Abstract
Autophagy has been implicated in a number of physiological processes important for human heath and disease. Autophagy involves the formation of a double-membrane cytosolic vesicle, an autophagosome. Central to the formation of the autophagosome is the ubiquitin-like protein autophagy-related (Atg)8 (microtubule-associated protein 1 light chain 3/LC3 in mammalian cells). Following autophagy induction, Atg8 shows the greatest change in expression of any of the proteins required for autophagy. The magnitude of autophagy is, in part, controlled by the amount of Atg8; thus, controlling Atg8 protein levels is one potential mechanism for modulating autophagy activity. We have identified a negative regulator of ATG8 transcription, Ume6, which acts along with a histone deacetylase complex including Sin3 and Rpd3 to regulate Atg8 levels; deletion of any of these components leads to an increase in Atg8 and a concomitant increase in autophagic activity. A similar regulatory mechanism is present in mammalian cells, indicating that this process is highly conserved.
Keywords: lysosome, membrane biogenesis, phagophore, stress, vacuole
Macroautophagy, hereafter referred to as autophagy, is an evolutionarily conserved process used by eukaryotic cells for the bulk degradation of intracellular proteins and organelles (1). Autophagy is not only vital for cell survival in nutrient-poor conditions (2) but is also linked to various physiological processes, including immune defense, tumor suppression, and prevention of neurodegeneration (3). Whereas autophagy plays a primarily protective role, it can also contribute to cell death; thus, the magnitude of autophagy must be carefully regulated.
Central to autophagy is the formation of autophagosomes (4), double-membrane–bound structures that engulf and deliver cytoplasmic materials to the vacuole/lysosome for degradation. During autophagosome formation, the autophagy-related ubiquitin-like protein Atg8/microtubule-associated protein 1 light chain 3 (LC3) covalently modifies phosphatidylethanolamine (PE). Almost one-fourth of the characterized autophagy-related (Atg) proteins in yeast are involved in the formation or stability of Atg8–PE, which plays a critical role in controlling expansion of the phagophore (the initial sequestering membrane), and in determining autophagosome size, thereby regulating autophagy activity (5, 6). Upon starvation, the level of ATG8 mRNA sharply increases, leading to a subsequent induction of the Atg8 protein level (7, 8). The increase in the amount of Atg8 during autophagy is critical for supplying a sufficient amount of this protein to maintain normal levels of autophagy; yeast strains deficient in Atg8 induction generate abnormally small autophagosomes (5). Thus, characterization of how Atg8 protein levels are modulated is of tremendous importance both in understanding the regulation of autophagy and for the elucidation of potential therapeutic targets. However, little is known about the mechanisms regulating ATG8 transcription.
Results
Ume6-Sin3-Rpd3 Complex Represses Atg8 Expression.
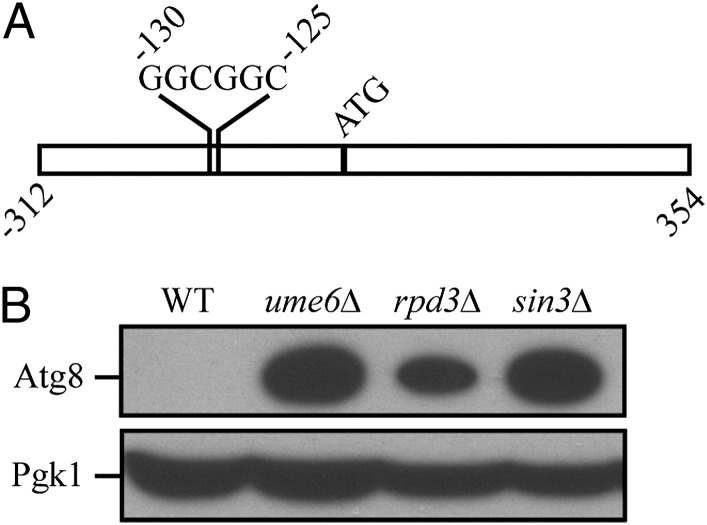
To identify candidate transcriptional regulators of ATG8, we analyzed its promoter region and identified an upstream regulatory sequence, URS1, which is a consensus binding site for the transcription factor Ume6 (Fig. 1A), which was previously identified during a whole-genome microarray analysis (9–11). The URS1 consensus site consists of two invariant GGC repeats, which tend to be immediately preceded by a C and several T nucleotides and followed by a T and two A nucleotides, although some variability exists in these positions (9). We examined the promoter regions of other ATG genes and note that the gene encoding Atg23 also contains a potential URS1 site. Ume6 is a zinc cluster protein that both represses and activates transcription of a diverse set of genes involved in meiosis and metabolism in response to nutritional cues such as glucose, nitrogen, and inositol (9, 12–14). If Ume6 regulates Atg8, then a ume6Δ strain should have altered Atg8 protein levels. We examined the protein level of Atg8 in a ume6Δ strain in vegetative (growing) conditions and found that it was substantially induced relative to the wild type (Fig. 1B).
