Abstract
Pericentromeric heterochromatin formation is mediated by repressive histone H3 lysine 9 methylation (H3K9Me) and its recognition by HP1 proteins. Intriguingly, in many organisms, RNAi is coupled to this process through poorly understood mechanisms. In Schizosaccharomyces pombe, the H3-K9 methyltransferase Clr4 and the heterochromatin protein 1 (HP1) ortholog Swi6 are critical for RNAi, whereas RNAi stimulates H3K9Me. In addition to the endoribonuclease Dcr1, RNAi in S. pombe requires two interacting protein complexes, the RITS complex, which contains an Argonaute subunit, and the RDRC complex, which contains an RNA-dependent RNA polymerase subunit. We previously identified Ers1 (essential for RNAi-dependent silencing) as an orphan protein that genetically acts in the RNAi pathway. Using recombinant proteins, we show here that Ers1 directly and specifically interacts with HP1/Swi6. Two-hybrid assays indicate that Ers1 also directly interacts with several RNAi factors. Consistent with these interactions, Ers1 associates in vivo with the RITS complex, the RDRC complex, and Dcr1, and it promotes interactions between these factors. Ers1, like Swi6, is also required for RNAi complexes to associate with pericentromeric noncoding RNAs. Overexpression of Ers1 results in a dominant-negative phenotype that can be specifically suppressed by increasing levels of the RDRC subunit Hrr1 or of Dcr1, further supporting a functional role for Ers1 in promoting the assembly of the RNAi machinery. Through the interactions described here, Ers1 may promote RNAi by tethering the corresponding enzyme complexes to HP1-coated chromatin, thereby placing them in proximity to the nascent noncoding RNA substrate.
Heterochromatin is a specialized form of DNA packaging that plays numerous critical roles in chromosome biology. However, the mechanisms that drive its assembly remain poorly understood. In fission yeast Schizosaccharomyces pombe, heterochromatin is assembled on pericentromeric repeats, subtelomeric regions, and the silent mat2/3 mating type locus (1, 2). Establishment and maintenance of heterochromatin at these sites depend on histone-modifying enzymes, histone marks, and histone-binding proteins. Among the histone-modifying enzymes is the histone methyltransferase, Clr4, the ortholog of Drosophila melanogaster and mammalian SU(VAR)3-9 enzymes (1, 2). Clr4 catalyzes methylation of histone H3 lysine 9 (H3K9me), an essential heterochromatin mark that serves as a docking site for the chromodomain-containing HP1 proteins Swi6 and Chp2 (1, 2). Surprisingly, many sites of heterochromatin formation correspond to sites that serve as templates for RNAi. Pericentromeric dh and dg repeats, for instance, are transcribed by RNA polymerase II, converted into double-stranded RNA by an RNA-dependent RNA polymerase (Rdp1), and processed into siRNAs by a Dicer enzyme (Dcr1) (1, 2). Remarkably, RNAi requires the formation of heterochromatin. Loss of Clr4 or of Swi6 results in a loss of siRNAs (3–5). RNAi also promotes, but is not essential for, H3K9Me (6). Precisely how the formation of heterochromatin promotes RNAi is unclear. A complex called RITS was identified, which contains an Argonaute protein, Ago1, a chromodomain protein, Chp1, which recognizes the H3K9Me mark, and a GW protein called Tas3, which links the two proteins together (5, 7). The RITS complex interacts with a second three-protein complex called RDRC, which contains an RNA-dependent RNA polymerase subunit (Rdp1), a helicase subunit (Hrr1), and a noncanonical polyA polymerase subunit (Cid12) (3). It was suggested that the recruitment of Ago1 to sites of H3K9Me mediated by the Chp1 subunit of RITS could explain the coupling between H3K9Me and RNAi (5, 7). However, although the components of RITS are each individually required for RNAi, disruption of the RITS complex by mutating the interaction interface of Ago1 and Tas3 revealed that a dramatic reduction in the interaction does not impact RNAi (8). These data, together with the known requirement for the HP1 protein Swi6 in siRNA accumulation, suggest that additional links between heterochromatin and the RNAi machinery likely exist.
Previously, we genetically identified a factor required for S. pombe silencing that we named Ers1, a 957-amino-acid protein with no obvious homologs outside the genus Schizosaccharomyces and no identifiable domains (9). This factor was independently identified by others and named Rsh1 (10) (for simplicity, we will refer to it as Ers1 here). We demonstrated that Ers1 is required for silencing at centromeres, but not at the mating type locus and telomeres, consistent with a role in RNAi-dependent heterochromatin formation (9). Ers1 is needed for proper levels of H3K9 methylation as well as the recruitment of the RITS complex to centromeres (9). Deletion of ers1+ also triggers accumulation of noncoding centromeric transcripts and a defect in siRNA production (9). High-throughput analysis demonstrated that ers1+ behaves genetically in a manner similar to silencing factors (10). However, whether Ers1 functions directly in the RNAi-dependent pathway of heterochromatic silencing has not been established.
Here we show that Ers1 plays a direct role in silencing by interacting with Swi6 and with several components of the RDRC and RITS complexes. We also demonstrate specific in vivo associations between Ers1 and pericentromeric noncoding RNAs, the RITS complex, the RDRC complex, and Dcr1. Moreover, we find that Ers1 is required for previously described associations between these complexes and for their associations with pericentromeric noncoding RNAs. Finally, we describe genetic studies that point to a functional role for Ers1 in activating the machinery responsible for the production of small RNAs. Through its interactions with Swi6 and RNAi factors, Ers1 may promote RNAi by tethering the corresponding complexes to HP1-coated chromatin, thereby placing the RNAi machinery in proximity to its nascent noncoding RNA substrate.
