For many decades, scientists have studied the cell’s ability to sense and respond to mechanical force. Recent advances in assaying cells using increasingly complex mechanical environments have lead to new insights into cellular function (1). Mechanical force plays a critical role within cells and has the capacity to affect diverse problems ranging from environmental change (2) to malaria (3). The importance of biomechanical forces is particularly apparent in many human diseases, such as heart disease and cancer (1). In PNAS, Hur et al. (4) describe an elegant, 3D force analysis approach they use to investigate cell mechanics that have important implications in atherosclerosis.
A central tenet in the study of cell mechanics holds that cells remain in constant communication with each other while thriving in environments that are ultimately 3D. Although the link between cellular communication and 3D forces has been appreciated, the vast majority of experiments that use cultured animal cells continue to be conducted on planar substrates (e.g., Petri dishes) that are effectively 2D environments, primarily because robustly controlling the 3D microenvironment of cells is not trivial. Hur et al. use a unique method, 3D inter/intracellular force microscopy (IFM). Building off their recent results in expanding 2D traction force microscopy (TFM) to analyze 3D stresses, the authors use IFM to examine 3D tension across individual cell bodies and at the junction between cells. In these experiments, endothelial cells—the type of cells that line blood vessels—are grown both sparsely and also in confluent monolayers, where communication between many cells must occur, to examine differences in each condition.
Using their IFM approach, the authors recover information normally lost in 2D systems, with profound implications when examining inter- and intracellular tension in these multicellular systems. In addition, the authors expose endothelial cells to shear stress by flowing fluid on their apical surface and couple these experiments with IFM. The resulting experiments expose cells to two flow profiles (laminar steady flow or oscillatory flow) that generate different modes of shear force and mimic specific locations in the vasculature that produce strikingly different responses relevant to atherosclerosis. In addition to generating insight into cellular mechanics, endothelial cell function, and vascular pathologies, this study points to the broad need to develop 3D experimental methods that mimic different physiological environments.
Assessing 3D Mechanical Force in Cells
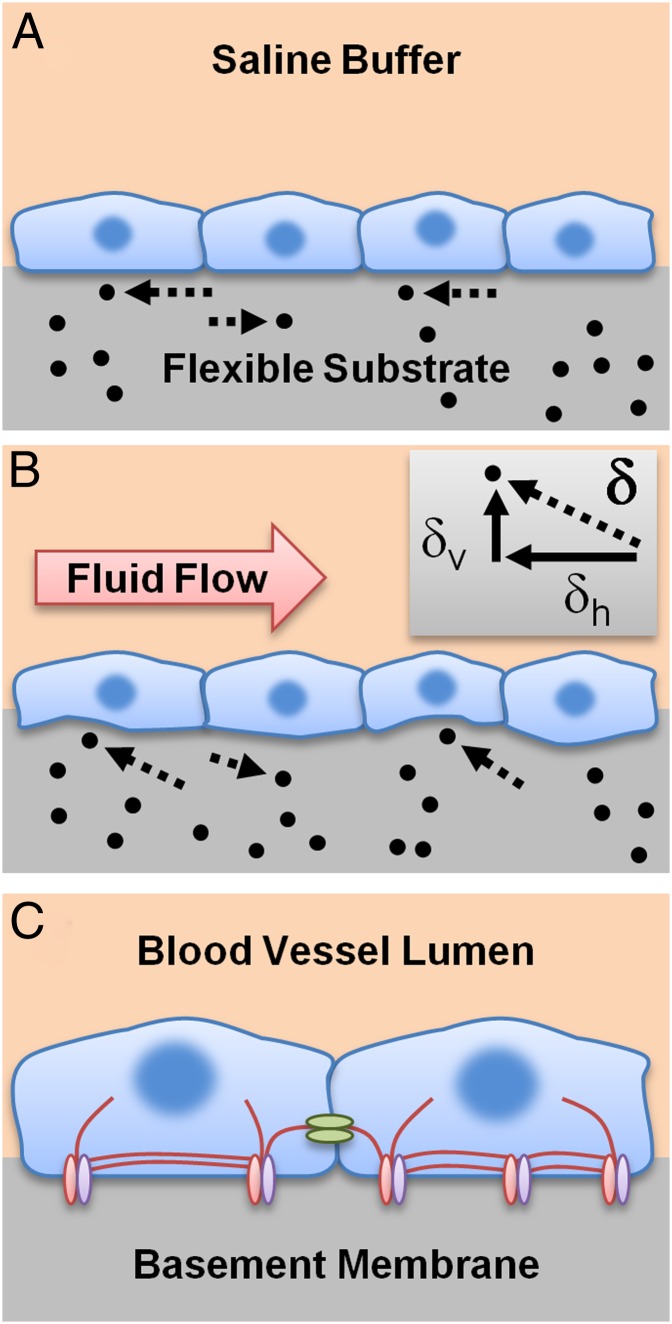
Developing new 3D experimental approaches is essential because organisms generate diverse physiological functions by specializing in three dimensions. Recent efforts include imparting localized control of apical–basal 3D stimulation, developing chip-based microsystems for probing mechanically activated calcium signaling, using 3D traction methods in cell migration, and examining 3D tractions of cells fully encapsulated in gels (5–8). Although our ability to control a cell’s micromechanical environment has advanced, assessing mechanical forces within a cellular monolayer remains a challenge. In their study, the authors assay these forces by adapting the fundamental principles of TFM, a method that measures 2D (planar) forces (9) at the point where the cell’s ventral surface contacts a flexible substrate. By optically noting the displacement of beads placed in these flexible substrates and applying Newton’s first law, traction forces applied by cells to their environment can be inferred (Fig. 1A). Unlike previous studies, the authors recently expanded this technique to perform 3D TFM (10) by measuring not only the planar, horizontal movements of beads, but also vertical movements (Fig. 1B).