Fig. 1.
(A) Diagram depicting the URS1 site in the ATG8 promoter. (B) The Ume6-Sin3-Rpd3 complex represses Atg8 expression. Wild-type (BY4742), ume6Δ, sin3Δ, and rpd3Δ yeast cells were grown in rich medium to midlog phase. Protein extracts from cells were prepared and subjected to immunoblotting with anti-Atg8 and anti-Pgk1 antiserum (the latter as a loading control).
Ume6 exerts control of transcription by forming a complex with the corepressor Sin3 and the histone deacetylase Rpd3 (15). Accordingly, we extended our analysis by examining the level of Atg8 in rpd3Δ and sin3Δ strains. Similar to the result with the ume6Δ strain, both the sin3Δ and rpd3Δ strains displayed a substantial induction of Atg8 expression in nutrient-rich conditions (Fig. 1B). Together, our data suggest that the Ume6-Sin3-Rpd3 complex negatively regulates Atg8 expression and, consequently, the amount of Atg8 available during autophagy.
Ume6 Binds the ATG8 Promoter and Negatively Regulates ATG8 Transcription.
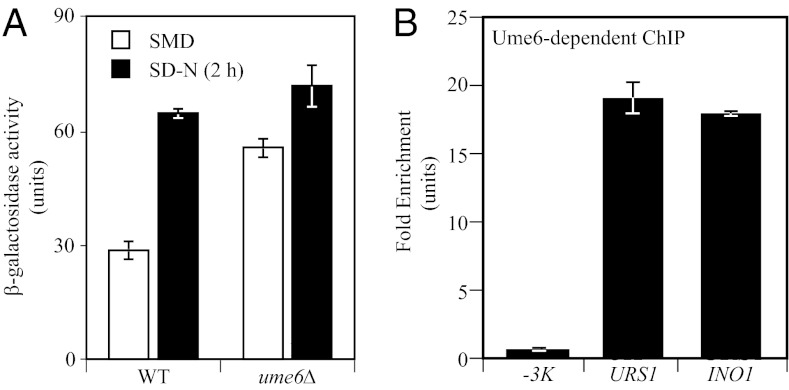
To determine whether Ume6 regulates ATG8 at a transcriptional level, we examined β-galactosidase activity in wild-type and ume6Δ strains expressing LacZ under the control of the ATG8 promoter in nutrient-rich and nitrogen starvation conditions. The β-galactosidase activity in the ume6Δ strain was substantially higher than that seen in the wild-type strain under growing conditions (Fig. 2A), suggesting that Ume6 negatively regulates ATG8 transcription, rather than exerting its effect directly on the Atg8 protein. The β-galactosidase activity in the ume6Δ strain increased only slightly under starvation conditions, suggesting that the transcription of ATG8 in growing conditions was close to the maximal level seen when fully induced (Fig. 2A). To further test whether Ume6 binds the ATG8 promoter, we conducted a chromatin immunoprecipitation (ChIP) analysis in a strain expressing Ume6 tagged with protein A (Ume6-PA). We examined the binding of Ume6-PA to the ATG8 URS1 region and a sequence 3 kb upstream of the ATG8 start codon (-3K), which served as a negative control; binding at the promoter of INO1 was examined as a positive control (16). The quantitative PCR results showed that Ume6-PA binding to the URS1 region was ∼19 times higher than that seen in the -3K control and was at a level similar to that detected for the INO1 promoter (Fig. 2B), suggesting that Ume6 actually bound to the URS1 element of the ATG8 promoter.
Fig. 2.
Ume6 binds the ATG8 promoter and negatively regulates ATG8 transcription. (A) Expression of ATG8p-LacZ in a UME6 deletion strain. Wild-type and ume6Δ cells containing LacZ driven by the ATG8 promoter were grown to midlog phase and switched to nitrogen starvation medium (SD-N) for 2 h. β-galactosidase activity was measured from protein extracts. (B) Protein A–tagged Ume6 binds the ATG8 promoter. ChIP analysis was conducted on two regions of the ATG8 promoter: the URS1 region and a region −3 kb upstream of the ATG8 start codon (-3K), which was used as a negative control. The URS1 region in the INO1 promoter served as a positive control. The ChIP results were normalized to the input DNA and calibrated to the -3K PCR product, which was set to 1.0. Error bars represent the SD of at least three independent experiments.
Rim15 Promotes Ume6 Phosphorylation and Functions As a Positive Regulator of Atg8 Induction.
