Abstract
The primary hormone-binding surface of the insulin receptor spans one face of the N-terminal β-helix of the α-subunit (the L1 domain) and an α-helix in its C-terminal segment (αCT). Crystallographic analysis of the free ectodomain has defined a contiguous dimer-related motif in which the αCT α-helix packs against L1 β-strands 2 and 3. To relate structure to function, we exploited expanded genetic-code technology to insert photo-activatable probes at key sites in L1 and αCT. The pattern of αCT-mediated photo–cross-linking within the free and bound receptor is in accord with the crystal structure and prior mutagenesis. Surprisingly, L1 photo-probes in β-strands 2 and 3, predicted to be shielded by αCT, efficiently cross-link to insulin. Furthermore, anomalous mutations were identified on neighboring surfaces of αCT and insulin that impair hormone-dependent activation of the intracellular receptor tyrosine kinase (contained within the transmembrane β-subunit) disproportionately to their effects on insulin binding. Taken together, these results suggest that αCT, in addition to its hormone-recognition role, provides a signaling element in the mechanism of receptor activation.
Keywords: nonstandard mutagenesis, alanine scanning, affinity
Insulin plays a central role in the control of vertebrate metabolism. Despite its long-standing use in the treatment of diabetes mellitus, how insulin binds to and activates the insulin receptor (IR) poses a major unsolved problem (1). Recent advances in the structural characterization of the IR ectodomain have provided a foundation for reinvestigation of this classic problem (2, 3). Here, we have used structure-based photo–cross-linking (PCL) and mutagenesis to identify a dynamic signaling element at the hormone-binding surface of the ectodomain.
Studies of insulin derivatives containing photo-activatable substitutions (para-azido-Phe; Pap) suggested that insulin undergoes a change in conformation on IR binding (4), partially exposing the hydrophobic core on displacement of the C-terminal segment of the B-chain (5). Such induced fit expands its nonpolar contact surface (1, 6). We hypothesized that induced fit of the IR could likewise expand its contact surface and in turn trigger receptor tyrosine kinase (TK) activation. Our studies focused on a tandem hormone-binding element comprising dimer-related structural motifs in the N-terminal β-helix of the α-subunit (L1 domain residues 1–158) and its C-terminal segment (αCT residues 704–715) (7, 8). Introduction of Pap was accomplished by orthogonal tRNA/amber suppression (9). Guided by PCL, we have characterized a unique class of mutations in insulin and the IR ectodomain that impair TK activation disproportionately to effects on binding. Taken together, our results identify cognate recognition α-helices in insulin and its receptor that participate in transmission of the insulin signal.
Our strategy had three parts. First, photo-probes were introduced by nonstandard mutagenesis (9–12) based on an ectodomain crystal structure (2, 3) and prior Ala scanning mutagenesis (8). PCL studies were then undertaken to validate structural relationships in the holoreceptor and test their relevance to the complex. Finally, surfaces adjoining critical ligand-receptor contacts were scanned in search of “signaling mutations,” defined as substitutions that impair activation disproportionately to binding. We anticipated that signaling elements in a receptor would exhibit intrinsic mobility and flexible linkage to other structural modules. Application to the IR focused on αCT, which (i) exhibits higher crystallographic thermal factors than the neighboring L1 β-helix, and (ii) is flexibly linked to the preceding type III fibronectin-homology domain (Fn3 in Fig. 1 A and B). Prior PCL studies of insulin established contacts between αCT and an A-chain α-helix (residues A1–A8) (4).
Fig. 1.
IR domain organization and ectodomain structure. (A) IR dimer: disulfide-bridged αβ monomer (Left) and ectodomain (Right), comprising (αβΔ)2 dimer wherein βΔ represents a fragment lacking transmembrane α-helix and intracellular domains. L1 and αCT are highlighted in green and red (asterisk at Left), respectively. Domains (gray scale) are otherwise designated cysteine-rich (CR), second Leu-rich repeat domain (L2), type III fibronectin-homology domains (Fn1-3), insert domain (segments ID-N and ID-C), transmembrane and juxtamembrane regions (TM/JM), tyrosine kinase (TK), and C-terminal segment (CT; 704-FEDYLHNVVFV-715; α-helix bold, disordered underlined). Disulfide bridges are shown as horizontal lines. (B) Crystal structure of ectodomain. One αβΔ protomer is shown as a ribbon; the other as sticks. L1 and αCT in each protomer are highlighted in green and red. Electron density of ID-N is incomplete. (C and D) Interaction between αCT α-helix (red) and L1 β-helix (green). (C) Side view of αCT (stick) and L1 (ribbon). L1 side chains pertinent to PCL are shown: Leu36, Leu37, Leu62, and Phe64 (black), and Arg47 and Phe51 (negative control sites, blue). Also shown is negative control site for apo receptor dimerization (Thr704; magenta). Tyr714, positive control site for insulin PCL, is not well ordered in structure. (D) Side view of αCT (CPK model). B–D are based on Protein Data Bank entry 3LOH. D is shown in stereo in SI Appendix, Fig. S1.
