studies have identified HDA6, a histone deacetylase that removes acetyl groups from the histone tail, as a key regulator that is not required for RNA-directed de novo methylation but is required for maintaining TGS by enhancing DNA methylation (28–30). HDA6 also regulates the TGS of both 35S-NPTII and RD29A-LUC (11). HDA6 is responsible for removing acetyl groups from H3K14, H4K5, and H4K12 (31). HDT1, another plant-specific histone deacetylase, is required for H3K9 deacetylation (32). Histone H3 (H3K9, H3K14) and H4 (H4K5, H4K8, H4K12, H4K16) acetylations are catalyzed by different HATs (33). ROS4 encodes a putative histone acetyltransferase that shows high similarity in the acetyltransferase domain to GNAT families (27). ROS4 is a single-copy gene in Arabidopsis, and its orthologs are found in both dicot and monocot plants, but not in animals. The ros4 mutation specifically causes a reduction in the acetylation of H3K18 and H3K23 but not of H3K9 and H3K14, suggesting that ROS4 might mainly acetylate H3K18 and H3K23 in vivo. We have also provided evidence that H3K18ac and H3K23ac are reduced by the ros4 mutation in the 35S promoter and in the promoters of four endogenous target genes. Because H3K18 and H3K23 acetylation are only reduced but not completely absent in ros4, we suspect that other histone acetyltransferases might work redundantly with ROS4 in acetylating H3K18 and H3K23. ROS4 appears to preferentially associate with chromatin lacking methylated H3K4, suggesting that unmethylated H3K4 is a common epigenetic feature for ROS4 target loci.
Materials and Methods
Seeds were sterilized and sown onto MS medium plates containing 2% (wt/vol) sucrose and 0.8% (wt/vol) agar. After 4 d at 4 °C, the plates were transferred to a growth chamber at 22 °C under long-day (23-h light/1-h dark) conditions. Seven-day-old seedlings were transferred to soil and kept in a growth room at 20 °C under 16-h light/8-h dark.
C24 carrying transgenes RD29A-LUC and 35S-NPTII (9) was used as the wild type. Except for the ddm1 mutant, which was described before (21), all other mutants in this study were screened from the EMS-mutagenized population of the wild type. Wild-type seeds can germinate and grow on MS medium containing 50 mg/L kanamycin. M2 seeds were sown on MS medium containing 50 mg/L kanamycin. After seeds germinated and grew in a growth chamber for 1 wk, the kanamycin-sensitive seedlings were transferred to MS medium without kanamycin to recover. The putative mutants were confirmed on 50 mg/L kanamycin medium in the next generation and then backcrossed with the C24 wild type for genetics analysis. If F1 seedlings exhibited the C24 phenotypes and F2 progeny from self-fertilized F1 plants showed an approximate 3:1 segregation of kanamycin-resistant to -sensitive phenotypes, the mutants were selected and crossed with Col-0. The F2 plants that showed kanamycin sensitivity and also carried the NPTII gene (as indicated by PCR) were used for mapping. Map-based cloning was performed as described (23).
For the complementation assay, the region from −2499 to −1 of ROS4 and the ROS4 full-length cDNA-fused MYC tag were in-sequence–cloned into pCAMBIA1300 (relevant primers are listed in Table S1). Then, Agrobacterium tumefaciens strain GV3101 carrying the constructs was transformed into ros4. The vector containing 35S::ROS1, which was described before (9), was used to transform other mutants. Transgenic plants that were resistant to both 30 mg/L hygromycin and 50 mg/L kanamycin in the T2 (second generation of transgenic plants) generation were selected. Two independent lines were used for further study.
RNA Analysis.
Real-time PCR was carried out to detect transcription levels of different genes. Total RNA was extracted from 5-d-old seedlings using TRIzol reagent (Invitrogen). After the RNA was incubated with RNase-free DNase I (New England BioLabs) at 37 °C to remove remaining DNA, 4 μg of RNA was reverse-transcribed to cDNA with M-MLV reverse transcriptase (Promega). The cDNAs were then diluted 10-fold, and 1 μL of the dilution was added to each 20-μL reaction system in SYBR Green Master Mix (TaKaRa). The PCR procedure was performed as described (34). Specific primers of each gene are listed in Table S1. EF1a was used as an internal control.
For Northern blotting, 15 μg of total RNA of each sample was used for detecting NPTII expression following the method previously described (23), except that 5-d-old seedlings were used. Total RNA used in RD29A and COR47 expression assays was extracted after the seedlings were treated in 300 mM NaCl for 3.5 h. Primers are listed in Table S1.
Bisulfite Sequencing.