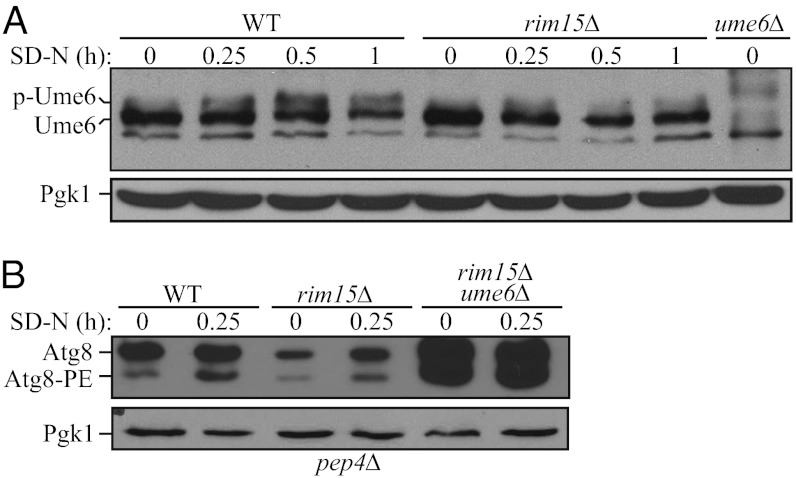
During meiosis, removal of Sin3 and Rpd3 from the Ume6 complex is regulated by the protein kinase Rim15 in response to nitrogen and glucose limitation. For example, when cells are grown on acetate as the sole carbon source in conditions of nitrogen limitation, Rim15 promotes Ume6 phosphorylation and disrupts the association of Sin3 and Rpd3 with the complex, thus relieving transcriptional repression of the target genes (12). In addition, we have shown that Rim15 is a positive regulator of autophagy (17, 18), although its relevant target(s) had not been identified. Therefore, we decided to investigate a potential role for Rim15 in Ume6-regulated Atg8 induction. Accordingly, we first tested whether Rim15 promotes Ume6 phosphorylation during autophagy. In wild-type cells, upon nitrogen starvation, Ume6 exhibited a slower migration, which is consistent with the previous finding (12) that Ume6 is subject to phosphorylation (Fig. 3A). However, RIM15 deletion caused a block in Ume6 phosphorylation in starvation conditions, suggesting that Rim15 promotes Ume6 phosphorylation during autophagy (Fig. 3A).
Fig. 3.
Rim15 promotes Ume6 phosphorylation and functions as a positive regulator of Atg8 induction. (A) Rim15 is required for Ume6 phosphorylation in starvation conditions. Wild-type (WT, BY4742) and rim15Δ cells were grown in rich medium and starved in SD-N for up to 1 h. Cells were collected, and protein extracts were analyzed with anti-Ume6 and anti-Pgk1 (loading control) antisera. (B) Wild-type (WT, YZD005), rim15Δ (YZD006), and rim15Δ ume6Δ (YZD007) cells in a pep4Δ background were grown in rich medium and shifted to SD-N for starvation. Cells were collected at the indicated time points and subjected to immunoblotting with anti-Atg8 and anti-Pgk1 antisera.
To further investigate the role of Rim15 in regulating Atg8 induction, we decided to examine Atg8 levels in the presence and absence of this kinase. Accurate measurement of Atg8 levels is complicated by the continuous degradation of Atg8–PE in the vacuole during autophagy. (Atg8 is one of the few Atg proteins that remains associated with the completed autophagosome, and a portion of the protein is delivered into the vacuole lumen where it is degraded.) Thus, we used a pep4Δ background strain. The hydrolase activity of Pep4, a vacuolar aspartyl protease, is required for the breakdown of autophagic bodies (the single-membrane intralumenal vesicles that result from fusion of autophagosomes with the vacuole) and the subsequent degradation of Atg8–PE. Therefore, we examined Atg8 levels in wild-type, rim15Δ, and rim15Δ ume6Δ strains in which the PEP4 locus was deleted to prevent the turnover of Atg8–PE. Before nitrogen starvation, wild-type cells displayed a basal level of Atg8, and even after a short 15-min period of nitrogen starvation, an increase in Atg8–PE could be detected (Fig. 3B). In rim15Δ cells, the basal level of Atg8 was clearly lower, and there was a lag in the generation of Atg8–PE, indicating that Rim15 functions as a positive regulator of Atg8 induction. Deletion of UME6 in the rim15Δ strain rescued the induction defect in response to nitrogen starvation, suggesting that Rim15 acts upstream of Ume6 to regulate Atg8 synthesis during autophagy.
Ume6 Negatively Regulates Autophagy Activity.
