Abstract
Asparagine-linked glycans (N-glycans) are crucial signals for protein folding, quality control, and endoplasmic reticulum (ER)-associated degradation (ERAD) in yeast and mammals. Although similar ERAD processes were reported in plants, little is known about their biochemical mechanisms, especially their relationships with N-glycans. Here, we show that a missense mutation in the Arabidopsis EMS-mutagenized bri1 suppressor 3 (EBS3) gene suppresses a dwarf mutant, bri1-9, the phenotypes of which are caused by ER retention and ERAD of a brassinosteroid receptor, BRASSINOSTEROID-INSENSITIVE 1 (BR1). EBS3 encodes the Arabidopsis ortholog of the yeast asparagine-linked glycosylation 9 (ALG9), which catalyzes the ER luminal addition of two terminal α1,2 mannose (Man) residues in assembling the three-branched N-glycan precursor [glucose(Glc)]3(Man)9[N-acetylglucosamine(GlcNAc)]2. Consistent with recent discoveries revealing the importance of the Glc3Man9GlcNAc2 C-branch in generating an ERAD signal, the ebs3-1 mutation prevents the Glc3Man9GlcNAc2 assembly and inhibits the ERAD of bri1-9. By contrast, overexpression of EBS4 in ebs3-1 bri1-9, which encodes the Arabidopsis ortholog of the yeast ALG12 catalyzing the ER luminal α1,6 Man addition, adds an α1,6 Man to the truncated N-glycan precursor accumulated in ebs3-1 bri1-9, promotes the bri1-9 ERAD, and neutralizes the ebs3-1 suppressor phenotype. Furthermore, a transfer (T)-DNA insertional alg3-T2 mutation, which causes accumulation of an even smaller N-glycan precursor carrying a different exposed α1,6 Man, promotes the ERAD of bri1-9 and enhances its dwarfism. Taken together, our results strongly suggest that the glycan signal to mark an ERAD client in Arabidopsis is likely conserved to be an α1,6 Man-exposed N-glycan.
Asparagine (Asn or N)-linked glycosylation is an important protein-modification process in all three domains of life (1). In animals, fungi, and plants, N-glycan is formed by transfer of a preassembled tetradecasaccharide Glc3Man9GlcNAc2 (where Glc, Man, and GlcNAc represent glucose, mannose, and N-acetylglucosamine, respectively) from its lipid carrier, dolichylpyrophosphate (Dol-PP), to selective Asn residues on nascent polypeptides (2) (Fig. S1). The biosynthesis of the three-branched Glc3Man9GlcNAc2 is a highly ordered assembly pathway at the endoplasmic reticulum (ER) membrane by sequential addition of sugars to the Dol-PP carrier involving highly specific glycosyltransferases that include ALG3, ALG9, and ALG12 catalyzing the ER luminal addition of an α1,3 Man, two α1,2 Man, and an α1,6 Man, respectively (Fig. S1) (3). The N-linked Glc3Man9GlcNAc2 glycan is subsequently processed in the ER and Golgi by extensive deglycosylation and sugar additions.
Extensive studies in yeast and mammals revealed that ER-processed N-glycans serve as important signals for protein folding, quality control (QC), degradation, and sorting (4). Rapid sequential trimming of two Glc residues on the A-branch by glucosidase I (GI) and GII generates Glc1Man9GlcNAc2, which interacts with calnexin (CNX) and its soluble homolog calreticulin (CRT), which recruit additional chaperones to facilitate protein folding, whereas removal of the last Glc by GII liberates glycoproteins from CNX/CRT (5) (Fig. S1). An incompletely/misfolded glycoprotein is recognized and reglucosylated by the ER-localized folding sensor UDP-Glc:glycoprotein glucosyltransferase (UGGT) for additional rounds of CNX/CRT-assisted folding, known as the CNX/CRT cycle (6). By contrast, slow trimming of the B-branch α1,2 Man by the ER α1,2 mannosidase I (ERManI) (7) (Fig. S1) was previously thought to create a critical N-glycan signal (Man8GlcNAc2) that interrupts futile CNX/CRT cycles of terminally misfolded proteins to deliver them for ER-associated degradation (ERAD) that involves retrotranslocation and cytosolic proteasomes (8). However, recent studies showed that whereas the B-branch α1,2 Man-trimming is a critical ERAD event, the actual N-glycan signal to mark an ERAD client is an exposed α1,6 Man generated by removing the terminal α1,2 Man from the C-branch (9, 10) (Fig. S1).
Although many studies reported the existence of similar ERQC and ERAD processes in plants (11, 12), little is known about their biochemical mechanisms, especially their relationship with N-glycan biosynthesis. Two Arabidopsis leucine-rich-repeat receptor-like kinases, BRASSINOSTEROID-INSENSITIVE 1 (BRI1) and EF-Tu Receptor (EFR), have recently emerged as model proteins to study ERQC and ERAD in plants (13). BRI1 functions as a cell surface receptor for the plant steroid hormone brassinosteroids (BRs) (14, 15), whereas EFR recognizes the bacterial translation elongation factor EF-Tu to initiate plant immunity responses (16). A Cys69Tyr mutation disrupting a highly conserved N-terminal disulfide bond and a Ser662Phe mutation in the BR-binding domain result in ER retention and ERAD of two structurally imperfect but biochemically competent BR receptors, bri1-5 and bri1-9, respectively, explaining their BR-insensitive dwarf phenotypes (17–19). A genetic screen looking for suppressors that restore the wild-type (WT) morphology to bri1-9 led to identification of EMS-mutagenized bri1 suppressor 1 (EBS1) and EBS2, encoding the Arabidopsis UGGT and a plant-specific CRT3, respectively (17, 18).
To identify additional factors of the plant ERQC and ERAD systems, we isolated and studied additional ebs mutants. We recently discovered that EBS4 encodes the Arabidopsis ortholog of the yeast/human ALG12 that catalyzes the ER luminal addition of an α1,6 Man residue of Glc3Man9GlcNAc2 and concluded that transfer of a completely assembled N-glycan precursor is required for successful ERAD of bri1-5 and bri1-9 (20). Here, we report that the Arabidopsis EBS3 gene encodes the Arabidopsis ortholog of the yeast/human ALG9 catalyzing the luminal addition of two α1,2 Man residues in assembling Glc3Man9GlcNAc2 (21), further confirming our previous conclusion. Our studies using an Arabidopsis alg3-T2 mutant that accumulates Man5GlcNAc2 (22, 23) and EBS4-overexpressing transgenic ebs3-1 bri1-9 lines revealed that the glycan signal to mark ERAD clients in Arabidopsis is likely conserved to be an exposed α1,6 Man residue on N-glycans.
Results
ebs3-1 Mutant Is Defective in the ERAD of bri1-9.