Results
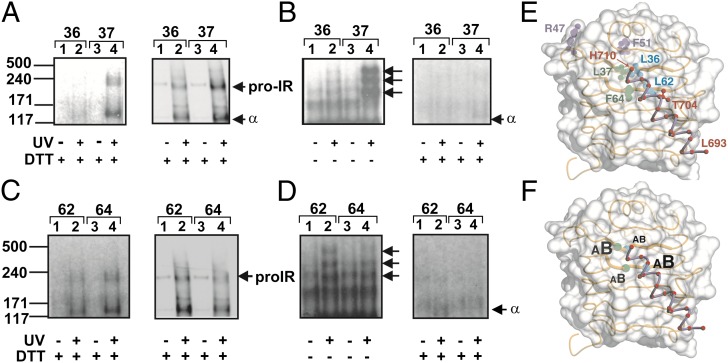
Pap was singly incorporated at IRα sites 36, 37, 46, 51, 62, 64, 704, or 714 (designated IRX36, IRX37, and so forth) (Fig. 1 C and D and SI Appendix, Fig. S1). Binding assays revealed decreased hormone affinities, with the exception of control constructs IRX36 and IRX37 (Fig. 2A and SI Appendix, Table S1). Initial studies focused on insulin-binding site F714 in αCT. As expected, PCL between IRX714 and 125I-TyrA14-insulin was observed. Photo-adducts (P-A), resolved by reducing SDS/PAGE, included a unique 125I-labeled band of apparent mass 134 kDa (Fig. 2B), consistent with an A-chain/IRα adduct. Nonreducing SDS/PAGE revealed major bands corresponding to an insulin/IR complex (ins/α2β2) and its partially reduced forms (ins/α2β and ins/αβ, presumably because of contaminating reductants). Precipitation of P-As by an anti-FLAG IgG confirmed the presence of the IR variant. Specificity of IRX714 PCL was confirmed by its disappearance in the presence of unlabeled insulin and attenuation in the presence of IGF-I (each 100 nM). Control studies were undertaken of IRX47 and IRX51 (peripheral to the L1-αCT hormone-binding surface) (Fig. 1 C and D and SI Appendix, Fig. S1) (8, 13). Following nonreducing SDS/PAGE, no P-As were detected (Fig. 2C).
Fig. 2.
IR-directed PCL. (A) Insulin binding assays by variant receptors. Wild-type receptor (●), IRX714 ( ), IRX62 (
), IRX62 ( ), and (
), and ( ) IRX714. (B) SDS/PAGE of positive control studies: PCL of 125I-TyrA14-insulin (125I-ins) to IRX714. Gel position of cross-linked 125I-ins/IR complex is labeled α. In lanes 4 and 5, 125I-TyrA14-insulin/IR complexes were preincubated in presence of insulin and IGF1 (100 nM) before PCL. In lanes 6 and 7, cross-linked 125I-ins/IR complexes were precipitated with anti-FLAG M2 monoclonal IgG (lane 6) or nonimmune IgG (lane 7). (C and D) Control PCL studies. (C) Cross-linked 125I-ins /IR complexes of IRX714, IRX47, and IRX51 were resolved by nonreducing SDS/PAGE; residues 47 and 51 are located on opposite side of L1 from insulin-binding sites (Fig. 1). Unreduced and partially reduced P-A are designated by arrows (Left); estimated masses 460, 320, and 240 K correspond to IR-α2β2, IR-α2β, and IR-αβ. (D) Reduced cross-linked α subunits of IRX704, IRX47, and IRX51 were blotted with anti-IR α-subunit antibody IαN (also designated N-20); residue 704 is located on opposite side of αCT/L1 interface (Fig. 1). α-Subunit and uncleaved proreceptor are designated α and pro-IR, respectively. (E) Role of residues 36 and 37 in α−α dimerization. Reduced cross-linked α-subunits of IRX36 and IRX37 were blotted with antibody IαN; α-subunit monomers and dimers are designated m and d. (F) Role of residues 62 and 64 in α−α subunit dimerization. Reduced crosslinked α subunits of IRX62 and IRX64 were blotted with antibody IαN; α-monomers and -dimers are designated m and d. Vertical white lines in panels separate images from different gels or different parts of same gel.
