Abstract
Activation of p53 upon DNA damage induces an array of target genes, leading to cell cycle arrest and/or apoptosis. However, the mechanism by which the cell fate is controlled by p53 remains to be clarified. Previously, we showed that DEC1, a basic helix–loop–helix transcription factor and a target of p53, is capable of inducing cell cycle arrest and mediating DNA damage-induced premature senescence. Here, we found that ectopic expression of DEC1 inhibits, whereas knockdown of DEC1 enhances, DNA damage-induced cell death. Surprisingly, we showed that the anti–cell-death activity of DEC1 is p53 dependent, but DEC1 does not directly modulate p53 expression. Instead, we showed that DEC1 inhibits the ability of p53 to induce macrophage inhibitory cytokine-1 (MIC-1), but not other prosurvival/proapoptotic targets, including p21 and Puma. Importantly, we showed that upon binding to their respective response elements on the MIC-1 promoter, DEC1 and p53 physically interact on the MIC-1 promoter via the basic helix–loop–helix domain in DEC1 and the tetramerization domain in p53, which likely weakens the DNA-binding activity of p53 to the MIC-1 promoter. Finally, we found that depletion of MIC-1 abrogates the ability of DEC1 to attenuate DNA damage-induced cell death. Together, we hypothesize that DEC1 controls the response of p53-dependent cell survival vs. cell death to a stress signal through MIC-1.
Keywords: Stimulated by retinoic acid 13, growth differentiation factor 15
In response to genotoxic stress, p53 is activated, leading to induction of cell death regulators, such as Puma, Bax, FDXR, and PolH, and cell survival regulators, such as p21, Mdm2, 14-3-3σ, and GADD45 (1–3). However, a central puzzle is how p53 determines the cell fate in response to a specific stimulus. Previously, we and others have shown that the expression level and functional domains of p53 are involved in the cell fate determination (4–7) as well as several cofactors that alter the ability of p53 to transcriptionally regulate its targets. For examples, the ASPP family proteins (8), p53β (9), and p90 (10) preferentially affect p53 to regulate proapoptotic targets, such as Bax, but not p21. Conversely, BRCA1 (11) and HZF (12) cooperate with p53 to transactivate p21 but not Bax. Interestingly, hCAS/CSE1L is able to selectively affect a set of p53 targets and p53-dependent apoptosis without directly binding to p53 (13).
DEC1 belongs to a subfamily of basic helix–loop–helix (bHLH) transcription factors and is a critical regulator of the circadian rhythm (14). DEC1 is defined as a senescence marker because it is up-regulated in premalignant tumors (15). Consistent with this, our previous studies showed that DEC1 is a target of the p53 family and mediates G1 cell cycle arrest and DNA damage-induced premature senescence (16, 17). Evidence also showed that DEC1 plays a role in cell death. For examples, overexpression of DEC1 inhibits serum starvation-induced apoptosis in HEK293 cells (18), and DEC1 mediates TGF-β–induced cell survival in breast cancer cells (19). However, other studies showed that loss of DEC1 leads to defective apoptosis of activated lymphocytes and confers thymocytes resistant to γ-irradiation–induced apoptosis (20). Thus, it is likely that DEC1 has both prosurvival and proapoptotic activities in cell and tissue context-dependent manners.
Macrophage inhibitory cytokine-1 (MIC-1), a divergent member of the TGF-β superfamily (21), is identified to play a role in many cellular responses and thus also called growth differentiation factor 15 (GDF15) (22), nonsteroidal anti-inflammatory drug-activated gene 1 (NAG1) (23), prostate-derived factor (PDF) (24), placental TGF-β (PTGFB) (25), and placental bone morphogenetic protein (PLAB) (26). MIC-1 is directly regulated by p53 and contributes to the response of cancer cells to chemotherapeutic agents by suppressing cell growth and/or promoting apoptosis (25, 27).