Results
Ers1 Functions in the RNAi Pathway.
To solidify the notion that Ers1 functions in the RNAi pathway, we examined its role in silencing at the mat2/3 locus, where RNAi-dependent mechanisms act redundantly with RNAi-independent mechanisms mediated by the Atf1–Pcr1 DNA-binding regulator (11). Like a dcr1Δ knockout, the ers1Δ mutation produces a silencing defect in cells lacking Pcr1 (Fig. S1). Ers1 is also required, like Dcr1, for silencing produced by tethering of Tas3 to ura4 transcripts (12) (Fig. S1). Unlike the case with Tas3, tethering of Ers1 was not sufficient to trigger silencing, although a reproducible increase in H3K9Me was induced by Ers1 in this context (Fig. S1). Presumably, the increase in H3K9Me does not reach a threshold required to trigger a robust reporter gene phenotype.
Two-Hybrid Analysis Reveals Prominent Interactions Between Ers1 and Swi6 and Between Ers1 and Tas3.
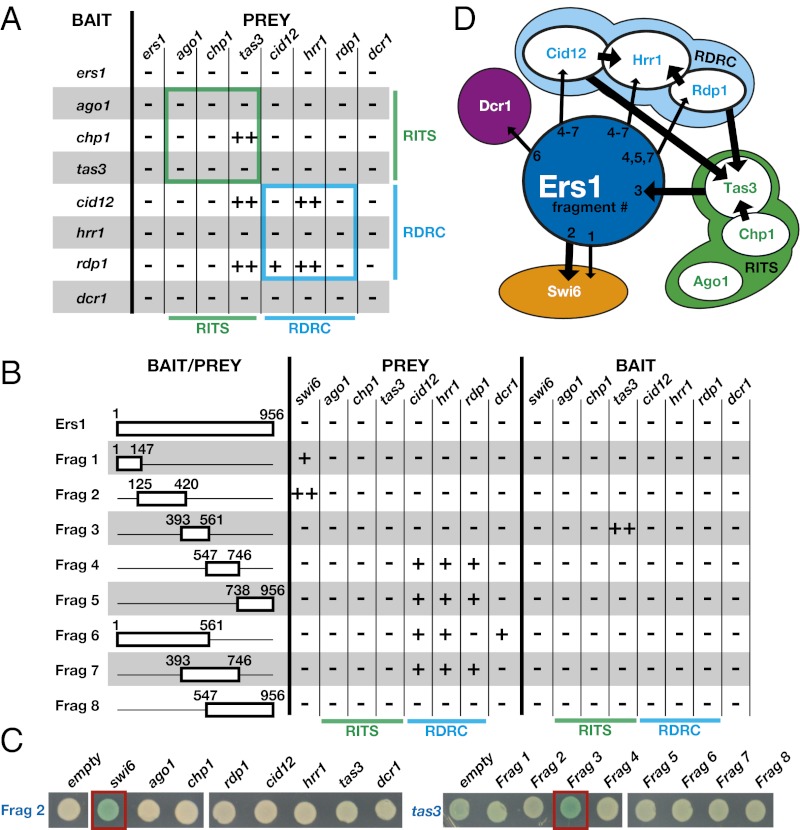
To test whether Ers1 might interact with any of the six components of RITS and RDRC complexes, we cloned full-length cDNAs for each of these factors as both baits and preys for LexA-based Saccharomyces cerevisiae two-hybrid assays (13). We assayed the interactions using quantitative β-galactosidase assays and expressed the results as fold change over the empty-prey control (Table S1). Using a stringent fourfold cutoff, we detected interactions within the RITS and RDRC complexes (Fig. 1 A and D and Table S1). Specifically, we detected interactions between Chp1 and Tas3 (RITS) and between Hrr1 and both Cid12 and Rdp1 (RDRC) (Fig. 1 A and D). We also detected interactions between Tas3 and Cid12 and between Tas3 and Rdp1 (Table S1 and Fig. 1A). These new interactions may help to explain previous studies indicating an association between these two complexes (3). As is not uncommon with two-hybrid assays, most interactions are only apparent in one bait–prey configuration. Unfortunately, we found no evidence for interactions involving Ers1 (Fig. 1A). Reasoning that such interactions may be masked in the full-length protein (a common cause of false-negatives in two-hybrid assays), we constructed eight bait and eight prey clones coding for fragments of the Ers1 protein (Fig. 1B) and assayed these against the RNAi factors as well as Swi6 and each component of the Clr4 complex (CLRC) (14–17), which are required for RNAi (Tables S2 and S3). Using the same cutoff, we identified an interaction between fragment 2 (amino acids 125–420) and full-length Swi6 and between fragment 3 (amino acids 393–561) and Tas3 (Fig. 1 B–D, indicated by ++). Several reproducible albeit weaker (below the fourfold threshold) interactions were identified between fragments of Ers1 and components of the RDRC or Dcr1 (indicated by + in Fig. 1 B and D).
Fig. 1.
Yeast two-hybrid analysis identifies Swi6, Tas3, and components of RDRC as Ers1 interaction partners. (A) Comprehensive two-hybrid analysis of RITS, RDRC, and Dcr1. ++ indicates a greater than fourfold activity over empty bait. + indicates a two- to fourfold activity over empty prey. (B) Analysis of interactions between Ers1 fragments and factors required for RNAi. Ers1 fragments are used as baits or prey to detect interactions with RITS, RDRC, Dcr1, and Swi6 by yeast two-hybrid. In the bait/prey column, Ers1 fragments are drawn to scale and the start and end positions of each fragment are denoted in amino acids. (C) Plate assays. β-Galactosidase plate assays of the strongest interactions with the Ers1 fragments are shown. Black denotes the prey; blue denotes the bait. Red boxes highlight the strong interactions between Swi6 and Ers1 fragment 2 and between Ers1 fragment 3 and Tas3. (D) Summary of interactions. Arrows represent yeast two-hybrid interactions. Thick arrows are interactions greater than fourfold over the empty-prey control; thin arrows represent interactions greater than twofold between the bait (base of the arrow) and the prey (point of the arrow).