) IRX714. (B) SDS/PAGE of positive control studies: PCL of 125I-TyrA14-insulin (125I-ins) to IRX714. Gel position of cross-linked 125I-ins/IR complex is labeled α. In lanes 4 and 5, 125I-TyrA14-insulin/IR complexes were preincubated in presence of insulin and IGF1 (100 nM) before PCL. In lanes 6 and 7, cross-linked 125I-ins/IR complexes were precipitated with anti-FLAG M2 monoclonal IgG (lane 6) or nonimmune IgG (lane 7). (C and D) Control PCL studies. (C) Cross-linked 125I-ins /IR complexes of IRX714, IRX47, and IRX51 were resolved by nonreducing SDS/PAGE; residues 47 and 51 are located on opposite side of L1 from insulin-binding sites (Fig. 1). Unreduced and partially reduced P-A are designated by arrows (Left); estimated masses 460, 320, and 240 K correspond to IR-α2β2, IR-α2β, and IR-αβ. (D) Reduced cross-linked α subunits of IRX704, IRX47, and IRX51 were blotted with anti-IR α-subunit antibody IαN (also designated N-20); residue 704 is located on opposite side of αCT/L1 interface (Fig. 1). α-Subunit and uncleaved proreceptor are designated α and pro-IR, respectively. (E) Role of residues 36 and 37 in α−α dimerization. Reduced cross-linked α-subunits of IRX36 and IRX37 were blotted with antibody IαN; α-subunit monomers and dimers are designated m and d. (F) Role of residues 62 and 64 in α−α subunit dimerization. Reduced crosslinked α subunits of IRX62 and IRX64 were blotted with antibody IαN; α-monomers and -dimers are designated m and d. Vertical white lines in panels separate images from different gels or different parts of same gel.
Studies of Free IRα.
The αCT/L1 interface in the free ectodomain (3) contains L1 residues Leu36, Leu37, Leu62, and Phe64 (black in Fig. 1 C and D and SI Appendix, Fig. S1). To probe this interface in the free IR, we investigated corresponding Pap variants in the absence of insulin. SDS/PAGE of IRX36, IRX37, IRX62, and IRX64 was undertaken before or after UV irradiation; P-As resolved under reducing conditions were probed by anti-IRα antiserum IαN. Whereas unirradiated samples yielded only a single band of apparent mass 134 kDa (Fig. 2 E and F), consistent with IRα, irradiation gave rise to new bands of apparent masses 261 and 290 kDa, consistent with covalent α2 complexes (Fig. 2 E and F). This doublet presumably arises as a consequence of heterogeneity of cross-linking, leading to heterogeneous SDS binding and electrophoretic mobility as has previously been described with Pap-mediated PCL of proteins (14). To demonstrate the specificity of α2 PCL, negative controls were provided by IRX47, IRX51, and IRX704; covalent α2 complexes were not detected (Fig. 2D).
Studies of the Insulin–IR Complex.
Ala substitutions at the conserved L1 sites Leu36, Leu37, or Phe64 (15) were previously found to impair insulin binding (8). However, our recent refinement of the crystal structure of the IR ectodomain indicated that these L1 residues are buried beneath the αCT helix (3), with relative surface accessibilities less than 1% (3) (SI Appendix, Table S2). Such impairment could be indirect (secondary to disruption of L1/αCT packing) or direct (if hormone binding displaces αCT). To distinguish between these models, we first tested whether IRX36, IRX37, IRX62 [also predicted to contact αCT (Fig. 1 C and D, and SI Appendix, Fig. S1)], and IRX64 exhibited PCL despite their low affinities. Surprisingly, PCL was in each case observed and yielded covalent 125I-labeled A-chain/IRα and biotin-labeled B-chain/IRα complexes (Fig. 3 A and C). PCL was mediated more efficiently by IRX37 than by IRX36, IRX62, or IRX64. As expected under nonreducing conditions, 125I was incorporated into the same bands as observed in control studies of IRX714 (Fig. 2C). To determine which insulin chain was the predominant PCL target, equal aliquots of each 125I-labeled cross-linked lysate were analyzed by nonreducing and reducing SDS/PAGE to exploit the A-chain–specific 125I label (Fig. 3 B and D). Whereas A-chain–specific PCL would retain the label irrespective of reduction, B-chain–specific PCL would lose the label following reduction. At each L1 site, the extent of radio-labeling was attenuated by reduction (Fig. 3 B and D), implying that such PCL predominantly involved the B-chain in accord with prior PCL studies of insulin (4, 16). Sites of Pap substitution are summarized in Fig. 3E and their relative PCL efficiencies to the A- and B-chains of insulin are shown in schematic form in Fig. 3F. These findings indicate that αCT must undergo conformational change to enable access of insulin to these side chains, consistent with insulin-induced reduction in α2 dimer formation (Fig. 2 E and F).
Fig. 3.