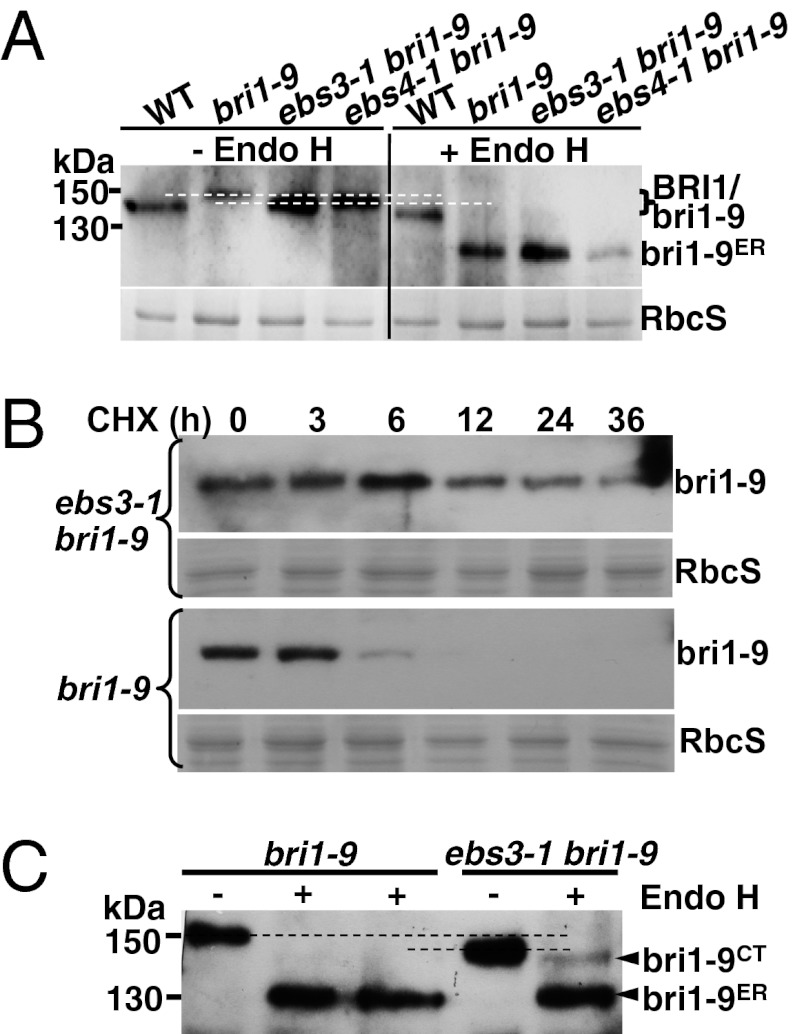
A previous genetic screen for extragenic suppressors of bri1-9 isolated more than 80 ebs mutants, including ebs1 and ebs2 mutants defective in retaining bri1-9 in the ER (17, 18). A secondary screen looking for ebs mutants with increased bri1-9 abundance identified several potential ERAD mutants including ebs3, ebs4 (20), and ebs5 (24). As shown in Fig. 1A, both ebs3-1 and ebs4-1 accumulate more bri1-9 than bri1-9. To test if the increased bri1-9 abundance is caused by increased synthesis or reduced degradation, we treated 3-wk-old seedlings with cycloheximide (CHX), a widely used protein synthesis inhibitor, and analyzed the bri1-9 abundance by immunoblot. Fig. 1B shows that CHX caused a rapid disappearance of the mutant BR receptor in bri1-9 but had a much weaker effect on the bri1-9 abundance in ebs3-1 bri1-9. A significant amount of bri1-9 was still present in ebs3-1 bri1-9 36 h after CHX treatment, whereas no bri1-9 was detectable after 12 h of CHX treatment in bri1-9. We, thus, concluded that the ebs3-1 mutation inhibits the bri1-9 ERAD.
Fig. 1.
ebs3-1 mutation inhibits the ERAD of bri1-9. (A) Immunoblot analysis of bri1-9 in ebs3-1 bri1-9. (B) Immunoblot analysis of bri1-9 stability in bri1-9 and ebs3-1 bri1-9. (C) Endo H analysis of bri1-9 in bri1-9 and ebs3-1 bri1-9. For A and C, total proteins from 4-wk-old leaves were treated with or without Endo H, separated by SDS/PAGE, and analyzed by immunoblot with anti-BRI1 antibody. Equal amounts of total proteins were used in A, whereas five times more proteins in bri1-9 than ebs3-1 bri1-9 were used in C, which also contains technical duplicates of Endo H-treated bri1-9 samples. For B, 3-wk-old seedlings were transferred from 1/2 MS-agar medium into liquid 1/2 MS medium containing 180 μM CHX. Equal amounts of seedlings were removed at different time points to extract total proteins into 2× SDS sample buffer, which were subsequently separated by SDS/PAGE and analyzed by immunoblot with anti-BRI1 antibody. The numbers on the left in A and C indicate molecular mass, bri1-9ER denotes Endo H-sensitive form, and bri1-9CT represents bri1-9 carrying C-type N-glycans. Dashed lines in A and C show the mobility difference between the bri1-9 band in bri1-9 and that of ebs3-1 bri1-9. Coomassie blue staining of the small subunit of Rubisco (RbcS) serves as the loading control.
We also treated protein extracts with Endo H, an endoglycosidase that cleaves high-mannose-type (H-type) N-glycans of ER-localized proteins but cannot remove Golgi-processed complex-type (C-type) N-glycans (25). Fig. 1A shows that the vast majority of bri1-9 was Endo H-sensitive in bri1-9 and two ebs mutants, indicating its predominant ER localization in all three genotypes. However, loading more proteins of ebs3-1 bri1-9 detected a small amount of bri1-9 carrying C-type N-glycans indicative of ER escape, whereas increased loading failed to detect the C-type N-glycan-containing BR receptor in bri1-9 (Fig. 1C). We suspected that the presence of a low level of the C-type N-glycan-containing bri1-9 in ebs3-1 bri1-9, likely attributable to saturation of the ER retention system by overaccumulated bri1-9 and its consequent leakage from the ER, is responsible for the suppressor phenotype because our earlier study showed that overexpression of bri1-9 could also suppress the bri1-9 mutation (20). Consistently, expression of a genomic EBS2 transgene, which encodes a rate-limiting factor for retaining bri1-9 in the ER (18, 24), neutralized the ebs3-1 suppressive effect on bri1-9 likely by preventing the ER leakage of a small amount of bri1-9 to the cell surface (Fig. S2).
ebs3-1 Mutation Weakly Suppresses bri1-9 with Partially Regained BR Sensitivity.
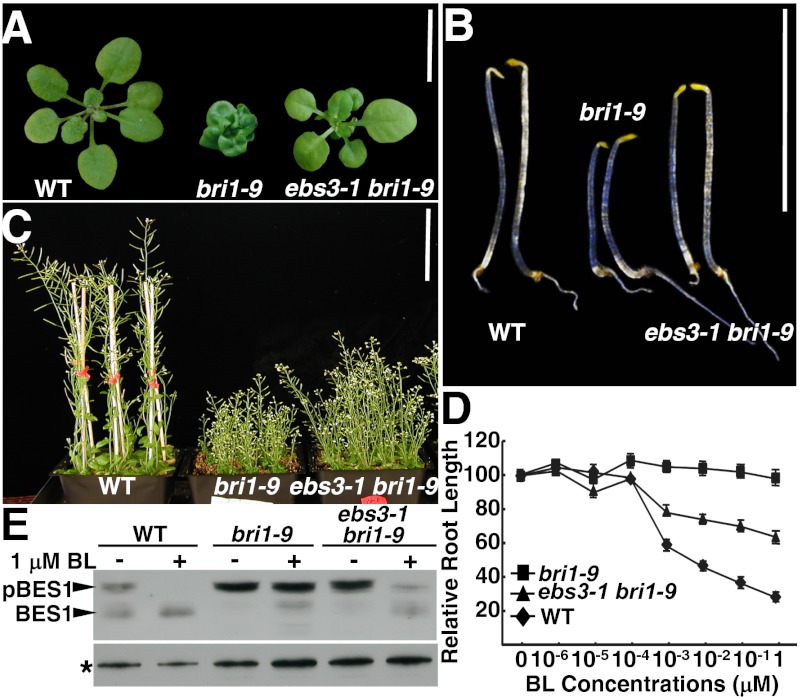
In line with accumulation of a low level of the C-type N-glycan–carrying bri1-9, ebs3-1 is a weak suppressor of bri1-9. The rosette of ebs3-1 bri1-9 is larger than that of bri1-9 (Fig. 2A), and its etiolated hypocotyl and inflorescence stem are longer than those of bri1-9 when grown in dark and soil, respectively (Fig. 2 B and C). The ebs3-1 also weakly suppresses the bri1-5 mutant that produces another ER-retained mutant BR receptor (19) (Fig. S3 A and B) but has no detectable effect on plant growth in a BRI1+ background (Fig. S3C). As expected from the morphological and immunoblot analyses, ebs3-1 bri1-9 partially regained BR sensitivity. Similar to what was previously reported (26), increasing concentrations of brassinolide (BL) (the most active BR) had little effect on the root growth of bri1-9 but inhibited that of the WT, as well as the ebs3-1 bri1-9 mutant, albeit to a lesser extent (Fig. 2D). The regained BR sensitivity was also detected by immunoblot analysis of the BL-induced dephosphorylation of BRI1 EMS SUPPRESSOR1 (BES1), a very robust biochemical marker for BR signaling (27). Fig. 2E reveals that BL had little effect on the BES1 phosphorylation status in bri1-9 but led to rapid dephosphorylation of BES1 in both the WT and ebs3-1 bri1-9.