Ers1 Interacts with Swi6 in Vitro and in Vivo.
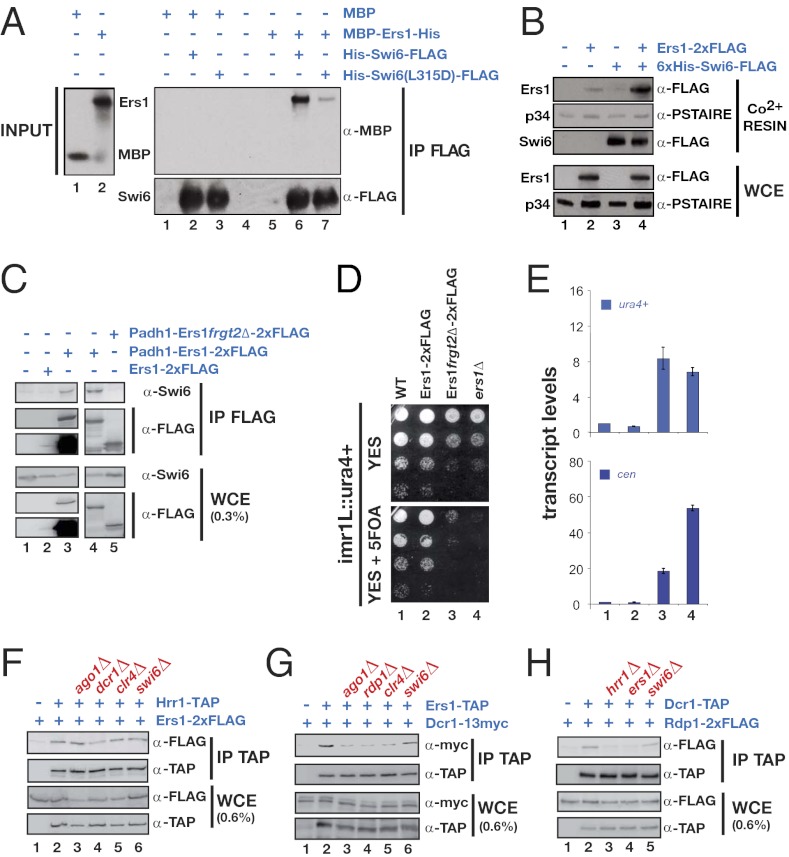
Because Swi6 can be made in recombinant form in Escherichia coli, we sought to test the Ers1–Swi6 interaction using purified proteins. We successfully expressed fragment 2 as a soluble maltose-binding protein (MBP) fusion. MBP–Ers1 fragment 2, but not MBP alone, specifically bound to beads containing His-Swi6-FLAG (Fig. 2A). Moreover, a mutation in the chromoshadow domain (CSD) that blocks Swi6 dimerization (18) resulted in reduced binding to Ers1, further demonstrating the specificity (Fig. 2A). We also attempted to use immobilized recombinant Swi6 to select Ers1 from S. pombe cell extracts overexpressing full-length, FLAG-tagged Ers1, and found that Swi6 beads but not control beads could select Ers1, but not a control protein (p34) from the extract (Fig. 2B). Thus, Ers1 is a direct Swi6-binding protein that functions in the RNAi pathway.
Fig. 2.
Ers1 interacts with Swi6 both in vitro and in vivo via its fragment 2 and associates with RNAi factors in vivo. (A) Swi6 affinity pull-down assay: MBP–Ers1. Purified recombinant 6×His-Swi6-FLAG (lanes 2 and 6 of IP FLAG blot) or mutant 6×His-Swi6(L315D)-6×His (lanes 3 and 7 of IP FLAG blot) was immobilized and used to test for binding to recombinant MBP (lanes 1–3 of IP FLAG blot) or recombinant MBP–Ers1 fragment 2–6×His (lanes 5–7 of IP FLAG blot). (B) Swi6 affinity pull-down assay: S. pombe lysate. Shown are pull-down assays using S. pombe lysates overexpressing Ers1 (lanes 1 and 3) or Ers1–2×FLAG (lanes 2 and 4). Immobilized recombinant Swi6, or resin alone, was used to affinity select proteins from the indicated S. pombe whole-cell lysate. Endogenous p34 is used as a negative control. (C) Coimmunoprecipitation experiment. Western blots showing that Swi6 coprecipitates with overexpressed Ers1-CBP-2×FLAG (lanes 3 and 4) but not with overexpressed Ers1fragment2Δ-CBP-2×FLAG (lane 5). (D) Effect of Ers1 fragment 2 deletion. Silencing of a ura4+ reporter gene inserted at the centromeric inner repeats (imr1L::ura4+) is abolished in a strain that expresses Ers1fragment2Δ-CBP-2×FLAG. (E) RT-qPCR assay. Measurement of ura4+ (Upper) and cen (Lower) transcript levels normalized to a control transcript act1+ and relative to wild type (lane 1). Ers1fragment2Δ-CBP-2×FLAG cells accumulate both transcripts (lane 3), similarly to an ers1Δ mutant (lane 4). Error bars represent deviation from the mean of two independent experiments. (F–H) Coimmunoprecipitation experiments. Shown are Western blots of immune pellets or whole-cell extracts of cells of the indicated genotypes.