IR-directed PCL to insulin A- or B-chains. (A and B) Cross-linking of L1 β-strand 2 IRX mutants. (A) Reducing SDS/PAGE of insulin cross-linking to IRX36 and IRX37. (Left) Autoradiograph of cross-linked 125I-TyrA14-insulin/IRX36 and /IRX37 complexes. (Right) Blot of cross-linked insulin-BxB/IRX36 and /IRX37 complexes probed with streptavidin. (B) Comparison of cross-linking of insulin A-chain and whole insulin molecule to IRX36 and IRX37. Equal aliquots of cross-linked 125I-ins/IRX36 and /IRX37 complexes were resolved in absence or presence of reductant (DTT−/+) and processed identically for autoradiography. Positions of unreduced and partially reduced α-subunit P-A are indicated by arrows as in Fig. 2C. (C and D) Analysis of cross-linking of L1 β-strand 3 IR-Pap mutants. (C) Reducing SDS analysis of insulin cross-linking to IRX62 and IRX64. (Left) Autoradiograph of cross-linked 125I-ins/IRX62 and /IRX64 complexes. (Right) Blot of cross-linked insulin-BxB/IRX62 and /IRX64 complexes probed with streptavidin. (D) Comparison of cross-linking of insulin A-chain and whole insulin molecule to IRX62 and IRX64. Equal aliquots of cross-linked 125I-ins/IRX62 and /IRX64 complexes were resolved in absence or presence of reductant (DTT−/+) and processed identically for autoradiography. Positions of unreduced and partially reduced α-subunit P-A are indicated by arrows as in B. (E and F) Schematic summary of Pap-directed PCL as mapped against surface of L1 and bound αCT α-helix (ribbon) (3). (E) Sites of Pap substitutions in engineered receptors. (F) Comparative cross-linking to insulin A- and B-chains is represented by respective sizes of lettering.
Role of αCT in IR Activation.
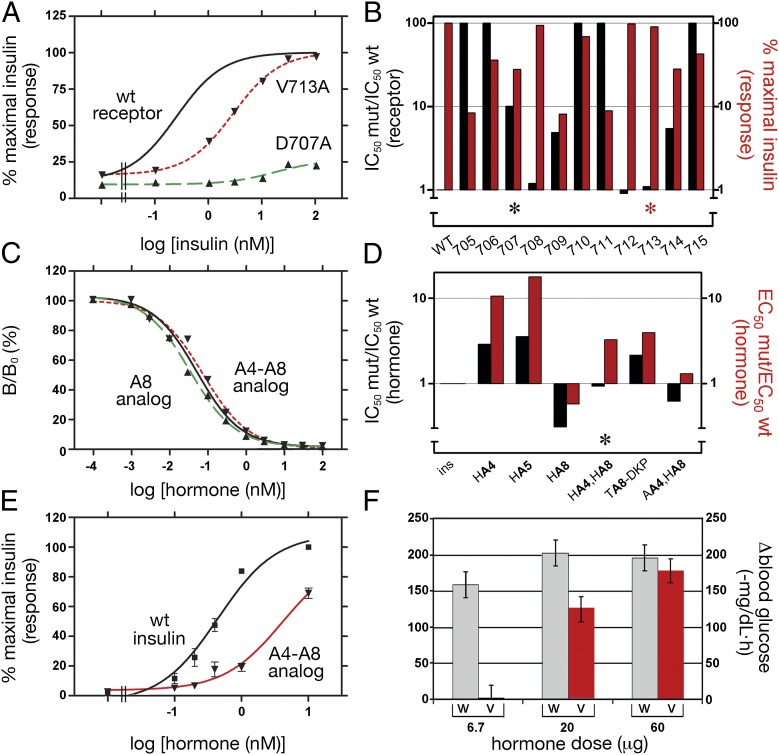
To test the functional role of αCT, we examined effects of Ala substitutions (8) on hormone binding and TK activation at equilibrium. Insulin-binding assays provided IC50 values (and so extent of IR occupancy). The TK assays by contrast measured cumulative Tyr autophosphorylation after 30 min (a kinetic probe of IR activation); in the absence of phosphatases, such activation reflects the sum of basal and hormone-stimulated rates. Under these conditions, binding of insulin enhanced Tyr autophosphorylation by 6- to 10-fold (Fig. 4A). Five possible outcomes were anticipated: (i) no perturbation of insulin binding or IR autophosphorylation may occur; (ii) Ala substitutions at conventional hormone-binding sites may impair insulin binding with no change in maximal IR autophosphorylation; (iii and iv) Ala substitutions at signaling sites may either exhibit a right-shift in the dependence of autophosphorylation on IR occupancy or a decreased maximal IR autophosphorylation; or (v) Ala substitutions may both impair hormone binding and abolish signaling without clear interpretation. Possible outcomes because of failure to reach equilibrium are excluded, as the kinetics of receptor association are only altered by mutations perturbing interactions between residues in the insulin hexamer surface and the receptor FnIII-1 and -2 domains (17).
Fig. 4.
Functional studies of variant receptors and insulin analogs. (A) Insulin dose-response curves for IR autophosphorylation for wild-type IR (solid line), D707A (dashed line, green), and V713A (dotted line, magenta). (B) Effects of Ala substitutions in αCT (704-FEDYLHNVVFV-715; α-helix bold, disordered underlined) on hormone binding (black bars and left vertical axis; ratio of variant:wild-type IC50 values) and insulin-stimulated autophosphorylation (red bars and right vertical axis; extent of modification relative to wild-type receptor). Higher black bars indicate greater impairment of hormone binding; higher red bars indicate greater retention of native autophosphorylation. Values at positions 707 and 713 are highlighted by black and red asterisks. Numerical values for these data are in SI Appendix, Table S2. (C) Competitive binding of insulin analogs: insulin (black solid line), HisA8-insulin (▲) and HisA4-HisA8-insulin (▼). (D) Histogram showing receptor-binding affinities and IR activation potencies of insulin analogs: ins, insulin; HA4, HisA4-insulin; HA5, HisA5-insulin; HA8, HisA8-insulin; HA4-HA8, HisA4-HisA8-insulin; TA8-DKP, ThrA8-DKP-insulin; and AA4-HA8, AlaA4-HisA8-insulin. (E) Dose-response of insulin-stimulated Akt phosphorylation: wild-type insulin (black squares) and HisA4-HisA8-insulin (▼). (F) Hypoglycemic potencies in diabetic rats: wild-type insulin (gray bars) and HisA4-HisA8-insulin (red bars) at successive doses.