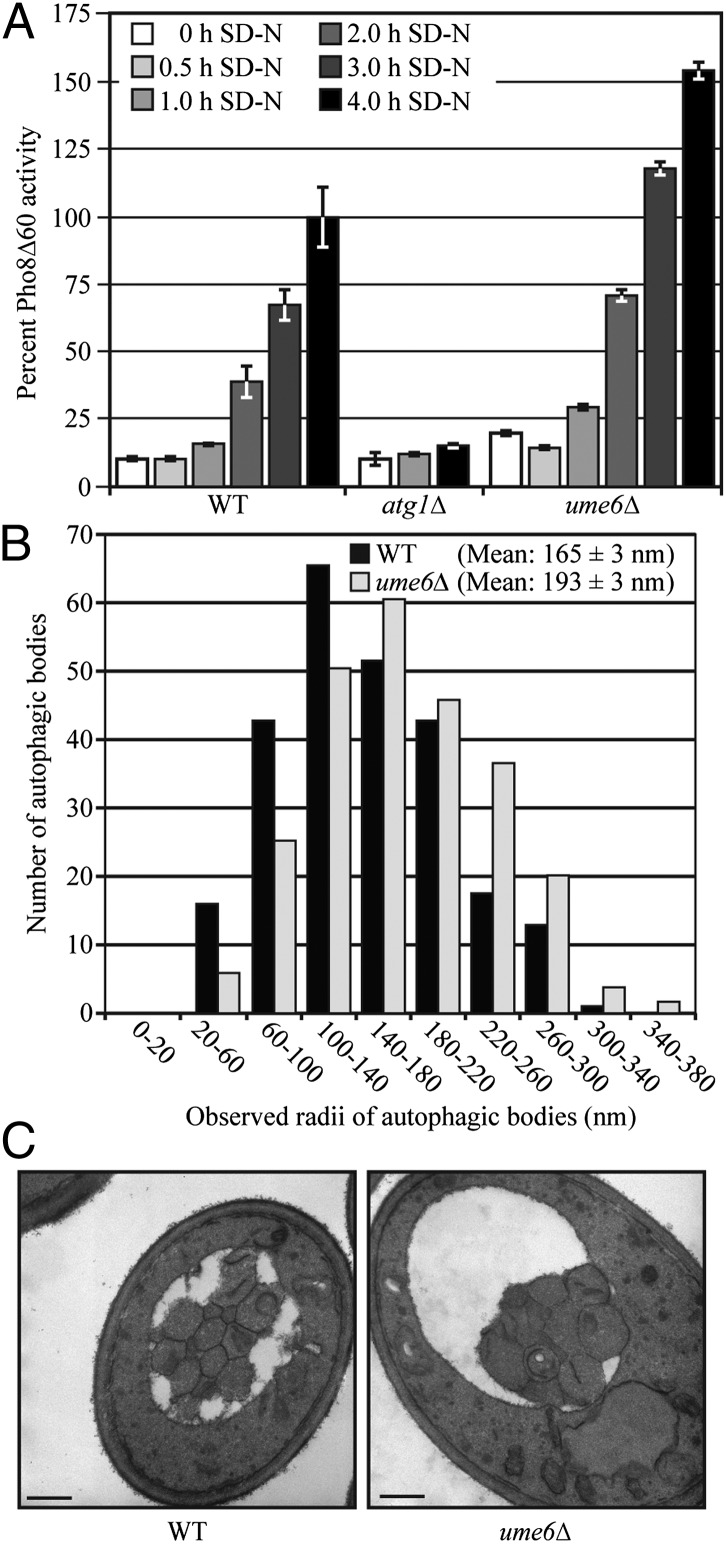
To determine whether modulation of Atg8 levels by Ume6 has a physiological effect on autophagy, we measured autophagy activity using the Pho8Δ60 assay (19). This assay measures the autophagy-dependent alkaline phosphatase activity of Pho8Δ60, a modified vacuolar alkaline phosphatase precursor that remains in the cytosol; Pho8Δ60 can only be delivered to the vacuole via autophagy, in which case, a C-terminal propeptide is subsequently removed, resulting in enzymatic activation. Therefore, the alkaline phosphatase activity of Pho8Δ60 reflects the magnitude of nonselective autophagic cargo delivery.
In growing conditions, the wild-type strain displayed a basal level of Pho8Δ60-dependent alkaline phosphatase activity, whereas the ume6Δ strain displayed an increase in the basal level of autophagy consistent with a role for Ume6 in negatively regulating autophagy by limiting the amount of Atg8 [Fig. 4A; 0 h in nitrogen starvation medium (SD-N)]. Upon nitrogen starvation, Pho8Δ60 activity increased in the wild-type cells but remained at the background level in an atg1Δ mutant, which is defective for autophagy. In ume6Δ cells, autophagy was induced more rapidly and to a higher level, as indicated by Pho8Δ60 activity compared with that seen in the wild type (Fig. 4A). Thus, Ume6 acts as a negative regulator of autophagy activity.