The EZ Methylation-Gold Kit (Zymo Research) was used for DNA methylation analysis. A 500-ng quantity of DNA was added in each reaction system, and all operations followed the protocol supplied with the kit. Approximately 75 ng of bisulfite-treated DNA was used in the PCR, which was carried out with primers that were specific for each region (Table S1). PCR products were cloned using the pMD18-T vector (TaKaRa), and at least eight independent clones of each sample were sequenced.
Histone Peptide-Binding Assay.
The following biotinylated histone peptides were purchased from Millipore and used in this binding assay: H2A (residues 12–406), H2B (12–407), H3 (1–21) (12–403), H3 (21–44) (12–404), H3K4me1 (12–563), H3K4me2 (12–460), H3K4me3 (12–564), H3K9me1 (12–569), H3K9me2 (12–430), H3K9me3 (12–568), H3K27me1 (12–567), H3K27me2 (12–566), H3K27me3 (12–565), and H4 (12–372). The PHD domain (residues 723–769) was cloned into the vector pGEX4T-2, expressed in E. coli, and purified by the glutathione Sepharose TM 4B column (GE Healthcare; 17-0756-01). Briefly, 0.5 μg of the peptides was incubated overnight at 4 °C with 5 μg of the GST-fused PHD-finger protein in a binding buffer [20 mM Hepes, pH 7.0, 250 mM NaCl, 0.1% (vol/vol) Triton X-100, 1 mM phenylmethylsulfonyl fluoride]. Streptavidin beads (Millipore) were then added and incubated for 1 h. After they were washed three times, the beads bound with proteins were analyzed by SDS/PAGE and stained with Coomassie blue.
Chromatin Immunoprecipitation Assay.
ChIP was performed as described previously (35) using 5-d-old seedlings grown in a growth chamber at 22 °C under 23-h light/1-h dark conditions. The antibodies used in the ChIP assays were anti-H3K4me2 (Millipore; 17-677), anti-H3K9me2 (Millipore; 17-648), anti-H3K18ac (Millipore; 07-354), anti-H3K23ac (Millipore; 07-355), and anti-MYC (Sigma; F3165). Finally, ChIP products were dissolved in 50 μL of water, and 0.5 μL of the solution was used in each qPCR reaction. Specific primers are listed in Table S1.
Histone Extraction and Western Blotting.
Seedlings were ground to powder in a mortar cooled with liquid nitrogen, and the powder was suspended in NIB buffer (250 mM sucrose, 60 mM KCl, 15 mM NaCl, 5 mM MgCl2, 1 mM CaCl2, 15 mM Pipes, pH 6.8, 0.8% Triton X-100). The suspension was centrifuged, and the pellet was resuspended in 0.4 M H2SO4 and incubated for at least 1 h at 4 °C. The preparation was centrifuged again before acetone was added to precipitate the histone protein. The preparation was kept at −20 °C overnight before the proteins were dissolved in 4 M urea and analyzed by Western blotting. The antibodies used in the Western blotting were anti-H3K9ac (Millipore; 07-352), anti-H3K14ac (Millipore; 07-353), anti-H3K18ac (Millipore; 07-354), and anti-H3K23ac (Millipore; 07-355).
Histochemical GUS Staining.
A DNA fragment containing the region from −2499 to −1 of ROS4 was cloned into the pCAMBIA1391 vector. The recombinant plasmid was introduced into A. tumefaciens strain GV3101, followed by transformation into C24 wild-type plants. Histochemical GUS staining was performed on transgenic T2 plants that were resistant to hygromycin as previously described (23).
Localization of the ROS4-GFP Fusion Protein.
The full-length cDNA of ROS4 without TAA (encoding a stop codon) was cloned into the modified vector pCAMBIA1300 at the site just behind the 35S promoter. Then GFP was fused in-frame with ROS4 at its C terminus. Transient expression in tobacco and GFP image acquisition were described previously (34).
Supplementary Material
Acknowledgments
This work was supported by grants from the National Nature Science Foundation of China (31121002).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1208557109/-/DCSupplemental.