Negative pattern (i) was observed at sites 708 and 712 (Fig. 4B and SI Appendix, Table S2). Binding-site pattern (ii) was observed at sites 706, 710, and 715 (Fig. 4B and SI Appendix, Table S2). E706 is orthogonal to the α-helix; H710 sits atop the α-helix; no density was observed for V715 (2, 3). Signaling pattern (iii) was observed at site 713 (Fig. 4B, red asterisk). Dose-response studies of IR autophosphorylation demonstrated a right-shift with unchanged maximal response (Fig. 4A). V713 is disordered in the crystal structure of the free ectodomain (2, 3). Signaling pattern (iv) was closest to that observed at site 707 (Fig. 4B, asterisk, and SI Appendix, Table S2). D707A (a clinical mutation associated with insulin resistance) led to 10-fold reduction in binding with a 4-fold decrease in autophosphorylation at saturating insulin concentrations (Fig. 4 A and B); D707 sits atop the αCT α-helix. This pattern was also observed with F714A; F714 is disordered in the crystal structure (2, 3). Representative of pattern (v) is F705A (Fig. 4B), which impaired binding by >250-fold (SI Appendix, Table S2) and abolished autophosphorylation (<10% of maximal wild-type response). This mixed pattern was also observed at sites 709 and 711 (Fig. 4B); F705 is completely and L709 partially buried at the L1-αCT interface; no density was observed for N711 (2, 3).
To test whether insulin side chains adjoining αCT [i.e., within the A1–A8 α-helix (4)] might also contribute to signaling, we prepared insulin analogs with substitutions at GluA4, GlnA5, and ThrA8, at which receptor binding is tolerant of Ala substitution (6). A-chain modifications were identified that exhibited disproportionate impairment of signaling (Fig. 4 C and D). In particular, the double-mutant HisA4-HisA8-insulin (Fig. 5A) exhibited native receptor binding (Fig. 4C) but a rightward shift in IR autophosphorylation with fivefold increased EC50 value (Fig. 4D, asterisk). These in vitro findings were extended to cell culture as probed by insulin-dependent serine phosphorylation of the downstream Akt kinase. The dose-response of Akt phosphorylation by HisA4-HisA8-insulin was right-shifted relative to wild-type insulin (Fig. 4E). Studies of hypoglycemic potency in a rat model demonstrated a corresponding reduction in activity at submaximal dose (Fig. 4F); full potency was elicited at a supraphysiological dose. Studies of analogs AlaA4-HisA8-insulin and HisA8-insulin suggested that the critical signaling-related residue is GluA4 (red in Fig. 5B); the A8 substitution in HisA4-HisA8-insulin serves to restore native affinity. Because the structure of HisA4-HisA8-insulin is essentially identical to wild-type (18), these findings suggest that an Asp707-αCT/GluA4-insulin related joint surface functions to trigger conformational changes in the IR leading to TK activation and downstream signaling.
Fig. 5.
Structure of HisA4-HisA8-insulin and dual role of A1-A8 α-helix. (A) Model of receptor-bound conformation of insulin and key structural features based on Rf-state protomer of mutant insulin as extracted from T3Rf3 zinc hexamer (Protein Database entry 3KQ6). A- and B-chain ribbons are shown in dark and light gray. Dashed segments in B-chain indicate proposed sites of conformational change: GlyB8 (22) and preceding N-terminal segment (Upper Left, ●); PheB24 (5) with detachment of C-terminal segment (Lower Left). Such detachment exposes IleA2 and ValA3 (green) for binding to αCT. Substitution of GluA4 by His (red) and ThrA8 by His (dark gray) yield an analog with native binding but partial defect in hormone-dependent IR autophosphorylation. Cystines (A6-A11, A7-B7, and A20-B19) are shown in gold and sulfur atoms as gold spheres. (B) Cylinder model of amphipathic A1-A8 α-helix proposed to play dual roles in receptor binding (IleA2 and ValA3;  ) and signaling (GluA8;
) and signaling (GluA8;  ). Cystine A6-A11 (gold) buttresses the α-helix. A1–A8 sequence and sites of substitution (↓) are shown (Lower).
). Cystine A6-A11 (gold) buttresses the α-helix. A1–A8 sequence and sites of substitution (↓) are shown (Lower).