Fig. 1.
IFM reveals 3D intracellular and junction tensions in endothelial cells. (A) Traditional 2D TFM detects only horizontal displacements in beads embedded in a flexible substrate of known stiffness, allowing the determination of the traction forces applied by cells to their substrate. (B) 3D TFM determines both the horizontal (δh) and vertical (δv) components of the displacement vector (δ), allowing calculation of a 3D traction force vector. For IFM, mathematically balancing forces across a collection of adjacent cells allows intracellular and junction tensions to be estimated. Additionally, fluid flow (red arrow) can be combined with IFM to apply shear force to cells cultured on flexible substrates. (C) In the body, endothelial cells sense shear force generated by flow in the blood vessel lumen and balance stress using a complex collection of structural protein components, including cytoskeletal stress fibers such as actin (red lines), focal adhesion complexes (red and purple ellipses) that attach to their substrate (basement membrane), and adherens junction complexes (green ellipses) that connect neighboring cells. Although this study has implications for understanding atherosclerosis, the underlying concepts are broadly applicable to fields ranging from environmental mechanics to cancer treatment.
In the present study, the authors perform 3D TFM and apply Newton’s third law to determine tension within cells and at the junctions between cells in a monolayer. Their study suggests that a type of mechanical interconnection between cells—called adherens junctions—could play a significant role in vascular physiology. These interconnections are one part of a complicated mechanical sensing scheme, and other factors affecting the mechanical response of the cells include substrate stiffness and the ability of the cell to attach itself to its substrate using organized protein complexes called focal adhesions (Fig. 1C). In the future, their approach could be integrated into systems for active control of cell physiology; for example, by coupling IFM with active control of substrates through light-activated polymers (11) or assessing changes with IFM when particular focal adhesion proteins (such as syndecans) dominate the formed attachments (12).
Analyzing Mechanics Within Confluent Monolayers
A significant impact of the work by Hur et al. is an increased understanding of how endothelial cells in confluent monolayers control their morphology and internal tension. The authors examine the biomechanical responses of cells grown in monolayers, representative of in vivo cellular conditions, as well as cells grown in two-cell groups. Cells within a monolayer had dramatically different tension formation response in comparison with cells growing in two-cell groups, particularly when stimulated by a shear flow. In two-cell groups, tangential intracellular tension was highest at the cell–cell junction, yet at distances away from the cell–cell junction, tension decreased.
On the other hand, for confluent monolayers exposed to a shear flow, tension distributions at cell–cell junctions were not statistically different from tension away from the junctions. The cells in a confluent monolayer generated similar mechanical tension throughout the cellular monolayer, which was not the case for the two-cell groups. The difference in mechanical sensing and response seen between single cell groups and confluent monolayers is important in other ways as well. For example, a recent study showed that cell motility is very different for single cells in comparison with monolayers, particularly because in monolayers a “tug-of-war” state exists (13). These types of cell–cell interactions will be especially important in understanding physiology as the field of cell mechanics continues to expand its use of 3D approaches.
Steady and Oscillatory Flow Encode Different 3D Mechanical Responses
Just as the authors are able to approach new physiological questions by exploring
A type of mechanical interconnection between cells—called adherens junctions—could play a significant role in vascular physiology.
3D tension within a multicellular monolayer, the mode (steady or oscillatory) of shear flow they apply to monlayers also makes a difference. Although the reaction of cells to laminar, steady flow is very relevant to endothelial cell physiology, it is especially interesting to examine their responses to oscillatory flow because this flow mode most clearly corresponds to flow in areas of the vasculature (i.e., branches) where pathologies like atherosclerosis can occur. It is well known that endothelial cells respond to different modes of shear flow; for example, by altering intracellular calcium signaling (14), biosynthesis of cartilage (15), and regulation of cell cycle (16).
However, investigators had not previously attempted to monitor internal, 3D traction forces in endothelial cells exposed to these two modes of shear-generating flow. By doing so, Hur et al. find that when applying laminar steady flow to a monolayer to mimic atheroprotective vascular flow—resulting in 12 dyne/cm2 of constant shear stress—endothelial cells adopt an elongated morphology that results in maximum intracellular tension in the flow direction. In contrast, when they apply oscillatory flow to mimic conditions in areas particularly susceptible to atherosclerosis, cells in monolayers maintain their preflow morphology and generate no significant change in tension in any direction. Determining what these results imply for the initiation of vasculature pathologies will be a challenge, and as previously noted, the authors suggest a critical role for adherens junctions in the process. Ultimately, this work highlights the need to integrate new 3D approaches with existing experimental modalities to gain richer physiological insights.