Fig. 2.
ebs3-1 is a weak suppressor of the bri1-9 mutant. (A–C) Images of 3-wk-old soil-grown plants (A), 5-d-old etiolated seedlings (B), and 7-wk-old soil-grown mature plants (C) of WT, bri1-9, and ebs3-1 bri1-9. [Scale bars: 1 cm (A and B) and 10 cm (C).] (D) Quantitative analysis of BR sensitivity. Root length of 10-d-old seedlings grown on BL-containing medium were measured and presented as the relative value of average root length of BL-treated seedlings to that of mock-treated seedlings. Each data point represents the average of ∼40 seedlings of duplicated experiments. Error bars denote SE. (E) Immunoblot analysis of BL-induced BES1 dephosphorylation. Total proteins were extracted in 2× SDS buffer from 2-wk-old seedlings treated with or without 1 μM BL for 1 h, separated by SDS/PAGE, and analyzed by immunoblot using an anti-BES1 antibody (27). Star indicates a nonspecific band for loading control.
ebs3-1 Mutation Affects Assembly of the N-Glycan Precursor.
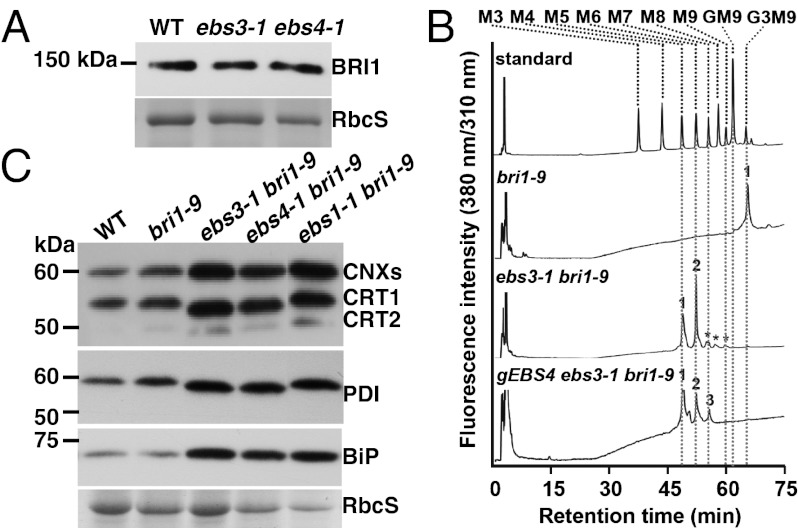
As indicated in Fig. 1 A and C, ebs3-1 increases not only the abundance but also the electrophoretic mobility of bri1-9. Because the mobility of deglycosylated bri1-9 band of ebs3-1 bri1-9 is the same as that of bri1-9 (Fig. 1 A and C), the mobility difference of bri1-9 is attributable to different glycoforms of bri1-9 in the two mutants. The bri1-9 is either hypoglycosylated at fewer sites or fully glycosylated with smaller glycans. Because hypoglycosylation often leads to multiple isoforms of an affected glycoprotein each carrying different numbers of N-glycans (21), our detection of a single bri1-9 band in ebs3-1 bri1-9 on immunoblot suggested that the observed mobility shift was caused by transfer of truncated N-glycan precursors to bri1-9. Further support for the presence of fully glycosylated bri1-9 with truncated glycans in ebs3-1 bri1-9 came from our observation that no obvious mobility difference of the WT BRI1 was detected between the WT and the two ebs mutants (Fig. 3A). A hypoglycosylated BRI1 should move faster than a fully glycosylated BRI1 on protein gels, whereas a fully glycosylated BRI1 in WT, ebs3-1, and ebs4-1 should exhibit the same mobility because BRI1 is not retained in the ER in all three genotypes and the Arabidopsis ER/Golgi-mediated glycan processing can effectively convert truncated H-type N-glycans, which are initially transferred in the ER to BRI1 in the two ebs mutants, to the same C-type N-glycans in the wild type (22, 23).
Fig. 3.
ebs3-1 likely affects assembly of the N-glycan precursor. (A) Immunoblot analysis of BRI1 in WT and ebs3-1 BRI1+. (B) Analysis of LLOs in bri1-9, ebs3-1 bri1-9, and a transgenic gEBS4 ebs3-1 bri1-9 line. LLOs of mature plants were extracted, acid-hydrolyzed, fluorescently labeled with PA, and analyzed using SF-HPLC by comparing their elution profiles with that of PA-sugar chain standards (Mn for MannGlcNAc2-PA and G3M9 for Glc3Man9GlcNAc2-PA). Asterisks indicate minor contaminants derived from the labeling process. (C) ebs3-1 affects the electrophoretic mobility of several ER-localized glycoproteins. For A and C, equal amounts of total proteins from 4-wk-old leaves were separated by SDS/PAGE and analyzed by immunoblot with antibodies against BRI1, maize CRT, PDI, or BiP. Numbers on the left indicate molecular mass. Coomassie blue staining of RbcS served as a loading control.
Further confirmation of a glycan-assembly defect came from size-fractionation (SF)-HPLC analysis of lipid-linked oligosaccharides (LLOs) of bri1-9 and ebs3-1 bri1-9, which were fluorescently labeled with 2-pyridylamino (PA) following acidic hydrolysis of the Dol-PP linker. As shown in Fig. 3B, whereas bri1-9 seedlings clearly accumulated the mature Glc3Man9GlcNAc2 [with its PA-labeled derivative having an identical elution position with the Glc3Man9GlcNAc2 (G3M9)-PA standard], the corresponding peak was absent in the ebs3-1 bri1-9 sample. Instead, two major peaks comigrating with M5 and M6 standards were detected in ebs3-1 bri1-9. The identity of the M6 peak was determined by reverse-phase (RP)-HPLC as Man6GlcNAc2ER, whereas the M5 peak was found to be a mixture of a small amount of Man5GlcNAc2ER and several minor contaminants (Fig. S4 A and B).
The effect of ebs3-1 on N-glycan assembly was also examined for several other ER-localized proteins, including a protein disulfide isomerase (PDI) (28), an ER-localized heat shock protein 70 (known as BiP) (29), and members of the CRT/CNX family (30). As shown in Fig. 3C, the mobility of PDI and CRT1 in ebs3-1 bri1-9 is slightly faster than that in bri1-9 and is similar to that in ebs4-1 (20). Fig. 3C also shows that ebs3-1 slightly activates the unfolded protein response (UPR), a master ER-surveillance system that stimulates synthesis of many ER chaperones in response to ER stresses (12). The protein abundance of CRTs, PDI, and BiP was slightly increased in ebs3-1 compared with the WT and bri1-9 and was similar to that in ebs1-1 and ebs4-1.