Discussion
This study has probed structure-function relationships at the hormone-binding surface of IR. We focused on two insulin-binding elements, L1 and αCT, at a unique dimer interface (3). First, receptor-based PCL, enabled by amber-suppression technology (9), was used to establish the presence of this interface in the holoreceptor and investigate its role in insulin binding. Experimental design focused on αCT and contiguous side chains in L1 (Fig. 1 D–F). Second, we identified an anomalous class of mutations in the cognate recognition α-helices of αCT and insulin required for IR TK activation.
PCL Studies.
PCL fidelity was assessed in control studies of IRX714. Disordered in the crystal structure (2), Phe714 was proposed to contact insulin based on its conservation, mutagenesis and key role in a minimal model of the hormone-IR complex (19). As predicted, IRX714 cross-linked to insulin whereas photo-probes at the edge of L1 do not. Such cross-linking involves the insulin A chain in accord with photo-scanning of insulin (4). We next investigated conserved sites in L1 that lie beneath αCT in the free ectodomain: Leu36, Leu37, Leu62, and Phe64 (β-strands 2 and 3). Ala substitutions at sites 36, 37, 62, or 64 each impaired insulin binding (SI Appendix, Table S1 and ref. 8). As predicted by the ectodomain structure (3), IRX36, IRX37, IRX62, and IRX64 in the absence of insulin gave rise to covalent α−α complexes. Dimer-related PCL was not observed in control studies of IR variants containing Pap distant from this or other dimer interfaces (IRX47, IRX51, and IRX704). These findings indicate that key structural relationships in the ectodomain extend to the holoreceptor.
Variants IRX36, IRX37, IRX62, and IRX64 were designed to probe whether the L1/αCT motif undergoes a change in conformation on insulin binding. Because αCT electron density is less well defined than that of L1 and not continuously connected across ID-N, we imagined that αCT may move on insulin binding, exposing the underlying L1 surface. Such movement would presumably be coupled to induced fit of insulin (5), which is likewise proposed to expose nonpolar surfaces in its classic core (1). The present studies provide evidence that these L1 residues indeed engage insulin. Just as conserved nonpolar side chains in insulin play dual roles in self-assembly and IR binding (1), we envisage that conserved nonpolar side chains in L1 contribute to both α–α dimerization (in the free receptor) and hormone binding (specific complex).
αCT Functions as an IR Signaling Element.
To test whether αCT contributes to IR activation, we characterized Ala substitutions in αCT and probed the cognate IR-binding surface of insulin (A1–A8 α-helix). αCT substitutions were associated with variable effects on insulin binding and autophosphorylation. Of particular interest, D707A [originally detected in a patient with extreme insulin resistance (20)] almost completely blocked activation of IR autophosphorylation, even at saturating concentrations of insulin. In the cognate surface of insulin pairwise substitution of GluA4 and ThrA8 by His has no net effect on insulin affinity but produced a rightward shift in dose-response of IR autophosphorylation. This shift was also seen in studies of Akt phosphorylation and in rat studies of hormone potency. Taken together, these results suggest that a polar composite αCT/A-chain surface functions to transmit the insulin signal to the IR β-subunit. Our findings are in accord with the partial agonism of αCT-mimetic peptides (19, 21).
Induced Fit of Insulin.
Previous studies of insulin analogs have suggested that, on receptor binding, the B-chain undergoes respective changes in conformation involving its N- and C-terminal segments (Fig. 5A). (i) In the TR transition among insulin hexamers, the conformation of the N-terminal segment (extended or α-helical) is coupled to the sign of the ϕ dihedral angle of GlyB8 (1). Whereas the free hormone resembles the classic T state (1), studies of d- and l-amino acid substitutions at B8 suggest that the bound hormone adopts an R-like B8 conformation (22). (ii) Evidence for detachment of the C-terminal segment of the B-chain from the α-helical core has likewise been provided by studies of destabilizing d-substitutions at PheB24 (5). Such detachment would expose IleA2 and ValA3 for αCT binding. We speculate that induced fit of insulin is coupled to changes in IR conformation associated with TK activation.
Limitations of PCL.
It is important to highlight the general limitations of PCL. Pap substitution and its photo-activation could perturb the structure of an interface, such that aberrant cross-links form. Furthermore, relative PCL efficiencies may reflect the chemical nature of neighboring functional groups. Finally, because of the prolonged time scale of UV irradiation (seconds), PLC may trap transient or intermediate contacts unrepresentative of the ground-state structure (23). Nonstandard Pap mutagenesis (11), nonetheless, provides a general approach to characterize membrane proteins as exemplified by studies of intracellular protein docking at the EGF receptor (12) and ligand binding by G protein-coupled receptors (10, 11).
Concluding Remarks.
Our results provide evidence that αCT functions as both an insulin-binding element and internal “trigger” in transmembrane signaling. Efforts are in progress to obtain crystals of hormone-receptor complexes to permit analysis of αCT position relative to L1 and insulin. Such structures will be of mechanistic interest and may aid design of nonpeptidic agonists for the treatment of diabetes.