Fig. 4.
Ume6 negatively regulates autophagy. (A) Autophagy as measured by the Pho8Δ60 assay is increased in ume6Δ cells. Wild-type (YCB193, SEY6210), atg1Δ (YCB194), and ume6Δ (YCB197) cells were grown in SMD medium and then starved for 0, 0.5, 1, 2, 3, and 4 h. The Pho8Δ60 activity was measured as described in Materials and Methods and normalized to the activity of the wild-type cells, which was set to 100%. Error bars indicate the SEM of three independent experiments. (B) Autophagosome size is increased in ume6Δ cells. Wild-type (FRY143, SEY6210) and ume6Δ (YCB234) strains with deletions of VPS4 and PEP4 to eliminate vesicles generated from the multivesicular body pathway and the breakdown of autophagic bodies, respectively, were grown in rich medium and starved in SD-N for 2 h. Samples were collected, prepared, and examined by TEM as described in Materials and Methods. The radius of each autophagosome was determined as described in Materials and Methods. The error represents the SEM for >400 autophagic bodies. (C) Representative TEM images of the cells in B. (Scale bars: 500 nm.)
We further sought to determine how deletion of UME6 caused an up-regulation of autophagy. An increase in the magnitude of autophagic cargo delivery suggested the possibility that more autophagosomes were being formed and/or the size of the autophagosomes were increased in the ume6Δ strain compared with the wild-type cells. After nitrogen starvation for either 1 or 2 h, the number and size of autophagic bodies per cell was examined by transmission electron microscopy (TEM). The mean number of autophagic bodies per cell section was slightly higher in the ume6Δ cells than wild-type at the 1 h (3.1 ± 0.4 and 2.3 ± 0.3, respectively) and 2 h (4.7 ± 0.4 and 4.0 ± 0.4, respectively) time points, but this difference was not statistically significant [P = 0.25 (1 h) and P = 0.19 (2 h)]. A highly significant difference (P < 5 × 10−8), however, was observed in the size of the autophagic bodies. The autophagic bodies of ume6Δ cells had an average cross-sectional radius that was 22% and 17% larger than that found in the wild-type cells at the 1 and 2 h time points, respectively (Fig. 4 B and C and Fig. S1). Notably, this translates into a substantial difference in average volume. To estimate the actual volume of the autophagic bodies from the observed cross-sectional radii, we used a statistical method previously developed for this purpose (20). The calculations indicated that the ume6Δ cells had autophagosomes that were 68% and 112% larger by volume than wild-type autophagosomes at the 1- and 2-h time points, respectively. This ∼2.1-fold increase in autophagosome volume at the 2-h time point is quite similar to the 1.8-fold increase in Pho8Δ60 activity at the same time point (Fig. 4 A and B); thus, a small increase in the diameter of autophagosomes has robust effects on the magnitude of bulk autophagy. We also noted that the average cross-sectional area of the wild-type and ume6Δ cells was 7.3 and 11.6 μm2, respectively, based on the measurements of over 300 cells each; however, no data suggest that autophagosome or autophagic body size is affected by cell size.
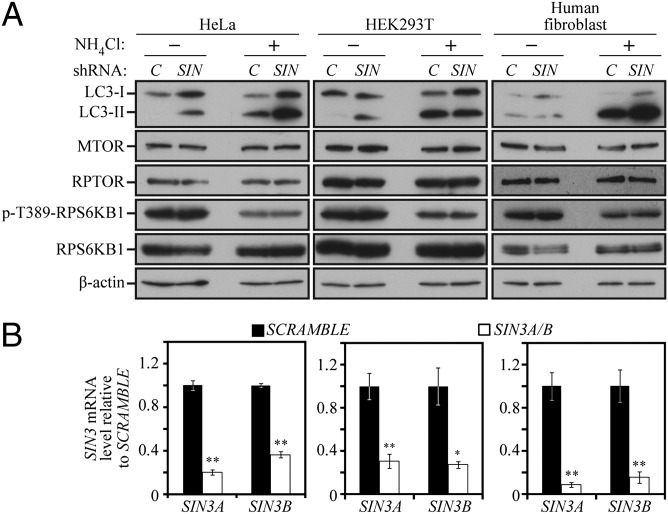
SIN3A and SIN3B Play Redundant Roles in Regulating LC3 Expression.