Cloning of the EBS3 Gene.
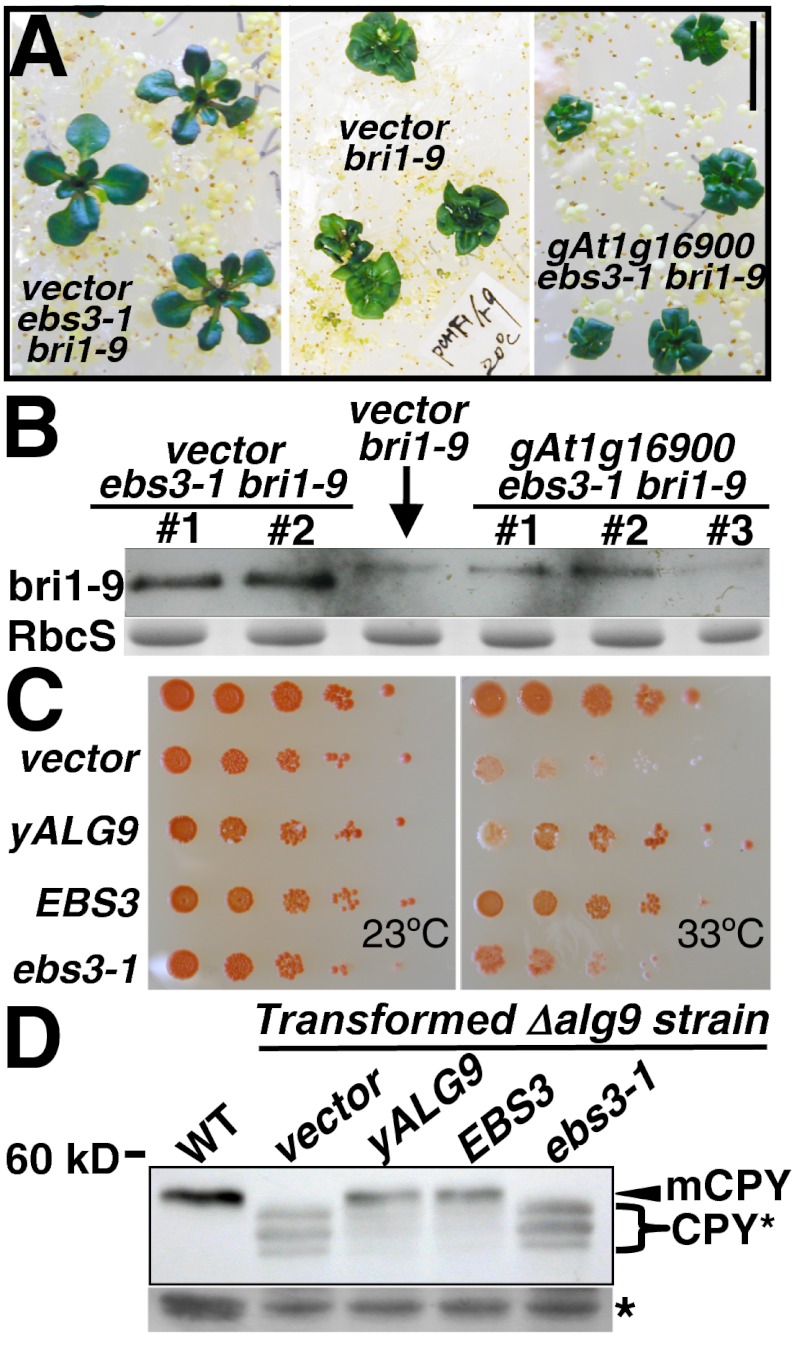
Based on the effects of ebs3-1 on the bri1-9 glycoforms and the LLO composition, we suspected that EBS3 might encode another ER-localized mannosyltransferase. PCR-based molecular mapping (SI Materials and Methods) located the EBS3 locus to a 200-kb region on the top of chromosome 1 that contains 59 annotated genes (Fig. S5 A and B). One of them, At1g16900, consisting of 10 exons and 9 introns (Fig. S5C), encodes a 570-aa protein highly similar to the yeast/human ALG9 (yALG9/hALG9) α1,2 mannosyltransferase and several predicted plant ALG9 homologs (Fig. S5D). The yALG9/hALG9 catalyzes the ER luminal addition of two α1,2 Man residues to assemble Glc3Man9GlcNAc2 (Fig. S1), and mutations in yALG9 result in accumulation of the Man6GlcNAc2ER glycan and inhibition of ERAD in yeast (21, 31–34). Sequencing analysis of PCR-amplified At1g16900 DNA from ebs3-1 bri1-9 revealed a single-nucleotide change that mutates Arg100 to Trp (Fig. S5D). This Arg residue is located near the end of the largest luminal loop between the first two predicted transmembrane segments (Fig. S5 D and E) and is absolutely conserved in all known ALG9s and two other α1,2 mannosyltransferases, phosphatidylinositol glycan anchor biosynthesis class B protein (PIG-B) and SMP3, involved in glycosylphosphatidylinositol synthesis (35). A further proof for At1g16900 being the EBS3 gene came from our rescue experiment showing that a genomic At1g16900 transgene rescued the morphological phenotype of ebs3-1 (Fig. 4A and Fig. S6) and its N-glycan and ERAD defects of bri1-9 (Fig. 4B).
Fig. 4.
EBS3 encodes the Arabidopsis ortholog of yALG9. (A) Images of 3-wk-old transgenic bri1-9 lines carrying an empty vector and transgenic ebs3-1 bri1-9 seedlings carrying a genomic EBS3 transgene (gEBS3) or an empty vector. (Scale bar: 1 cm.) (B) Immunoblot analysis of bri1-9. The numbers on top of the gel panel indicate different transgenic lines. (C) EBS3 complemented the yeast Δalg9 mutation. Growth efficiency of WT or Δalg9 wbp1-2 cells transformed with the vector or an expression plasmid of yALG9, EBS3 or ebs3-1 cDNA. Transformants were spotted in 10-fold serial dilution on synthetic medium and grown for 3 d at 23 °C or 33 °C. (D) Immunoblot analysis of CPY of the WT yeast strain and Δalg9 strain transformed with indicated plasmids. For B and D, equal amounts of total proteins extracted from 4-wk-old soil-grown seedlings (B) or yeast cells of midlog growth phase (D) were separated by SDS/PAGE and analyzed by immunoblot using an anti-BRI1 (B) or anti-CPY (D) antibody. Coomassie blue staining of RbcS served as a loading control in (B). mCPY indicates the mature CPY, CPY*s represent three isoforms of CPY carrying different numbers of truncated N-glycan in (D), and star denotes a cross-reacting band used for loading control.
EBS3 Is the Arabidopsis Ortholog of yALG9.