We were unable to detect association between Ers1 and Swi6 in soluble S. pombe cell extracts using standard coimmunoprecipitation protocols. Because the interaction may be transient in vivo and because of the relatively low abundance of Ers1, we attempted to detect an interaction between an overexpressed tagged allele of Ers1 and endogenous Swi6. Indeed, we detected a specific interaction (Fig. 2C). This interaction was resistant to DNase and RNase treatement of extracts before immunoprecipitation (see Fig. S3A). Importantly, this Ers1–Swi6 association is disrupted when the region corresponding to fragment 2 is deleted from Ers1 (Fig. 2C). Correspondingly, the Swi6 interaction-deficient Ers1 mutant is defective in centromeric silencing as determined by reporter gene assays and RNA measurements (Fig. 2 D and E).
Ers1 Associates with RNAi Factors in Vivo and Is Required for the Assembly of RNAi Supercomplexes.
Because we did not have recombinant RNAi factors available, we tested the association of Ers1 with these components using a coimmunoprecipitation strategy in which the endogenous proteins were differentially epitope tagged (Fig. S2). Using this assay, previous studies have shown that RITS, RDRC, and Dcr1 mutually interact but that these associations are highly cooperative, such that lack of any of these components reduces the interactions between these proteins (3, 19). In the case of Ers1, we found that it coimmunoprecipiates with Tas3 (RITS), Chp1 (RITS), Rdp1 (RDRC), Hrr1 (RDRC), and Dcr1 (Fig. 2 F and G and Fig. S3 C–E). In most cases, mutations that block RNAi (e.g., ago1Δ, dcr1Δ, and rdp1Δ) or heterochromatin (e.g., clr4Δ) reduce these interactions, consistent with a model in which the supercomplexes involved in this process assemble in a highly cooperative manner (Fig. 2G and Fig. S3 C–F). Strikingly, for Hrr1, the interaction with Ers1 is preserved in every mutant background tested and is resistant to DNase or RNase treatment of extracts, indicating a particularly robust interaction between Ers1 and this RNAi factor in vivo (Fig. 2F and Fig. S3B).
Like other factors involved in RNAi, Ers1 is required for the association of the RDRC with Dcr1 and for the association of RITS with RDRC (Fig. 2H and Fig. S3G), but does not affect interactions between components of individual complexes (Fig. S3 H and I). Swi6 is not required for the association of Ers1 with components of the RDRC or Dcr1. However, we did observe a partial dependence on Swi6 for the association of Ers1 with the RITS component Tas3 (Fig. S3D), indicating that Ers1 and Swi6 act cooperatively in this instance. Finally, Ers1 associates with Stc1 (Fig. S3F), another factor required for RNAi that has been proposed to physically link the RNAi machinery to the Clr4 complex (20).
Ers1 and RNAi Factors Associate with Pericentromeric Noncoding RNAs in a Mutually Dependent Manner.
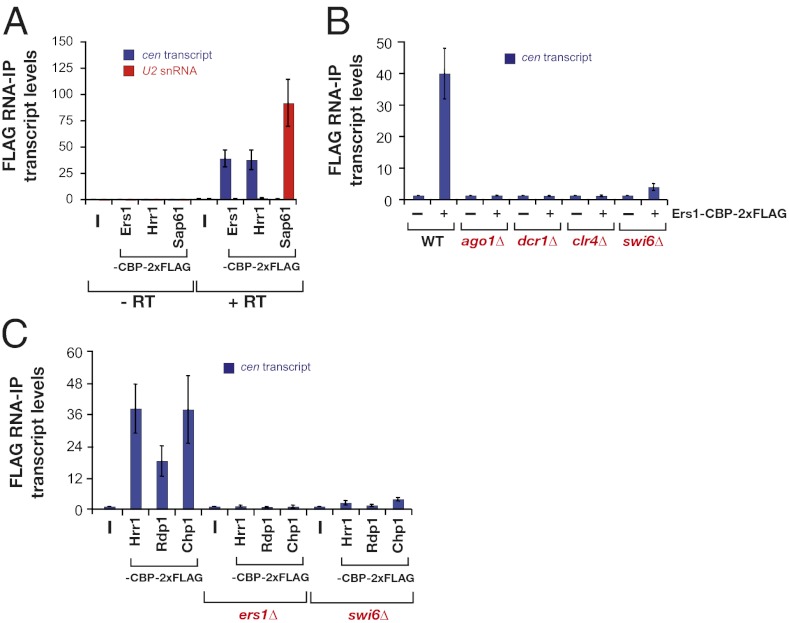
The association of RITS and RDRC with pericentromeric transcripts can be detected using an RNA immunoprecipitation assay (RIP), which is methodologically analogous to ChIP (19). Using this method, we found that Ers1, like the RDRC component Hrr1, associates with pericentromeric transcripts but not with a control RNA, U2 snRNA, a component of the spliceosome (Fig. 3A). In contrast, an abundant U2 snRNP protein, Sap61, associates with U2 snRNA but displays no enrichment for pericentromeric transcripts (Fig. 3A). Consistent with the cooperative nature of the assembly of RNAi supercomplexes, Ers1 does not associate with pericentromeric transcripts in cells lacking Ago1, Dcr1, or Clr4 (Fig. 3B). Likewise, the association of RITS and RDRC components with transcripts is abolished in ers1Δ cells (Fig. 3C). Significantly, we found that Swi6 is also required for Ers1, the RITS complex, and the RDRC to associate with pericentromeric transcripts (Fig. 3 B and C), supporting a pivotal role for Swi6 in the assembly of RNAi complexes in vivo.
Fig. 3.