We next explored the possibility that the mechanism of Atg8/LC3 regulation that we discovered here was conserved in higher eukaryotes. The transcription factor Ume6 has no clear homolog in mammalian cells, but 2 homologs of SIN3 (SIN3A and SIN3B) and 11 homologs of RPD3 exist in vertebrates. Accordingly, SIN3 was chosen for further study. SIN3A and SIN3B were knocked down by treating HeLa cells with three individual shRNA targeted to SIN3A and two individual shRNA targeted to SIN3B alone and in combination, with nearly identical results in terms of the degree of knockdown; SIN3A and SIN3B mRNA levels were reduced to ∼9–30% and 15–36% of the scrambled control, respectively (Fig. 5B). Knockdown of SIN3A and SIN3B individually had little or no effect on LC3 levels compared with the scrambled control (Fig. S2A). In contrast, when SIN3A and SIN3B were simultaneously knocked down, a robust increase in LC3 levels was readily apparent in various shRNA combinations under nutrient-rich conditions in HeLa cells (Fig. 5 A and B). The same results were also found for HEK293T and human fibroblast cell lines (Fig. 5A). The increase in LC3 protein levels was not attributable to indirect effects through protein level or activity of the mechanistic target of rapamycin (MTOR) complex, because the amount of MTOR and its associated protein regulatory associated protein of MTOR, complex 1 (RPTOR) were unchanged (Fig. 5A), nor was there an effect on the activity of MTORC1 or MTORC2, as determined by the phosphorylation state of their targets ribosomal protein S6 kinase, 70kDa, polypeptide 1 (RPS6KB1/S6K) and eukaryotic translation initiation factor 4E binding protein 1 (EIF4EBP1/4EBP1), or v-akt murine thymoma viral oncogene homolog 1 (AKT1), respectively (Fig. 5A and Fig. S2B). Furthermore, we examined autophagic flux by exposing the cells to NH4Cl, which raises the lysosomal pH and prevents the turnover of LC3 (similar, in effect, to deleting the PEP4 gene in yeast). The presence of NH4Cl resulted in elevated levels of LC3-II, indicating that the knockdown of SIN3A/B caused an increase in basal autophagy, and not just an increase in the level of LC3. Note that in human fibroblasts, the lack of a clear difference in LC3 levels in the absence, but not the presence, of NH4Cl indicates a rapid rate of lysosomal turnover of this protein (21).
Fig. 5.
SIN3A and SIN3B play redundant roles in regulating LC3 expression. (A) SIN3A- and SIN3B-targeted shRNA was prepared and used to generate viruses as described in Materials and Methods. The shRNA-expressing viruses were infected in combination into HeLa, HEK293T, and human fibroblast cells using scrambled DNA as a control (C). Cell lysates were analyzed by immunoblotting with the indicated antibodies. (B) SIN3A and SIN3B mRNA levels were monitored by quantitative PCR in shRNA-treated cells. The values for scrambled DNA were set to 1.0, and the other values were normalized. *P < 0.05; **P < 0.01. Error bars represent the SD.
Discussion
Our findings suggest that in response to nitrogen starvation, the kinase Rim15 phosphorylates Ume6. During meiosis, this phosphorylation causes the dissociation of Ume6 from Sin3-Rpd3, leading to transcriptional activation (12, 14). Rim15 plays an important role in integrating many nutrient-regulatory signals and, therefore, plays a central role in regulating autophagy (17, 22). Rim15 is negatively regulated through direct phosphorylation by cAMP-dependent protein kinase A (PKA) and Sch9 in the presence of glucose and nitrogen (23, 24) and is involved in the autophagy induction that occurs upon PKA-Sch9 inactivation (18). PKA and Sch9 are upstream sensors that act to negatively regulate autophagy; however, the downstream components in this signaling pathway are unknown, and how PKA and Sch9 signaling affects the autophagy machinery to regulate autophagosome formation has not been elucidated previously. Yeast Sch9 is homologous to mammalian ribosomal protein S6 kinase, 70kDa (RPS6KB/p70S6 kinase) or AKT1 (18). In mammalian cells, AKT1 phosphorylates and inactivates the forkhead box O (FOXO) family of transcription factors (25). During muscle atrophy, FOXO3 induces the expression of multiple autophagy genes including Lc3, Gabrapl1 (an Lc3 homolog), unc-51-like kinase 1 (Ulk1), Atg4, Atg12, phosphoinositide-3-kinase, class 3 (Pik3c3/Vps34), and beclin 1, autophagy related (Becn1) (26, 27); and in hepatic tissue, FOXO1 regulates the autophagy genes Gabarapl1, Pik3c2, and Atg12 (28). Although Ume6 is not conserved in mammalian cells, the regulation of the FOXO family by Sch9 and AKT1 suggests that the pathway regulating Atg8/LC3 may be conserved from yeast to human. Just as the knockdown of SIN3A and SIN3B promotes an increase in cellular LC3 levels, inhibition of RPD3 promotes a similar increase (29–31), although the detailed mechanism has not been determined.