To directly test whether EBS3 is an ortholog of yALG9, we took advantage of the existence of a yeast Δalg9 wbp1-2 double mutant that exhibits a temperature-sensitive growth phenotype (21). We replaced the ORF of yALG9 in the pYEp352-yALG9 expression plasmid with that of EBS3 to generate pYEp352-EBS3, introduced the Arg100Trp mutation to make pYEp352-ebs3-1, and individually transformed the three plasmids and a vector control into the Δalg9 wbp1-2 strain. As shown in Fig. 4C, transformation of the yALG9 or EBS3 plasmid but not the ebs3-1 or vector plasmid suppressed the 33 °C growth defect of the Δalg9 wbp1-2 mutant. We also transformed each plasmid into the Δalg9 single mutant and analyzed the glycosylation pattern of vacuolar carboxypeptidase Y (CPY) carrying 4 glycosylation sites (36). As shown in Fig. 4D, both yALG9 and EBS3 but not the vector or ebs3-1 plasmid rescued the glycosylation defect of CPY, converting three faster-moving hypoglycosylated CPY* bands into a single mature CPY band (mCPY) with identical mobility to that of WT cells. These results demonstrated that EBS3 is the Arabidopsis ortholog of yALG9 and that the Arg100Trp mutation destroys its α1,2 mannosyltransferase activity.
Exposed α1,6 Mannose Residue Is the Likely Glycan Signal for the bri1-9 ERAD.
Recent studies revealed that the ERAD signal in yeast and mammals is an exposed α1,6 Man on N-glycans generated by removing an α1,2 Man residue from the C-branch (9, 10) (Fig. S1). This unique demannosylation was catalyzed by the yeast homologous to mannosidase I (Htm1) and its mammalian homologs, ER degradation enhancing-mannosidase-like proteins (EDEMs) (37). We hypothesized that the most likely reason for the defective bri1-9 ERAD in ebs3-1 bri1-9 is lack of a correct ERAD glycan signal on bri1-9.
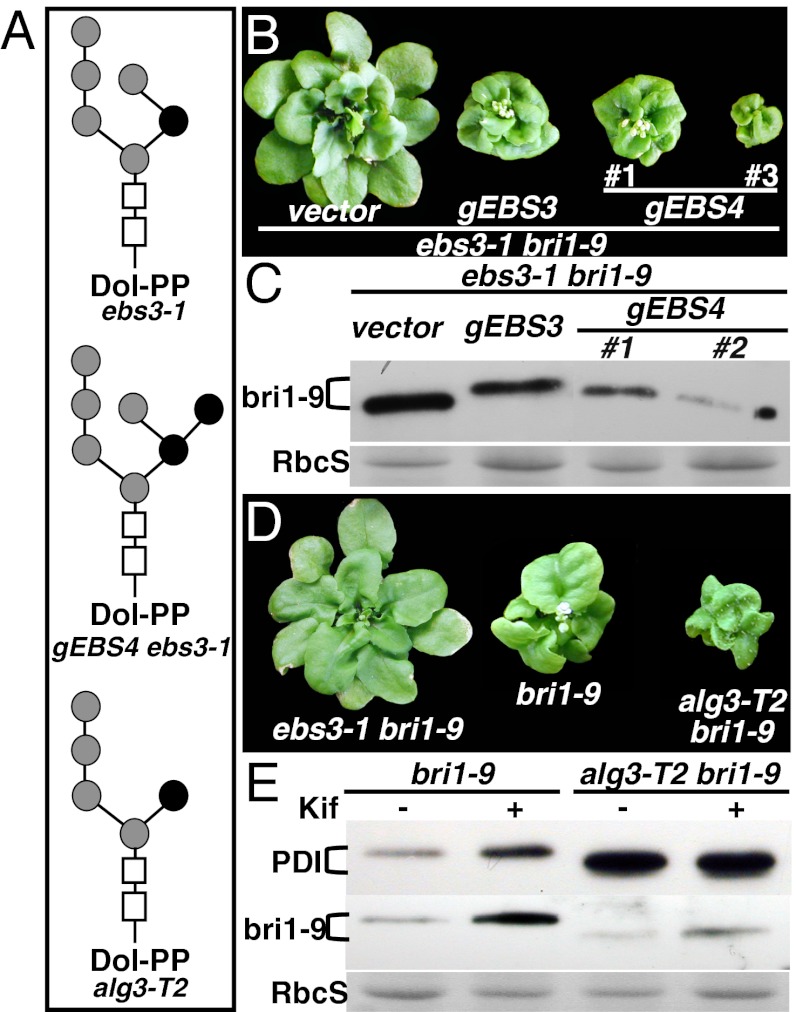
To test our hypothesis, we used a previously described genetic method for generating α1,6 Man-exposed N-glycans in ebs3-1. It was known that overexpressed yeast ALG12 could bypass the requirement of an intact B-branch for adding the second α1,6 Man to Man6GlcNAc2ER to generate a unique Man7GlcNAc2 glycan (38) (Fig. 5A). If an α1,6 Man-exposed N-glycan was the ERAD signal in Arabidopsis, we would expect that overexpression of EBS4 (encoding the Arabidopsis ALG12) should suppress the ebs3-1 bri1-9 phenotype and rescue the ERAD defect of bri1-9. Indeed, as shown in Fig. 5B, gEBS4 ebs3-1 bri1-9 transgenic mutants were morphologically similar to or even smaller than a typical EBS3-rescued ebs3-1 bri1-9 mutant. SF-HPLC analysis of LLOs of the gEBS4 ebs3-1 bri1-9 transgenic plants revealed the presence of a small peak at the position of a M7 standard (Fig. 3B), which was determined by RP-HPLC to contain the predicted Man7GlcNAc2 B-isoform with an exposed α1,6 Man (Fig. S4C). Although Man6GlcNAc2 remains the major LLO in the gEBS4 ebs3-1 bri1-9 line, a mere 7.14% (1/14) conversion efficiency of Man6GlcNAc2ER to Man7GlcNAc2ER is sufficient to have, on average, 1 α1,6 Man-exposing N-glycan on each bri1-9 (containing 14 predicted N-glycan sites) for marking the mutant receptor for ERAD. Indeed, an immunoblot analysis showed that EBS4 overexpression could reverse the inhibitory effect of ebs3-1 on the bri1-9 ERAD while causing little change in its mobility on protein gels (Fig. 5C).
Fig. 5.
An exposed α1,6 Man residue is likely the glycan signal for bri1-9 ERAD. (A) Schematic structures of suspected major LLOs in ebs3-1 bri1-9, alg3-T2 bri1-9, or transgenic gEBS4 ebs3-1 bri1-9 line. Rectangles denote the two GlcNAc residues and circles represent Man residues with black circles indicating α1,6-linked Man residues. (B) Images of 5-wk-old soil-grown transgenic ebs3-1 bri1-9 mutant containing pPZP212, a gEBS3, or gEBS4 genomic transgene. (C) Immunoblot analysis of the bri1-9 abundance. Numbers in B and C indicate independent transgenic lines. (D) Images of 5-wk-old soil-grown mutants of ebs3-1 bri1-9, bri1-9, and alg3-T2 bri1-9. (E) Immunoblot analysis of PDI and bri1-9. For C and E, equal amounts of total proteins extracted in 2× SDS buffer from 4-wk-old leaves were treated with or without 10 μM Kif, separated by SDS/PAGE, and analyzed by immunoblot with anti-PDI or anti-BRI1 antibody. Coomassie blue staining of RbcS served as a loading control.