References
- 1.Chinnusamy V, Zhu JK. RNA-directed DNA methylation and demethylation in plants. Sci China C Life Sci. 2009;52:331–343. doi: 10.1007/s11427-009-0052-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furner IJ, Matzke M. Methylation and demethylation of the Arabidopsis genome. Curr Opin Plant Biol. 2011;14(2):137–141. doi: 10.1016/j.pbi.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 3.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ. RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol. 2009;21:367–376. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 5.Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Q, Gong Z. The coupling of epigenome replication with DNA replication. Curr Opin Plant Biol. 2011;14(2):187–194. doi: 10.1016/j.pbi.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 8.Zhu JK. Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet. 2009;43:143–166. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gong Z, et al. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- 10.Huettel B, et al. Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J. 2006;25:2828–2836. doi: 10.1038/sj.emboj.7601150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He XJ, et al. NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation. Genes Dev. 2009;23:318–330. doi: 10.1101/gad.1765209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He XJ, et al. A conserved transcriptional regulator is required for RNA-directed DNA methylation and plant development. Genes Dev. 2009;23:2717–2722. doi: 10.1101/gad.1851809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He XJ, et al. An effector of RNA-directed DNA methylation in Arabidopsis is an ARGONAUTE 4- and RNA-binding protein. Cell. 2009;137:498–508. doi: 10.1016/j.cell.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng X, Zhu J, Kapoor A, Zhu JK. Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J. 2007;26:1691–1701. doi: 10.1038/sj.emboj.7601603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng X, et al. ROS3 is an RNA-binding protein required for DNA demethylation in Arabidopsis. Nature. 2008;455:1259–1262. doi: 10.1038/nature07305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee WY, Lee D, Chung WI, Kwon CS. Arabidopsis ING and Alfin1-like protein families localize to the nucleus and bind to H3K4me3/2 via plant homeodomain fingers. Plant J. 2009;58:511–524. doi: 10.1111/j.1365-313X.2009.03795.x. [DOI] [PubMed] [Google Scholar]
- 17.Shi X, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442(7098):96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lan F, et al. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007;448:718–722. doi: 10.1038/nature06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Org T, et al. The autoimmune regulator PHD finger binds to non-methylated histone H3K4 to activate gene expression. EMBO Rep. 2008;9:370–376. doi: 10.1038/embor.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanchez R, Zhou MM. The PHD finger: A versatile epigenome reader. Trends Biochem Sci. 2011;36:364–372. doi: 10.1016/j.tibs.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen J, Ren X, Cao R, Liu J, Gong Z. Transcriptional gene silencing mediated by a plastid inner envelope phosphoenolpyruvate/phosphate translocator CUE1 in Arabidopsis. Plant Physiol. 2009;150:1990–1996. doi: 10.1104/pp.109.139626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kapoor A, et al. Mutations in a conserved replication protein suppress transcriptional gene silencing in a DNA-methylation-independent manner in Arabidopsis. Curr Biol. 2005;15:1912–1918. doi: 10.1016/j.cub.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 23.Xia R, et al. ROR1/RPA2A, a putative replication protein A2, functions in epigenetic gene silencing and in regulation of meristem development in Arabidopsis. Plant Cell. 2006;18(1):85–103. doi: 10.1105/tpc.105.037507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gong Z, Zhu JK. Active DNA demethylation by oxidation and repair. Cell Res. 2011;21:1649–1651. doi: 10.1038/cr.2011.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qian W, et al. A histone acetyltransferase regulates active DNA demethylation in Arabidopsis. Science. 2012 doi: 10.1126/science.1219416. 10.1126/science.1219416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng B, et al. Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes Dev. 2009;23:2850–2860. doi: 10.1101/gad.1868009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pandey R, et al. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 2002;30:5036–5055. doi: 10.1093/nar/gkf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aufsatz W, Mette MF, van der Winden J, Matzke M, Matzke AJ. HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. EMBO J. 2002;21:6832–6841. doi: 10.1093/emboj/cdf663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murfett J, Wang XJ, Hagen G, Guilfoyle TJ. Identification of Arabidopsis histone deacetylase HDA6 mutants that affect transgene expression. Plant Cell. 2001;13:1047–1061. doi: 10.1105/tpc.13.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Probst AV, et al. Arabidopsis histone deacetylase HDA6 is required for maintenance of transcriptional gene silencing and determines nuclear organization of rDNA repeats. Plant Cell. 2004;16:1021–1034. doi: 10.1105/tpc.018754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Earley K, et al. Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes Dev. 2006;20:1283–1293. doi: 10.1101/gad.1417706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lawrence RJ, et al. A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol Cell. 2004;13:599–609. doi: 10.1016/s1097-2765(04)00064-4. [DOI] [PubMed] [Google Scholar]
- 33.Earley KW, Shook MS, Brower-Toland B, Hicks L, Pikaard CS. In vitro specificities of Arabidopsis co-activator histone acetyltransferases: Implications for histone hyperacetylation in gene activation. Plant J. 2007;52:615–626. doi: 10.1111/j.1365-313X.2007.03264.x. [DOI] [PubMed] [Google Scholar]
- 34.Liu Q, et al. DNA replication factor C1 mediates genomic stability and transcriptional gene silencing in Arabidopsis. Plant Cell. 2010;22:2336–2352. doi: 10.1105/tpc.110.076349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saleh A, Alvarez-Venegas R, Avramova Z. An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc. 2008;3:1018–1025. doi: 10.1038/nprot.2008.66. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.