Ers1 and RNAi factors associate with pericentromeric noncoding transcripts in a mutually dependent manner. (A) RNA-immunoprecipitation (RIP) experiments: wild-type cells. Shown are the enrichments normalized to levels of act1+ transcripts and relative to wild type. Reactions performed without reverse transcriptase (−RT) are indicated as a negative control. Error bars represent SD from three experiments. (B) Genetic requirements for Ers1 association with pericentromeric RNAs. RIPs were performed using strains of the indicated genotypes performed as in A. For Ers1-CBP-2×FLAG IPs, the associated transcript levels in ago1Δ, dcr1Δ, clr4Δ, and swi6Δ cells have been normalized to mutants expressing untagged Ers1, because loss of silencing in these strains results in higher levels of cen RNAs and therefore show higher background RNA levels in the IP experiments. Error bars represent SD from three experiments. (C) Requirement for Ers1 and Swi6 for RITS and RDRC association with pericentromeric RNAs. RIPs were performed as in A on strains of the indicated genotypes. Diagram shows the enrichment of cen transcripts normalized to act1+ RNA levels and relative to untagged strains. Error bars represent SD from three experiments.
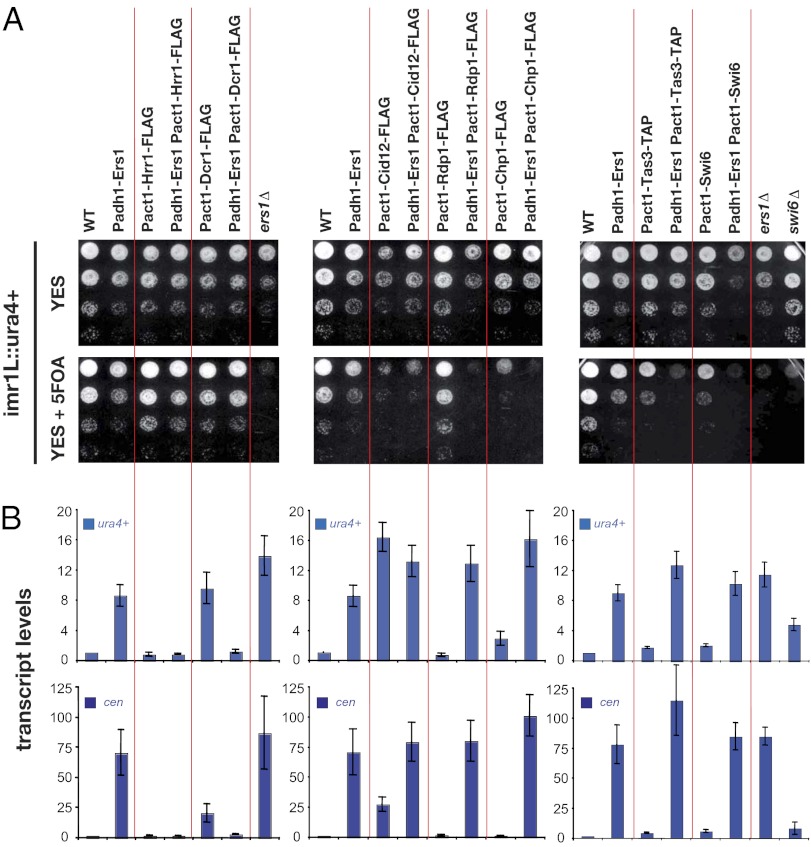
Overexpression of Ers1 Produces Dominant-Negative Phenotypes Suppressed by Overexpression of Dcr1 and Hrr1.
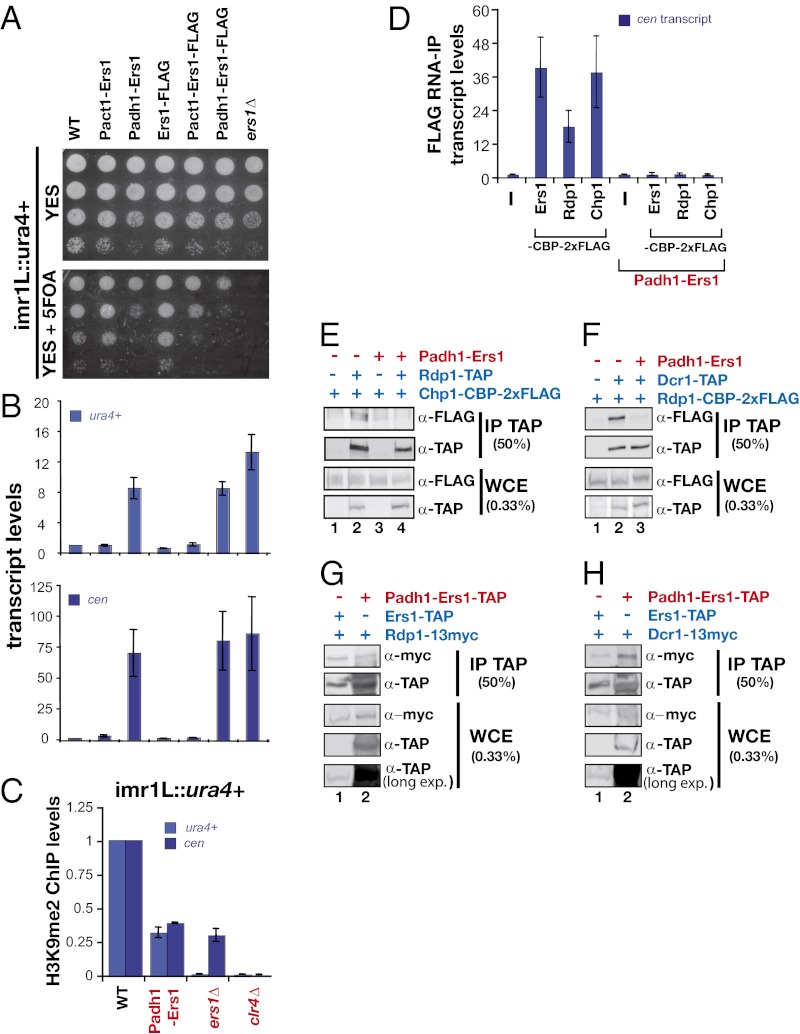
Overexpression of proteins that exhibit multiple protein–protein interaction domains can result in a dominant-negative phenotype in which mass action results in the loss of functional complexes at the expense of partially assembled complexes. Such behavior has been proposed to explain the dose sensitivity of signaling scaffolds. We observed that overexpression of Ers1 using the strong adh1 promoter (Fig. S4 A and B) produces a defect in pericentromeric reporter gene silencing (Fig. 4 A and B) and mat3 reporter gene silencing in pcr1Δ cells (Fig. S4C) and results in a decrease in pericentromeric H3K9Me (Fig. 4C) as well as in RITS binding to chromatin (Fig. S4D). Remarkably, overexpression of Ers1 not only blocks the association of Rdp1 and Chp1 with pericentromeric noncoding RNAs, it also inhibits its own association with these RNAs (Fig. 4D). Consistent with the thinking outlined above in which overexpression is anticipated to produce partially assembled complexes, overexpression of Ers1 disrupts the association of RITS with RDRC (Fig. 4E) and of RDRC with Dcr1 (Fig. 4F), yet Ers1 still associates with each complex when assayed individually (Fig. 4 G and H).
Fig. 4.
Overexpression of Ers1 triggers a dominant-negative phenotype for pericentromeric silencing. (A) Effect of Ers1 overexpression. Silencing of a ura4+ reporter gene inserted at the centromeric inner repeats (imr1L::ura4+) was assayed in cells overexpressing Ers1 with the strong adh1+ promoter or the act1+ promoter. (B) RT-qPCR assays. Shown are measurements of ura4+ (Upper) and cen (Lower) transcript levels normalized to a control transcript act1+ and relative to wild type. Error bars represent SD from three experiments. (C) ChIP analysis of H3K9me2 levels at the endogeneous cen repeats and at the imr1L::ura4+ locus. Signals have been normalized to act1+ and are shown relative to wild type. Error bars represent the deviation from the mean of two experiments. (D) RIP assay. Association of cen transcripts with Ers1-CBP-2×FLAG, Rdp1-CBP-2×FLAG, and Chp1-CBP-2×FLAG was assayed in cells overexpressing Ers1. Shown is the enrichment of cen transcripts normalized to act1+ RNA levels and relative to untagged strains. Error bars represent SD from three experiments. (E–H) Coimmunoprecipitation experiments. Shown are Western blots of immune pellets or whole-cell extracts of cells of the indicated genotypes.