Additional support for an α1,6 Man-exposed N-glycan being the critical ERAD signal came from our genetic analysis of an Arabidopsis alg3 mutant. Loss-of-function alg3 mutations block the formation of Glc3Man6GlcNAc2, resulting in transfer of Glc3Man5GlcNAc2 with a different free α1,6 Man from Dol-PP to proteins (Fig. 5A) (22, 23), whereas a recent study showed a normal ERAD process in an yeast Δalg3 mutant (9). If this α1,6 Man can also function as an ERAD signal in Arabidopsis, we would expect that an alg3 mutation should promote rather than inhibit the ERAD of bri1-9. We obtained a transfer (T)-DNA insertional alg3 mutant [The Arabidopsis Information Resource (TAIR) accession no. SALK_046061; previously named alg3-T2] (39) and crossed the alg3-T2 mutation into bri1-9. Fig. 5D shows that the alg3-T2 bri1-9 mutant is morphologically more severe than bri1-9. It is important to note that alg3-T2 itself has no detectable effect on plant growth (23), although it does activate UPR because the abundance of PDI in alg3-T2 bri1-9 is significantly higher than that in bri1-9 (Fig. 5E). By contrast, a T-DNA insertional mutation of the Arabidopsis stroma cell-derived factor 2-like protein (SDF2), required for the correct folding of EFR (40), failed to enhance the bri1-9 dwarfism (Fig. S7) even though the sdf2-2 mutant was hypersensitive to ER stresses (41), suggesting that the phenotypic enhancement of bri1-9 by alg3-T2 is not caused by an abnormal ER stress response but is likely attributable to a stimulatory effect of alg3-T2 on the bri1-9 ERAD. Indeed, immunoblotting with a BRI1 antibody revealed that whereas alg3-T2 increases the mobility of bri1-9 because of smaller N-glycans, it decreases the bri1-9 abundance, which is the exactly opposite of what was observed in ebs3-1 and ebs4-1 mutants that accumulate larger N-glycan precursors (Fig. 5E). Interestingly, the ERAD of bri-9 in alg3-T2 bri1-9 could still be inhibited by treatment with kifunensine (Kif), a well-known inhibitor of α1,2 mannosidase (42) that prevents ERAD of both bri1-5 and bri1-9 carrying fully assembled N-glycans (19, 20) (Fig. 5E), although its inhibitory effect on the bri1-9 ERAD is much weaker in alg3-T2 bri1-9 than in bri1-9, suggesting that the bri1-9 ERAD might require additional Man trimming. Taken together, these results strongly suggested that an exposed α1,6 Man is likely the glycan signal for a plant ERAD process.
Discussion
In this study, we demonstrated that EBS3 encodes the Arabidopsis ortholog of yALG9 that catalyzes the ER luminal addition of two α1,2 Man residues for assembling Glc3Man9GlcNAc2 (31). First, ebs3-1 contains a single-nucleotide change in At1g16900, and its bri1-9 suppressor phenotype was rescued by expression of a genomic At1g16900 transgene. Second, the predicted At1g16900 protein exhibits the highest sequence homology among all annotated Arabidopsis proteins to the yeast ALG9 protein (Fig. S8). The other two Arabidopsis proteins showing limited sequence homology are EBS4 and At5g14850 annotated to encode a putative homolog of the yeast PIG-B involved in glycosylphosphatidylinositol biosynthesis (35). Third, the WT At1g16900 was able to complement the growth phenotype of the yeast Δalg9 wbp1-2 double mutant and the N-glycosylation defect of CPY of the Δalg9 yeast mutant (Fig. 4 C and D). Consistent with sequence analysis suggesting a crucial catalytic role of the Arg100 residue for several mannosyltransferases (35), our yeast complementation assay showed that the R100W mutation destroys its α1,2 mannosyltransferase activity in yeast cells, suggesting that ebs3-1 is likely a null mutant. Thus, EBS3 works together with the recently discovered AtALG3 (22, 23) and EBS4 (20) as the three Arabidopsis ER-luminal mannosyltransferases to add four more Man residues to Dol-PP-Man5GlcNAc2 to assemble Dol-PP-Man9GlcNAc2 that will be triglucosylated before the fully assembled Glc3Man9GlcNAc2 can be transferred to nascent polypeptides (Fig. S1).
In yeast, the α1,2 mannosyltransferase ALG9 catalyzes the ER luminal addition of two terminal α1,2 Man residues to create the B- and C-dimannose branches (31), whereas the α1,6 mannosyltransferase ALG12 exhibits high substrate specificity toward Dol-PP-Man7GlcNAc2 but is very inefficient in adding the second α1,6 Man when the B-branch lacks the terminal α1,2 Man (43) (Fig. S1). Thus, null alg9 mutations in yeast result in accumulation of Dol-PP-Man6GlcNAc2 and protein hypoglycosylation because of reduced transfer efficiency of immature glycans by the yeast oligosaccharide transferase (OST) (21, 43). Consistent with our findings that EBS3 is an Arabidopsis ortholog of yALG9 and that the R100W mutation is either a null or very severe mutation, our HPLC analyses of LLOs indicated that ebs3-1 bri1-9 accumulates Dol-PP-Man6GlcNAc2. Surprisingly, ebs3-1 bri1-9 also accumulates Dol-PP-Man5GlcNAc2, the known substrate for ALG3 (22, 23). This is likely caused by feedback inhibition of ALG3 by overaccumulation of its product, Dol-PP-Man6GlcNAc2, because Dol-PP-Man5GlcNAc2 was also detected in a yeast alg9 mutant (21). Despite the glycan-assembly defect, neither the WT BRI1 nor bri1-9 is hypoglycosylated, which is likely attributable to the fact that the Arabidopsis OSTs can efficiently transfer truncated glycans from their Dol-PP linker to bri1-9 and other glycoproteins (22, 23), explaining no obvious growth defect of an ebs3-1 BRI1+ mutant (Fig. S3C).
A crucial decision in ERQC is when to stop futile refolding attempts to divert a terminally misfolded protein from the CNX/CRT cycle to ERAD. A previously popular “mannosidase timer” model posited that the slow α1,2 Man trimming of the B-branch by ERManI generates the ERAD glycan Man8GlcNAc2 that can be recognized by dedicated ERAD lectins (7). However, recent studies showed that although trimming the B-branch is a necessary ERAD step, the true ERAD signal is an exposed α1,6 Man created by trimming the C-branch (9, 10). In this study, we carried out two genetic experiments that suggested that α1,6 Man-exposed N-glycans likely function as a conserved glycan signal for a plant ERAD process. First, we showed that overexpression of EBS4 in ebs3-1 bri1-9 neutralized the suppressive effect of ebs3-1 on bri1-9 by promoting bri1-9 ERAD because overexpressed ALG12 in yeast could bypass the requirement of a complete B-branch for adding an α1,6 Man residue (Fig. 5A). SF-HPLC coupled with RP-HPLC analyses did detect the presence of a Dol-PP-Man7GlcNAc2 lacking the B- and C-branch terminal α1,2 Man residues in a gEBS4 ebs3-1 bri1-9 transgenic mutant (Fig. S4C). This unique α1,6 Man-exposing Man7GlcNAc2 glycan is likely transferred from its Dol-PP linker to bri1-9 in the gEBS4 ebs3-1 bri1-9 transgenic line because its electromobility on immunoblot is slightly reduced compared with that of bri1-9 in a transgenic ebs3-1 bri1-9 line carrying an empty vector (Fig. 5C). Second, we crossed a T-DNA insertional alg3-T2 mutation, which is known to create N-glycans carrying a different free α1,6 Man residue (Fig. 5A) (22, 23), to bri1-9 and discovered that the bri1-9 level in alg3-T2 bri1-9 is lower instead of higher than that in ebs3-1 bri1-9 and ebs4-1 bri1-9 despite the N-glycans on bri1-9 carrying fewer Man residues in alg3-T2 than that in the two ebs mutants. Taken together, these results suggested that the glycan signal to mark an ERAD client protein is likely conserved in plants. Given the recent discoveries showing that EFR is misfolded and degraded in the absence of UGGT, CRT3, and other ER proteins/chaperones (13), it will be interesting to determine whether ERAD of incompletely folded EFR also depends on such a conserved N-glycan signal.