One frequently overlooked method of regulating the magnitude of autophagy is the regulation of the size of the autophagosome. Research in yeast has shown that the average size of the autophagosome is modulated by the amount of available Atg8 (5). Our results provide strong evidence that transcriptional repression plays a major role in regulating Atg8/LC3 levels, and this up-regulation results in an increase in the size of the autophagosome. Basal autophagy is especially important in the liver and other cells such as neurons and myocytes, which, after differentiation, cease dividing. Modulation of LC3 levels through inhibition of histone deacetylation at the LC3 locus may be a viable option to increase basal autophagy in nondividing terminally differentiated cells.
Materials and Methods
Yeast.
Gene disruptions and PA tag integrations were performed using a standard method (32). Yeast cells were grown in rich medium [YPD; 1% yeast extract, 2% peptone, and 2% glucose (all wt/vol)] or synthetic minimal medium (SMD; 0.67% yeast nitrogen base, 2% glucose, supplemented with the appropriate amino acids and vitamins). Autophagy was induced in starvation medium (SD-N; 0.17% yeast nitrogen base without amino acids, containing 2% glucose). The yeast strains used in this study are listed in Table S1. Protein extraction, immunoblot, GFP-Atg8 processing, and alkaline phosphatase (Pho8Δ60) assays were performed as described previously (18, 19, 33). Yeast strains containing the β-galactosidase reporter plasmid ATG8p-LacZ(416) were grown in SMD or shifted to SD-N to induce autophagy and then examined with a β-galactosidase assay as described previously (34). ChIP was performed as described previously (35).
Samples for TEM were prepared as described previously (5). Sections (85 nm) were cut using a Leica Ultracut-E microtome at the University of Michigan Microscopy and Image Analysis Laboratory. Images were acquired on a Philips CM100 BioTwin electron microscope at the University of Michigan Molecular, Cellular and Developmental Biology departmental TEM facility. The observed radii of the autophagic body cross-sections were determined and used to estimate the actual radii as described (20), which were converted to volume. Statistical significance was determined using the Mann–Whitney U test.
Cell Culture.
Knockdown of SIN3 homologs was performed by cloning SIN3A- and SIN3B-targeted shRNA into the pLKO1 lentiviral expression vector, and these plasmids were cotransfected together with psPAX2 and pMP2 plasmids into actively growing cells. HeLa, HEK293T, and human fibroblast cells were infected with isolated viruses, selected for puromycin resistance, and analyzed on the seventh day after infection with scrambled DNA as a control. Cell lysates were suspended in Nonidet P-40 buffer and subjected to SDS/PAGE and Western blot analysis.
Supplementary Material
Acknowledgments
D.J.K., C.R.B., M.J., M.Z., M.A.L.-D., Z.D., M.U., and Z.X. were supported by National Institutes of Health (NIH) Grant GM53396. M.U. was also supported, in part, by the Japan Society for the Promotion of Science. K.I., T.S., and A.K. were supported by NIH Grant DK083491.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1200313109/-/DCSupplemental.
References
- 1.Xie Z, Klionsky DJ. Autophagosome formation: Core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 2.Yorimitsu T, Klionsky DJ. Autophagy: Molecular machinery for self-eating. Cell Death Differ. 2005;12:1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klionsky DJ, et al. A comprehensive glossary of autophagy-related molecules and processes (2nd edition) Autophagy. 2011;7:1273–1294. doi: 10.4161/auto.7.11.17661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell. 2008;19:3290–3298. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weidberg H, et al. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010;29:1792–1802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang W-P, Scott SV, Kim J, Klionsky DJ. The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J Biol Chem. 2000;275:5845–5851. doi: 10.1074/jbc.275.8.5845. [DOI] [PubMed] [Google Scholar]
- 8.Kirisako T, et al. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams RM, et al. The Ume6 regulon coordinates metabolic and meiotic gene expression in yeast. Proc Natl Acad Sci USA. 2002;99:13431–13436. doi: 10.1073/pnas.202495299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park HD, Luche RM, Cooper TG. The yeast UME6 gene product is required for transcriptional repression mediated by the CAR1 URS1 repressor binding site. Nucleic Acids Res. 1992;20:1909–1915. doi: 10.1093/nar/20.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strich R, et al. UME6 is a key regulator of nitrogen repression and meiotic development. Genes Dev. 1994;8:796–810. doi: 10.1101/gad.8.7.796. [DOI] [PubMed] [Google Scholar]