To further probe the mechanism behind the dominant-negative phenotype, we tested whether it could be suppressed by overexpression of another component, thereby presumably restoring the formation of functional RNAi complexes. We analyzed the effects of overexpressing Rdp1, Hrr1, Cid12, Dcr1, Chp1, Tas3, and Swi6. Among these, only overexpression of the RDRC component Hrr1 and Dcr1 suppresses the Ers1 dominant-negative phenotype (Fig. 5 A and B). The suppression by overexpression of Dcr1 is remarkable in that Dcr1 overexpression itself produces a silencing defect (most evident at the RNA level; Fig. 5 A and B). Thus, overexpression of Ers1 and Dcr1 mutually suppresses each other’s dominant-negative phenotype, suggesting a particularly close functional relationship between these proteins. In contrast, although overexpression of Cid12 using the act1 promoter produces a silencing defect, this phenotype is not suppressed by overexpression of Ers1 (Fig. 5 A and B). Overexpression of proteins was verified by immunoblotting (Figs. S5 and S6).
Fig. 5.
Overexpression of either the RDRC component Hrr1 or Dcr1 suppresses the dominant-negative phenotype associated with overexpression of Ers1. (A) Silencing assays. Examination of the expression state of a ura4+ reporter gene inserted at the centromeric inner repeats (imr1L::ura4+) in cells of the indicated genotypes. (B) RT-qPCR analysis. Shown are measurements of ura4+ (Upper) and cen (Lower) transcript levels normalized to a control transcript act1+ and relative to wild type. Error bars represent SD from three experiments.
Discussion
Since the exciting discovery nearly a decade ago that RNAi is linked to H3K9Me, there has been a great deal of progress in the identification of the core protein complexes required for both H3K9Me and RNAi. However, how these activities are coordinated is still not clear. A major breakthrough was the identification of the RITS complex, which contains both a high-affinity H3K9Me-binding module in the chromodomain of the Chp1 subunit and the Argonaute protein Ago1 (5, 7). However, subsequent work demonstrated that the disruption of the RITS complex by mutation of the Argonaute-binding GW hooks in Tas3 did not affect silencing or siRNA production (8). A defect for the Tas3–GW mutant was uncovered by disrupting the gene coding for the Clr4 histone methyltransferase and reintroducing the wild-type clr4 through a genetic cross (8). This phenotype revealed by this maneuver suggests a redundant role for RITS in connecting H3K9Me and RNAi that only becomes evident under sensitizing conditions. Consistent with this possibility, the HP1 protein Swi6 was shown to be required for RNAi, offering another avenue by which the H3K9Me and RNAi pathways could be connected (3, 4). Moreover, Swi6 was shown to be required for the recruitment of RDRC to chromatin (21). However, no physical link between Swi6 and the RNAi machinery had been identified. Our studies of Ers1 begin to clarify the connection between heterochromatin and RNAi.