Materials and Methods
Plant Materials and Growth Conditions.
Arabidopsis ecotype Col-0 is the parental line for mutants and transgenic plants except bri1-9 (Ws-2) for cloning EBS3 and bri1-5 (Ws-2) for genetic analysis. The T-DNA mutants of ALG3 (TAIR accession no. SALK_046061) and SDF2 (TAIR accession no. SALK_141321) were obtained from the Arabidopsis Biological Resource Center (ABRC) at Ohio State University. Methods for seed sterilization and conditions for plant growth were described previously (44).
Plasmid Construction and Generation of Transgenic Plants.
The p35S-EBS1 and the genomic constructs of EBS2 and EBS4 were described previously (18, 20). A 5-kb genomic fragment of At1g16900, including 1.5-kb promoter and 500-bp terminator sequences, was PCR-amplified from the BAC clone F17F16, cloned into pPZP212 (45) to make pPZP212-gEBS3, and sequenced to ensure no PCR error. Empty vectors and transgene constructs of EBS1, EBS2, EBS3, and EBS4 were individually transformed into bri1-9 or ebs3-1 bri1-9 mutants by the vacuum infiltration method (46).
Yeast Growth and Complementation Assay.
Standard growth medium and conditions were used to grow the wild-type yeast strain SS328, Δalg9 strain YG414, and Δalg9 wbp1-2 strain YG415 (21). The ORF of EBS3 was PCR-amplified from the At1g16900 cDNA clone U24181 to replace that of the yeast ALG9 from the pYEp352-yALG9 expression plasmid to create pYEp352-EBS3 using the strategy described previously (20). The Stratagene QuikChange XL Site-Directed Mutagenesis kit was used to generate pYEp352-ebs3-1 from pYEp352-EBS3 using the primers listed in Table S1. These plasmids were fully sequenced to ensure no PCR error and were individually transformed into YG414 or YG415 via a rapid transformation protocol (47).
Protein Extraction and Immunoblot Analysis.
Arabidopsis seedlings harvested from agar, soil, or liquid 1/2 Murashige and Skoog (MS) medium supplemented with or without BL (Chemiclones) or Kif (Toronto Research Chemicals) were ground in liquid N2, dissolved (50 mg seedlings/100 μL) in 2× SDS buffer [0.125 M Tris pH 6.8, 4% (wt/vol) SDS, 20% (vol/vol) glycerol, 0.2 M DTT, 0.02% (wt/vol) bromophenol blue] and boiled for 10 min. After centrifugation, supernatants were used for immunoblot analyses or incubated with or without 1,000 U of Endo Hf in 1× G5 buffer (New England Biolabs) for 1 h at 37 °C. To perform the CHX chase experiment, 3-wk-old seedlings were transferred from 1/2 MS agar plates into 1/2 MS medium containing 180 μM CHX (Sigma), and equal amounts of seedlings were removed at different time points to extract total proteins into 2× SDS sample buffer. After 10 min of boiling, equal amounts of total proteins, equivalent of 5 mg of seedlings, were separated by 7.5% SDS/PAGE and analyzed by immunoblot with anti-BRI1 antibody. To extract yeast proteins, cells of midlog phase grown in a 28 °C shaking incubator were collected by centrifugation, resuspended in 1× extraction buffer [0.3 M sorbitol, 0.1 M NaCl, 5 mM MgCl2, and 10 mM Tris (pH7.4)], lysed by violent vortexing with glass beads, mixed with equal volume of 2× SDS buffer, boiled for 10 min, and centrifuged to collect supernatants. Protein samples of plants or yeast cells were separated on 7% or 10% SDS/PAGE gel, transferred onto Immobilon-P membrane (Millipore), and analyzed by immunoblot with antibodies made against BRI1 (27), PDI (Rose Biotechnology), BiP (SPA-818; Stressgen), maize-CRTs (48), or a monoclonal anti-CPY antibody (10A5; Invitrogen).
Extraction and Analysis of LLOs.
The LLOs from bri1-9, bri1-9 ebs3-1, and gEBS4 ebs3-1 bri1-9 were extracted, hydrolyzed, pyridylaminated, and analyzed by SF-HPLC and RP-HPLC according to a previously reported protocol (23). The elution positions of PA-labeled oligosaccharides were compared with those of PA-sugar standards purchased from Masuda Chemical Industries.
Supplementary Material
Acknowledgments
We thank ABRC for supplying cDNA/BAC clones of At1g16900 and T-DNA insertional mutant of ALG3 (TAIR accession no. SALK_046061) and SDF2 (TAIR accession no. SALK_141321), F. Tax for seeds of bri1-9 (WS-2) and bri1-5, J. Chory for anti-BRI1 antibody, Y. Yin for anti-BES1 antiserum, R. Boston for anti-maize CRT antibody, A. Chang for anti-CPY antibody, M. Aebi for yeast strains and YEp352-yALG9 plasmid, and members of Li laboratory for stimulating discussion. This work was supported, in part, by National Natural Science Foundation of China Grant 31070246 (to Z.H.), National Institutes of Health Grant GM060519 (to J.L.), and Department of Energy Grant ER15672 (to J.L.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1119173109/-/DCSupplemental.
References
- 1.Abu-Qarn M, Eichler J, Sharon N. Not just for Eukarya anymore: Protein glycosylation in Bacteria and Archaea. Curr Opin Struct Biol. 2008;18:544–550. doi: 10.1016/j.sbi.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 2.Kelleher DJ, Gilmore R. An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology. 2006;16:47R–62R. doi: 10.1093/glycob/cwj066. [DOI] [PubMed] [Google Scholar]
- 3.Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- 4.Molinari M. N-glycan structure dictates extension of protein folding or onset of disposal. Nat Chem Biol. 2007;3:313–320. doi: 10.1038/nchembio880. [DOI] [PubMed] [Google Scholar]
- 5.Caramelo JJ, Parodi AJ. How sugars convey information on protein conformation in the endoplasmic reticulum. Semin Cell Dev Biol. 2007;18:732–742. doi: 10.1016/j.semcdb.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caramelo JJ, Parodi AJ. Getting in and out from calnexin/calreticulin cycles. J Biol Chem. 2008;283:10221–10225. doi: 10.1074/jbc.R700048200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lederkremer GZ, Glickman MH. A window of opportunity: Timing protein degradation by trimming of sugars and ubiquitins. Trends Biochem Sci. 2005;30:297–303. doi: 10.1016/j.tibs.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 8.Vembar SS, Brodsky JL. One step at a time: Endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clerc S, et al. Htm1 protein generates the N-glycan signal for glycoprotein degradation in the endoplasmic reticulum. J Cell Biol. 2009;184:159–172. doi: 10.1083/jcb.200809198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quan EM, et al. Defining the glycan destruction signal for endoplasmic reticulum-associated degradation. Mol Cell. 2008;32:870–877. doi: 10.1016/j.molcel.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ceriotti A, Roberts LM. Endoplasmic reticulum-associated protein degradation in plant cells. In: Robinson DG, editor. The Plant Endoplasmic Reticulum. Heidelberg: Springer; 2006. pp. 75–98. [Google Scholar]
- 12.Vitale A, Boston RS. Endoplasmic reticulum quality control and the unfolded protein response: Insights from plants. Traffic. 2008;9:1581–1588. doi: 10.1111/j.1600-0854.2008.00780.x. [DOI] [PubMed] [Google Scholar]
- 13.Saijo Y. ER quality control of immune receptors and regulators in plants. Cell Microbiol. 2010;12:716–724. doi: 10.1111/j.1462-5822.2010.01472.x. [DOI] [PubMed] [Google Scholar]
- 14.Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 15.Kinoshita T, et al. Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature. 2005;433:167–171. doi: 10.1038/nature03227. [DOI] [PubMed] [Google Scholar]