The rapid pace of discovery in 3D cell mechanics is resulting in dramatic increases in scientific understanding. These achievements will continue to influence biomechanics at all scales. One example is the emergence of organ-on-a-chip microdevices, a technology that combines knowledge from studies in cell biomechanics, tissue engineering, and microfabrication to create 3D microenvironments that better mimic tissues and organs (17). Exciting possibilities exist in adapting these biomimetic devices as model systems for testing drug efficacy among other biological responses, thus reducing the need for animal and human testing. A cornerstone of this technology is the integration of 3D cell mechanics that accurately mimic the in vivo tissue environment. We anticipate an exciting future in which experimental microsystems continue to address more complicated 3D biomechanical problems, spanning scales from the molecular to the ecological, and significantly impacting our understanding of biological systems.
Acknowledgments
We received support from Grants CMMI-0856187, CMMI-1160840, and CNS-1135850 from the National Science Foundation and Grant N000140910215 from the Office of Naval Research.
Footnotes
The authors declare no conflict of interest.
See companion article on page 11110.
References
- 1.Fletcher DA, Mullins RD. Cell mechanics and the cytoskeleton. Nature. 2010;463:485–492. doi: 10.1038/nature08908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waite JH, Broomell CC. Changing environments and structure—property relationships in marine biomaterials. J Exp Biol. 2012;215:873–883. doi: 10.1242/jeb.058925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suresh S, et al. Connections between single-cell biomechanics and human disease states: Gastrointestinal cancer and malaria. Acta Biomater. 2005;1:15–30. doi: 10.1016/j.actbio.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Hur SS, et al. Roles of cell confluency and fluid shear in 3-dimensional intracellular forces in endothelial cells. Proc Natl Acad Sci USA. 2012;109:11110–11115. doi: 10.1073/pnas.1207326109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim Y, Joshi SD, Davidson LA, LeDuc PR, Messner WC. Dynamic control of 3D chemical profiles with a single 2D microfluidic platform. Lab Chip. 2011;11:2182–2188. doi: 10.1039/c1lc20077a. [DOI] [PubMed] [Google Scholar]
- 6.Ruder WC, et al. Three-dimensional microfiber devices that mimic physiological environments to probe cell mechanics and signaling. Lab Chip. 2012;12:1775–1779. doi: 10.1039/c2lc21117c. [DOI] [PubMed] [Google Scholar]
- 7.Legant WR, et al. Measurement of mechanical tractions exerted by cells in three-dimensional matrices. Nat Methods. 2010;7:969–971. doi: 10.1038/nmeth.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maskarinec SA, Franck C, Tirrell DA, Ravichandran G. Quantifying cellular traction forces in three dimensions. Proc Natl Acad Sci USA. 2009;106:22108–22113. doi: 10.1073/pnas.0904565106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munevar S, Wang YL, Dembo M. Traction force microscopy of migrating normal and H-ras transformed 3T3 fibroblasts. Biophys J. 2001;80:1744–1757. doi: 10.1016/s0006-3495(01)76145-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hur SS, Zhao Y, Li YS, Botvinick E, Chien S. Live cells exert 3-dimensional traction forces on their substrata. Cell Mol Bioeng. 2009;2:425–436. doi: 10.1007/s12195-009-0082-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guvendiren M, Burdick JA. Stiffening hydrogels to probe short- and long-term cellular responses to dynamic mechanics. Nat Commun. 2012;3:792. doi: 10.1038/ncomms1792. [DOI] [PubMed] [Google Scholar]
- 12.Bellin RM, et al. Defining the role of syndecan-4 in mechanotransduction using surface-modification approaches. Proc Natl Acad Sci USA. 2009;106:22102–22107. doi: 10.1073/pnas.0902639106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tambe DT, et al. Collective cell guidance by cooperative intercellular forces. Nat Mater. 2011;10:469–475. doi: 10.1038/nmat3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helmlinger G, Berk BC, Nerem RM. Calcium responses of endothelial cell monolayers subjected to pulsatile and steady laminar flow differ. Am J Physiol. 1995;269:C367–C375. doi: 10.1152/ajpcell.1995.269.2.C367. [DOI] [PubMed] [Google Scholar]
- 15.Kim YJ, Sah RL, Grodzinsky AJ, Plaas AH, Sandy JD. Mechanical regulation of cartilage biosynthetic behavior: physical stimuli. Arch Biochem Biophys. 1994;311:1–12. doi: 10.1006/abbi.1994.1201. [DOI] [PubMed] [Google Scholar]
- 16.Zhou J, et al. Force-specific activation of Smad1/5 regulates vascular endothelial cell cycle progression in response to disturbed flow. Proc Natl Acad Sci USA. 2012;109:7770–7775. doi: 10.1073/pnas.1205476109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huh D, et al. Reconstituting organ-level lung functions on a chip. Science. 2010;328:1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]