Before this work, it was unclear whether or not Ers1 participated directly in RNAi. The in vivo physical associations of Ers1 with pericentromeric noncoding RNAs and RNAi components that we describe here strongly support a direct role in the process. Furthermore, our two-hybrid studies provide evidence that Ers1 participates in protein–protein interactions with factors required for RNAi, the strongest being with Swi6 and Tas3. Additional interactions were observed with fragments of Ers1 and components of the RDRC and Dcr1. We were able to reconstitute the Ers1–Swi6 interaction with recombinant proteins produced in E. coli, defining a unique direct interaction between an HP1 protein and an RNAi factor in S. pombe. The only other such example described in eukaryotes is the direct interaction between Drosophila HP1a and the Argonaute superfamily protein Piwi (22). A comparison with our results indicates that, whereas the link between RNAi complexes and HP1 is generally highly conserved, the biochemical details of the linkage can differ significantly between species. Strict biochemical validation of the directness of the interaction of Ers1 with components of RITS and the RDRC will require the production of the corresponding proteins in soluble, recombinant form. Nonetheless, the unique ability of Hrr1 and Dcr1 overexpression to suppress the effect of Ers1 overexpression, the two-hybrid interaction data, and the coimmunoprecipitation data are together consistent with the view that Ers1 directly interacts with these proteins in vivo. The ability of Ers1 to interact with Hrr1 in mutant cells lacking Ago1, Dcr1, or Clr4 (Fig. 2F) further supports this notion.
A parsimonious model would be that the Ers1–Swi6 interaction helps to tether the three key RNAi components (RITS, RDRC, and Dcr1) to chromatin, thereby bringing the RNAi machinery into proximity to the nascent noncoding RNA substrate. Because recruitment of the Chp1 component of RITS does not require Swi6 (6), the interactions between Ers1 and the RDRC/Dcr1 complex may be particularly important in terms of recruitment to chromatin. This view is particularly consistent with our genetic suppression results. In addition, while this work was under review, an independent study by Hayashi et al. (23) showed that tethering of Hrr1 to centromeric regions can bypass the requirement for Swi6 in heterochromatin formation, further highlighting a role for HP1 in recruiting RDRC to chromatin. The Ers1–Swi6 interaction documented here also likely explains the previously described role for Swi6 in RNAi and the observation that Swi6 physically associates with centromeric transcripts in vivo (4). The associations of Swi6 may be cell-cycle regulated because pericentromeric heterochromatin has been shown to be transiently disrupted to a degree and selectively transcribed during S phase at which time RNAi appears to be most active (24, 25). Curiously, mutation of a docking site for Swi6, lysine 9 of the H3 tail, has differential effects on RNAi that depend on the region examined (26). Whereas siRNAs templated by the dh repeat absolutely require the H3K9Me mark for their accumulation, those derived from the dg repeat are much less dependent on H3K9Me. A possible explanation for the latter observation is the finding that a transcript-associated RNA export factor, Mlo3, is directly methylated by Clr4 and acts in parallel with H3K9Me to promote siRNA accumulation (27). Intriguingly, Mlo3 was identified as a protein associated with Swi6 in vivo (28), raising the possibility that Swi6 may dock to either a methylated histone or to a methylated nonhistone protein to nucleate the assembly of RNAi complexes on the transcript. Finally, because Ers1, but not Swi6, is required for the recruitment of RITS to chromatin (9), Ers1 likely also plays Swi6-independent roles promoting RNAi. In agreement with this, centromeric silencing triggered by the tethering of Hrr1 can bypass a requirement for Swi6, but not Ers1, indicating that the function of Ers1 is not limited to bridging HP1 to RDRC (23).
An additional contact between the histone methylation and RNAi machineries is likely to be provided by the LIM domain protein Stc1. Stc1 has phenotypes similar to Ers1 in that it is required for the assembly of RNAi complexes and coimmunoprecipitates with the RITS complex (20). The direct binding partners of Stc1 are unknown as no recombinant protein or two-hybrid studies with Stc1 have been reported. However, Stc1 has been found to copurify with the CLRC complex, and, when artificially tethered to DNA, can trigger silencing independently of RNAi (20). On the basis of these observations, it has been proposed that Stc1 physically links RITS to the CLRC. Thus, at least three distinct adhesive links between the H3K9Me and RNAi pathways have emerged: one mediated by Stc1 that involves the CLRC complex, one mediated by RITS that requires the H3K9 methyl mark, and a third mediated by Ers1 that is mediated by direct binding to HP1/Swi6. Although much biochemical work remains to be done to flesh out the mechanistic and structural details, the existence of such a multilayered network of interactions between the H3K9Me and RNAi pathways may explain how siRNA production in S. pombe is tightly restricted to those regions of the genome that are stably packaged into heterochromatin. Such a coupling may prevent normal cellular mRNAs from being subjected to destruction by erroneous RNAi.
Methods
Fission yeast strain cultivation and strain construction was performed using standard methods. Two-hybrid analysis was performed by mating bait and prey strains and performing both plate and liquid β-galactosidase assays. His-tagged Swi6 was peformed in E. coli and purified using Cobalt-NTA affinity chromatography. Immunoprecipitation experiments were performed using moderate salt concentrations (200 mM KCl) on whole-cell extracts prepared by disruption of frozen cells. Standard ChIP, RNA immunoprecipitatoin, RT-qPCR, and immunoblotting methods were used. Fission yeast strains are available in Table S4, primers in Table S5, and yeast two-hybrid plasmids in Table S6.