Materials and Methods
Detailed materials and methods are available in SI Appendix.
Materials.
Plasmids encoding amber suppressor tyrosyl tRNA from Bacillus stearothermophilus and an engineered Pap-specific tRNA synthetase from Escherichia coli were provided by S.-X. Ye and T. P. Sakmar (Rockefeller University, New York, NY) (24). FLAG-tagged IR and insulin analogs were prepared as previously described (8, 18).
General Methods.
PCR-based subcloning used the In-Fusion Cloning System (Clontech); site-directed mutagenesis was performed using QuikChange (Agilent Technologies). Constructs were in each case verified by DNA sequencing. Insulin-binding and IR autophosphorylation assays were performed as previously described (8).
Transient Receptor Expression.
The 293PEAK rapid cells (ATCC) were maintained and cotransfected as previously described (8). Three days posttransfection, cells were harvested in the dark by lysis in 0.15 M NaCl and 0.1 M Tris-HCl (pH 8.0) containing Triton X-100 (1% vol/vol), glycerol (10% vol/vol), and a protease inhibitor mixture.
Receptor PCL.
Modified receptors were partially purified in the dark by wheat-germ agglutinin chromatography (8). Eluates (50 μL) were incubated overnight at 4 °C with 125I-TyrA14-insulin (240 pM). For PCL, binding reactions were irradiated on ice for 15 s at a lamp distance of 2 cm. Covalent complexes were resolved by SDS/PAGE and visualized by autoradiography.
PCL Mapping.
IR cross-linking to 125I-TyrA14-insulin was assigned to the A chain based on 125I-labeling of PCL bands as resolved under reducing conditions by SDS/PAGE. Cross-linking to the B-chain was detected based on the biotin moiety of B1-biotin-labeled insulin analog (insulin-BxB) by streptavidin blotting.
Biological Assays.
Insulin-stimulated Akt phosphorylation was assessed in R− fibroblasts stably expressing IR (isoform B) (25). Hypoglycemic potency was measured in male Sprague-Dawley rats rendered diabetic by streptozotocin (18). The protocol was approved by the Institutional Animal Care and Use Committee of Case Western Reserve University.
Supplementary Material
Acknowledgments
We thank S. Chen, K. Huang, S. Nakagawa, and B. Xu for advice; S.-X. Ye and T. P. Sakmar for Pap-related constructs; S. Wang and S.-Q. Hu for insulin synthesis; P. G. Katsoyannis for biotin-labeled insulin analog; Novo-Nordisk A/S for 125I-insulin; Aubree Ng (Oregon National Primate Research Center) for technical assistance; and C. W. Ward for encouragement. This work was supported by the American Diabetes Association (J.W. and M.A.W.); National Institutes of Health Grants DK040949 (to J.W. and M.A.W.) and OD011092 (to C.T.R.); and Australian National Health and Medical Research Council Grant 1005896 and the Hazel and Pip Appel Fund and his contribution made possible through Victorian State Government Operational Infrastructure Support and Australian Government National Health and Medical Research Independent Research Institutes Infrastructure Support Scheme (to M.C.L.).
Footnotes
Conflict of interest statement: J.W. is a consultant to Thermalin Diabetes and owns stock in Novo-Nordisk A/S; L.J.W. owns stock in Novo-Nordisk A/S; N.B.P. is a consultant to Thermalin Diabetes; F.I.-B. owns stock in Thermalin Diabetes; and M.A.W. owns stock in Thermalin Diabetes, serves as its Chief Scientific Officer, is a member of its Board of Directors, and served as a consultant to Merck and the DEKA Research and Development Corporation.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205681109/-/DCSupplemental.