- 16.Zipfel C, et al. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125:749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 17.Jin H, Yan Z, Nam KH, Li J. Allele-specific suppression of a defective brassinosteroid receptor reveals a physiological role of UGGT in ER quality control. Mol Cell. 2007;26:821–830. doi: 10.1016/j.molcel.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin H, Hong Z, Su W, Li J. A plant-specific calreticulin is a key retention factor for a defective brassinosteroid receptor in the endoplasmic reticulum. Proc Natl Acad Sci USA. 2009;106:13612–13617. doi: 10.1073/pnas.0906144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong Z, Jin H, Tzfira T, Li J. Multiple mechanism-mediated retention of a defective brassinosteroid receptor in the endoplasmic reticulum of Arabidopsis. Plant Cell. 2008;20:3418–3429. doi: 10.1105/tpc.108.061879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong Z, et al. Mutations of an alpha1,6 mannosyltransferase inhibit endoplasmic reticulum-associated degradation of defective brassinosteroid receptors in Arabidopsis. Plant Cell. 2009;21:3792–3802. doi: 10.1105/tpc.109.070284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burda P, et al. Stepwise assembly of the lipid-linked oligosaccharide in the endoplasmic reticulum of Saccharomyces cerevisiae: Identification of the ALG9 gene encoding a putative mannosyl transferase. Proc Natl Acad Sci USA. 1996;93:7160–7165. doi: 10.1073/pnas.93.14.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henquet M, et al. Identification of the gene encoding the alpha1,3-mannosyltransferase (ALG3) in Arabidopsis and characterization of downstream n-glycan processing. Plant Cell. 2008;20:1652–1664. doi: 10.1105/tpc.108.060731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kajiura H, Seki T, Fujiyama K. Arabidopsis thaliana ALG3 mutant synthesizes immature oligosaccharides in the ER and accumulates unique N-glycans. Glycobiology. 2010;20:736–751. doi: 10.1093/glycob/cwq028. [DOI] [PubMed] [Google Scholar]
- 24.Su W, Liu Y, Xia Y, Hong Z, Li J. Conserved endoplasmic reticulum-associated degradation system to eliminate mutated receptor-like kinases in Arabidopsis. Proc Natl Acad Sci USA. 2011;108:870–875. doi: 10.1073/pnas.1013251108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maley F, Trimble RB, Tarentino AL, Plummer TH., Jr Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal Biochem. 1989;180:195–204. doi: 10.1016/0003-2697(89)90115-2. [DOI] [PubMed] [Google Scholar]
- 26.Clouse SD, Langford M, McMorris TC. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111:671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mora-García S, et al. Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 2004;18:448–460. doi: 10.1101/gad.1174204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Houston NL, et al. Phylogenetic analyses identify 10 classes of the protein disulfide isomerase family in plants, including single-domain protein disulfide isomerase-related proteins. Plant Physiol. 2005;137:762–778. doi: 10.1104/pp.104.056507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sung DY, Vierling E, Guy CL. Comprehensive expression profile analysis of the Arabidopsis Hsp70 gene family. Plant Physiol. 2001;126:789–800. doi: 10.1104/pp.126.2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Persson S, et al. Phylogenetic analyses and expression studies reveal two distinct groups of calreticulin isoforms in higher plants. Plant Physiol. 2003;133:1385–1396. doi: 10.1104/pp.103.024943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frank CG, Aebi M. ALG9 mannosyltransferase is involved in two different steps of lipid-linked oligosaccharide biosynthesis. Glycobiology. 2005;15:1156–1163. doi: 10.1093/glycob/cwj002. [DOI] [PubMed] [Google Scholar]
- 32.Frank CG, et al. Identification and functional analysis of a defect in the human ALG9 gene: Definition of congenital disorder of glycosylation type IL. Am J Hum Genet. 2004;75:146–150. doi: 10.1086/422367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jakob CA, Burda P, Roth J, Aebi M. Degradation of misfolded endoplasmic reticulum glycoproteins in Saccharomyces cerevisiae is determined by a specific oligosaccharide structure. J Cell Biol. 1998;142:1223–1233. doi: 10.1083/jcb.142.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weinstein M, et al. CDG-IL: An infant with a novel mutation in the ALG9 gene and additional phenotypic features. Am J Med Genet A. 2005;136:194–197. doi: 10.1002/ajmg.a.30851. [DOI] [PubMed] [Google Scholar]
- 35.Oriol R, Martinez-Duncker I, Chantret I, Mollicone R, Codogno P. Common origin and evolution of glycosyltransferases using Dol-P-monosaccharides as donor substrate. Mol Biol Evol. 2002;19:1451–1463. doi: 10.1093/oxfordjournals.molbev.a004208. [DOI] [PubMed] [Google Scholar]
- 36.Kostova Z, Wolf DH. Importance of carbohydrate positioning in the recognition of mutated CPY for ER-associated degradation. J Cell Sci. 2005;118:1485–1492. doi: 10.1242/jcs.01740. [DOI] [PubMed] [Google Scholar]
- 37.Kanehara K, Kawaguchi S, Ng DT. The EDEM and Yos9p families of lectin-like ERAD factors. Semin Cell Dev Biol. 2007;18:743–750. doi: 10.1016/j.semcdb.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 38.Burda P, Jakob CA, Beinhauer J, Hegemann JH, Aebi M. Ordered assembly of the asymmetrically branched lipid-linked oligosaccharide in the endoplasmic reticulum is ensured by the substrate specificity of the individual glycosyltransferases. Glycobiology. 1999;9:617–625. doi: 10.1093/glycob/9.6.617. [DOI] [PubMed] [Google Scholar]
- 39.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 40.Nekrasov V, et al. Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. EMBO J. 2009;28:3428–3438. doi: 10.1038/emboj.2009.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schott A, et al. Arabidopsis stromal-derived Factor2 (SDF2) is a crucial target of the unfolded protein response in the endoplasmic reticulum. J Biol Chem. 2010;285:18113–18121. doi: 10.1074/jbc.M110.117176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tokunaga F, Brostrom C, Koide T, Arvan P. Endoplasmic reticulum (ER)-associated degradation of misfolded N-linked glycoproteins is suppressed upon inhibition of ER mannosidase I. J Biol Chem. 2000;275:40757–40764. doi: 10.1074/jbc.M001073200. [DOI] [PubMed] [Google Scholar]
- 43.Cipollo JF, Trimble RB. The accumulation of Man(6)GlcNAc(2)-PP-dolichol in the Saccharomyces cerevisiae Deltaalg9 mutant reveals a regulatory role for the Alg3p alpha1,3-Man middle-arm addition in downstream oligosaccharide-lipid and glycoprotein glycan processing. J Biol Chem. 2000;275:4267–4277. doi: 10.1074/jbc.275.6.4267. [DOI] [PubMed] [Google Scholar]
- 44.Li J, Nam KH, Vafeados D, Chory J. BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 2001;127:14–22. doi: 10.1104/pp.127.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hajdukiewicz P, Svab Z, Maliga P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- 46.Bechtold N, Pelletier G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol. 1998;82:259–266. doi: 10.1385/0-89603-391-0:259. [DOI] [PubMed] [Google Scholar]
- 47.Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 48.Pagny S, et al. Protein recycling from the Golgi apparatus to the endoplasmic reticulum in plants and its minor contribution to calreticulin retention. Plant Cell. 2000;12:739–756. doi: 10.1105/tpc.12.5.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.