Detailed methods can be found in the SI Methods.
Supplementary Material
Acknowledgments
We thank Danesh Moazed and Karl Ekwall for strains and protocols, members of the H.D.M. laboratory, and anonymous reviewers for helpful comments on the manuscript. M.R., S.B. and J.F.G. were supported by a Human Frontier Science Program postdoctoral fellowship, the German Research Foundation (BR 3511/1-1), and National Institutes of Health (NIH)–National Institute of General Medical Sciences–Initiative for Maximizing Student Development Predoctoral Fellowship (R25-GM56847), respectively. S.C. and R.S.I. are supported by a graduate research fellowship from the National Science Foundation. This work was supported by NIH Grant GM071801 (to H.D.M.). H.D.M. was a fellow of the Leukemia and Lymphoma Society.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1204947109/-/DCSupplemental.
References
- 1.Grewal SI. RNAi-dependent formation of heterochromatin and its diverse functions. Curr Opin Genet Dev. 2010;20:134–141. doi: 10.1016/j.gde.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lejeune E, Allshire RC. Common ground: Small RNA programming and chromatin modifications. Curr Opin Cell Biol. 2011;23:258–265. doi: 10.1016/j.ceb.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 3.Motamedi MR, et al. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004;119:789–802. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 4.Motamedi MR, et al. HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol Cell. 2008;32:778–790. doi: 10.1016/j.molcel.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noma K, et al. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat Genet. 2004;36:1174–1180. doi: 10.1038/ng1452. [DOI] [PubMed] [Google Scholar]
- 6.Sadaie M, Iida T, Urano T, Nakayama J. A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast. EMBO J. 2004;23:3825–3835. doi: 10.1038/sj.emboj.7600401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verdel A, et al. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Partridge JF, et al. Functional separation of the requirements for establishment and maintenance of centromeric heterochromatin. Mol Cell. 2007;26:593–602. doi: 10.1016/j.molcel.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 9.Rougemaille M, Shankar S, Braun S, Rowley M, Madhani HD. Ers1, a rapidly diverging protein essential for RNA interference-dependent heterochromatic silencing in Schizosaccharomyces pombe. J Biol Chem. 2008;283:25770–25773. doi: 10.1074/jbc.C800140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roguev A, et al. Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science. 2008;322:405–410. doi: 10.1126/science.1162609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jia S, Noma K, Grewal SI. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science. 2004;304:1971–1976. doi: 10.1126/science.1099035. [DOI] [PubMed] [Google Scholar]
- 12.Bühler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125:873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 13.Estojak J, Brent R, Golemis EA. Correlation of two-hybrid affinity data with in vitro measurements. Mol Cell Biol. 1995;15:5820–5829. doi: 10.1128/mcb.15.10.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong EJ, Villén J, Gerace EL, Gygi SP, Moazed D. A cullin E3 ubiquitin ligase complex associates with Rik1 and the Clr4 histone H3-K9 methyltransferase and is required for RNAi-mediated heterochromatin formation. RNA Biol. 2005;2:106–111. doi: 10.4161/rna.2.3.2131. [DOI] [PubMed] [Google Scholar]
- 15.Jia S, Kobayashi R, Grewal SI. Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat Cell Biol. 2005;7:1007–1013. doi: 10.1038/ncb1300. [DOI] [PubMed] [Google Scholar]
- 16.Li F, et al. Two novel proteins, dos1 and dos2, interact with rik1 to regulate heterochromatic RNA interference and histone modification. Curr Biol. 2005;15:1448–1457. doi: 10.1016/j.cub.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 17.Horn PJ, Bastie JN, Peterson CL. A Rik1-associated, cullin-dependent E3 ubiquitin ligase is essential for heterochromatin formation. Genes Dev. 2005;19:1705–1714. doi: 10.1101/gad.1328005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Canzio D, et al. Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly. Mol Cell. 2011;41:67–81. doi: 10.1016/j.molcel.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colmenares SU, Buker SM, Buhler M, Dlakić M, Moazed D. Coupling of double-stranded RNA synthesis and siRNA generation in fission yeast RNAi. Mol Cell. 2007;27:449–461. doi: 10.1016/j.molcel.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Bayne EH, et al. Stc1: A critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell. 2010;140:666–677. doi: 10.1016/j.cell.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sugiyama T, Cam H, Verdel A, Moazed D, Grewal SI. RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc Natl Acad Sci USA. 2005;102:152–157. doi: 10.1073/pnas.0407641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brower-Toland B, et al. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 2007;21:2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayashi A, et al. Heterochromatin protein 1 homologue Swi6 acts in concert with Ers1 to regulate RNAi-directed heterochromatin assembly. Proc Natl Acad Sci USA. 2012;109:6159–6164. doi: 10.1073/pnas.1116972109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen ES, et al. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- 25.Kloc A, Zaratiegui M, Nora E, Martienssen R. RNA interference guides histone modification during the S phase of chromosomal replication. Curr Biol. 2008;18:490–495. doi: 10.1016/j.cub.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerace EL, Halic M, Moazed D. The methyltransferase activity of Clr4Suv39h triggers RNAi independently of histone H3K9 methylation. Mol Cell. 2010;39:360–372. doi: 10.1016/j.molcel.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang K, et al. Clr4/Suv39 and RNA quality control factors cooperate to trigger RNAi and suppress antisense RNA. Science. 2011;331:1624–1627. doi: 10.1126/science.1198712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fischer T, et al. Diverse roles of HP1 proteins in heterochromatin assembly and functions in fission yeast. Proc Natl Acad Sci USA. 2009;106:8998–9003. doi: 10.1073/pnas.0813063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.