References
- 1.Baker EN, et al. The structure of 2Zn pig insulin crystals at 1.5 Ä resolution. Philos Trans R Soc Lond B Biol Sci. 1988;319:369–456. doi: 10.1098/rstb.1988.0058. [DOI] [PubMed] [Google Scholar]
- 2.McKern NM, et al. Structure of the insulin receptor ectodomain reveals a folded-over conformation. Nature. 2006;443:218–221. doi: 10.1038/nature05106. [DOI] [PubMed] [Google Scholar]
- 3.Smith BJ, et al. Structural resolution of a tandem hormone-binding element in the insulin receptor and its implications for design of peptide agonists. Proc Natl Acad Sci USA. 2010;107:6771–6776. doi: 10.1073/pnas.1001813107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu B, et al. Decoding the cryptic active conformation of a protein by synthetic photoscanning: Insulin inserts a detachable arm between receptor domains. J Biol Chem. 2009;284:14597–14608. doi: 10.1074/jbc.M900087200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hua QX, et al. Enhancing the activity of a protein by stereospecific unfolding: Conformational life cycle of insulin and its evolutionary origins. J Biol Chem. 2009;284:14586–14596. doi: 10.1074/jbc.M900085200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kristensen C, et al. Alanine scanning mutagenesis of insulin. J Biol Chem. 1997;272:12978–12983. doi: 10.1074/jbc.272.20.12978. [DOI] [PubMed] [Google Scholar]
- 7.Kurose T, et al. Cross-linking of a B25 azidophenylalanine insulin derivative to the carboxyl-terminal region of the α-subunit of the insulin receptor. Identification of a new insulin-binding domain in the insulin receptor. J Biol Chem. 1994;269:29190–29197. [PubMed] [Google Scholar]
- 8.Whittaker J, Whittaker L. Characterization of the functional insulin binding epitopes of the full-length insulin receptor. J Biol Chem. 2005;280:20932–20936. doi: 10.1074/jbc.M411320200. [DOI] [PubMed] [Google Scholar]
- 9.Young TS, Schultz PG. Beyond the canonical 20 amino acids: Expanding the genetic lexicon. J Biol Chem. 2010;285:11039–11044. doi: 10.1074/jbc.R109.091306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grunbeck A, Huber T, Sachdev P, Sakmar TP. Mapping the ligand-binding site on a G protein-coupled receptor (GPCR) using genetically encoded photocrosslinkers. Biochemistry. 2011;50:3411–3413. doi: 10.1021/bi200214r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coin I, Perrin MH, Vale WW, Wang L. Photo-cross-linkers incorporated into G-protein-coupled receptors in mammalian cells: A ligand comparison. Angew Chem Int Ed. 2011;50:8077–8081. doi: 10.1002/anie.201102646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hino N, et al. Protein photo-cross-linking in mammalian cells by site-specific incorporation of a photoreactive amino acid. Nat Methods. 2005;2:201–206. doi: 10.1038/nmeth739. [DOI] [PubMed] [Google Scholar]
- 13.Lou M, et al. The first three domains of the insulin receptor differ structurally from the insulin-like growth factor 1 receptor in the regions governing ligand specificity. Proc Natl Acad Sci USA. 2006;103:12429–12434. doi: 10.1073/pnas.0605395103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takimoto JK, Adams KL, Xiang Z, Wang L. Improving orthogonal tRNA-synthetase recognition for efficient unnatural amino acid incorporation and application in mammalian cells. Mol Biosyst. 2009;5:931–934. doi: 10.1039/b904228h. [DOI] [PubMed] [Google Scholar]
- 15.Hernández-Sánchez C, Mansilla A, de Pablo F, Zardoya R. Evolution of the insulin receptor family and receptor isoform expression in vertebrates. Mol Biol Evol. 2008;25:1043–1053. doi: 10.1093/molbev/msn036. [DOI] [PubMed] [Google Scholar]
- 16.Huang K, et al. How insulin binds: The B-chain α-helix contacts the L1 β-helix of the insulin receptor. J Mol Biol. 2004;341:529–550. doi: 10.1016/j.jmb.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 17.Sajid W, et al. Structural basis of the aberrant receptor binding properties of hagfish and lamprey insulins. Biochemistry. 2009;48:11283–11295. doi: 10.1021/bi901269j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phillips NB, et al. Supramolecular protein engineering: Design of zinc-stapled insulin hexamers as a long acting depot. J Biol Chem. 2010;285:11755–11759. doi: 10.1074/jbc.C110.105825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Menting JG, Ward CW, Margetts MB, Lawrence MC. A thermodynamic study of ligand binding to the first three domains of the human insulin receptor: Relationship between the receptor α-chain C-terminal peptide and the site 1 insulin mimetic peptides. Biochemistry. 2009;48:5492–5500. doi: 10.1021/bi900261q. [DOI] [PubMed] [Google Scholar]
- 20.Hart LM, et al. An insulin receptor mutant (Asp707 → Ala), involved in leprechaunism, is processed and transported to the cell surface but unable to bind insulin. J Biol Chem. 1996;271:18719–18724. doi: 10.1074/jbc.271.31.18719. [DOI] [PubMed] [Google Scholar]
- 21.Pillutla RC, et al. Peptides identify the critical hotspots involved in the biological activation of the insulin receptor. J Biol Chem. 2002;277:22590–22594. doi: 10.1074/jbc.M202119200. [DOI] [PubMed] [Google Scholar]
- 22.Hua QX, et al. Toward the active conformation of insulin: Stereospecific modulation of a structural switch in the B chain. J Biol Chem. 2006;281:24900–24909. doi: 10.1074/jbc.M602691200. [DOI] [PubMed] [Google Scholar]
- 23.Brunner J. New photolabeling and crosslinking methods. Annu Rev Biochem. 1993;62:483–514. doi: 10.1146/annurev.bi.62.070193.002411. [DOI] [PubMed] [Google Scholar]
- 24.Ye S, Huber T, Vogel R, Sakmar TP. FTIR analysis of GPCR activation using azido probes. Nat Chem Biol. 2009;5:397–399. doi: 10.1038/nchembio.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sohma Y, et al. Contribution of residue B5 to the folding and function of insulin and IGF-I: Constraints and fine-tuning in the evolution of a protein family. J Biol Chem. 2010;285:5040–5055. doi: 10.1074/jbc.M109.062992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.