- 12.Pnueli L, Edry I, Cohen M, Kassir Y. Glucose and nitrogen regulate the switch from histone deacetylation to acetylation for expression of early meiosis-specific genes in budding yeast. Mol Cell Biol. 2004;24:5197–5208. doi: 10.1128/MCB.24.12.5197-5208.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Washburn BK, Esposito RE. Identification of the Sin3-binding site in Ume6 defines a two-step process for conversion of Ume6 from a transcriptional repressor to an activator in yeast. Mol Cell Biol. 2001;21:2057–2069. doi: 10.1128/MCB.21.6.2057-2069.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao Y, Mitchell AP. Shared roles of yeast glycogen synthase kinase 3 family members in nitrogen-responsive phosphorylation of meiotic regulator Ume6p. Mol Cell Biol. 2000;20:5447–5453. doi: 10.1128/mcb.20.15.5447-5453.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- 16.Jackson JC, Lopes JM. The yeast UME6 gene is required for both negative and positive transcriptional regulation of phospholipid biosynthetic gene expression. Nucleic Acids Res. 1996;24:1322–1329. doi: 10.1093/nar/24.7.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Z, Geng J, Yen W-L, Wang K, Klionsky DJ. Positive or negative roles of different cyclin-dependent kinase Pho85-cyclin complexes orchestrate induction of autophagy in Saccharomyces cerevisiae. Mol Cell. 2010;38:250–264. doi: 10.1016/j.molcel.2010.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yorimitsu T, Zaman S, Broach JR, Klionsky DJ. Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:4180–4189. doi: 10.1091/mbc.E07-05-0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noda T, Matsuura A, Wada Y, Ohsumi Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1995;210:126–132. doi: 10.1006/bbrc.1995.1636. [DOI] [PubMed] [Google Scholar]
- 20.Xie Z, et al. Indirect estimation of the area density of Atg8 on the phagophore. Autophagy. 2009;5:217–220. doi: 10.4161/auto.5.2.7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swinnen E, et al. Rim15 and the crossroads of nutrient signalling pathways in Saccharomyces cerevisiae. Cell Div. 2006;1:3. doi: 10.1186/1747-1028-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pedruzzi I, et al. TOR and PKA signaling pathways converge on the protein kinase Rim15 to control entry into G0. Mol Cell. 2003;12:1607–1613. doi: 10.1016/s1097-2765(03)00485-4. [DOI] [PubMed] [Google Scholar]
- 24.Reinders A, Bürckert N, Boller T, Wiemken A, De Virgilio C. Saccharomyces cerevisiae cAMP-dependent protein kinase controls entry into stationary phase through the Rim15p protein kinase. Genes Dev. 1998;12:2943–2955. doi: 10.1101/gad.12.18.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salih DA, Brunet A. FoxO transcription factors in the maintenance of cellular homeostasis during aging. Curr Opin Cell Biol. 2008;20:126–136. doi: 10.1016/j.ceb.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mammucari C, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 27.Zhao J, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 28.Liu HY, et al. Hepatic autophagy is suppressed in the presence of insulin resistance and hyperinsulinemia: Inhibition of FoxO1-dependent expression of key autophagy genes by insulin. J Biol Chem. 2009;284:31484–31492. doi: 10.1074/jbc.M109.033936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park JH, et al. A new synthetic HDAC inhibitor, MHY218, induces apoptosis or autophagy-related cell death in tamoxifen-resistant MCF-7 breast cancer cells. Invest New Drugs. 2011 doi: 10.1007/s10637-011-9752-z. [DOI] [PubMed] [Google Scholar]
- 30.Ahn MY, Ahn SG, Yoon JH. Apicidin, a histone deaceylase inhibitor, induces both apoptosis and autophagy in human oral squamous carcinoma cells. Oral Oncol. 2011;47:1032–1038. doi: 10.1016/j.oraloncology.2011.07.027. [DOI] [PubMed] [Google Scholar]
- 31.Chen MY, et al. Decitabine and suberoylanilide hydroxamic acid (SAHA) inhibit growth of ovarian cancer cell lines and xenografts while inducing expression of imprinted tumor suppressor genes, apoptosis, G2/M arrest, and autophagy. Cancer. 2011;117:4424–4438. doi: 10.1002/cncr.26073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 33.Shintani T, Klionsky DJ. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J Biol Chem. 2004;279:29889–29894. doi: 10.1074/jbc.M404399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rose M, Botstein D. Construction and use of gene fusions to lacZ (β-galactosidase) that are expressed in yeast. Methods Enzymol. 1983;101:167–180. doi: 10.1016/0076-6879(83)01012-5. [DOI] [PubMed] [Google Scholar]
- 35.Aparicio O, et al. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr Protoc Mol Biol. 2005 doi: 10.1002/0471142727.mb2103s69. Chapter 21:Unit 21 23. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.