In this study, we explored how DEC1 modulates p53-dependent DNA damage response. We provided evidence that DEC1 inhibits DNA damage-induced cell death by attenuating p53 induction of MIC-1. This finding suggests that in response to DNA damage, p53 is activated and then induces DEC1, which in turn modulates the ability of p53 to regulate MIC-1, a proapoptotic target. Thus, DEC1-p53-MIC-1 constitutes a unique feedback loop to control the response of p53-dependent cell survival vs. cell death upon genotoxic stress.
Results
DEC1 Attenuates DNA Damage-Induced Cell Death.
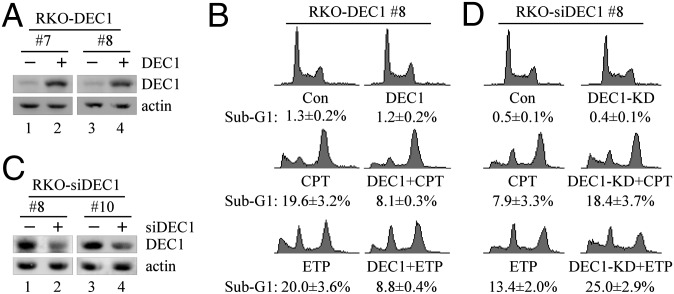
To examine whether DEC1 modulates DNA damage-induced cell death, colorectal carcinoma (RKO) cells were uninduced or induced to express DEC1, followed by treatment with camptothecin (CPT) and etoposide (ETP). As shown in Fig. 1A, upon addition of doxycycline (an analog of tetracycline), an equivalent level of DEC1 was expressed in two individual clones (7 and 8). Next, DNA histogram analysis was performed to measure the percentage of sub-G1 population using clone 8. We found that ectopic expression of DEC1 had no effect (Fig. 1B, Top; 1.3% in control vs. 1.2% in DEC1-producing cells), whereas upon treatment with CPT and ETP, the sub-G1 population was remarkably increased by CPT (1.3% in control vs. 19.6% in CPT treated) and ETP (1.3% in control vs. 20% in ETP treated). CPT is an inhibitor of topoisomerase I, whereas ETP is an inhibitor of topoisomerase II, both of which can induce double-strand DNA break. However, upon DEC1 induction, the percentage of sub-G1 population was markedly decreased to 8.1% by CPT and to 8.8% by ETP (Fig. 1B). Similar results were observed in clone 7 (Fig. S1A). In addition, short-term cell proliferation assay showed that upon DEC1 induction in MCF7 cells (Fig. S1B, compare lanes 1 and 3 with 2 and 4, respectively), the sensitivity of MCF7 cells to CPT was markedly decreased (Fig. S1C).
Fig. 1.
DEC1 inhibits DNA damage-induced cell death. (A) Western blots were prepared with extracts from RKO cells uninduced (−) or induced (+) to express DEC1 for 24 h. DEC1 and actin were detected by their respective antibodies. (B) DNA histogram analysis was performed with RKO cells uninduced or induced to express DEC1 for 12 h along with mock treatment or treatment with CPT (250 nM) or ETP (20 μg/mL) for 48 h. The assay is described in SI Materials and Methods (mean ± SD; n = 3). (C) Western blots were prepared with extracts from RKO cells uninduced (−) or induced (+) to express DEC1 shRNA-1 (RKO-siDEC1#8) or -2 (RKO-siDEC1#10) for 72 h along with treatment with CPT (250 nM) for 18 h. (D) DNA histogram analysis was performed with RKO cells uninduced or induced to express DEC1 shRNA for 72 h along with mock treatment or treatment with CPT (500 nM) or ETP (40 μg/mL) for 30 h.
To examine whether endogenous DEC1 is involved in DNA damage-induced cell death, RKO cells were uninduced or induced to knock down DEC1, followed by treatment with CPT and ETP. As showed in Fig. 1C, upon addition of doxycycline, the level of endogenous DEC1 was knocked down in two individual clones (8 and 10). Next, DNA histogram analysis showed that DEC1 knockdown (KD) alone had no effect on cell death (Fig. 1D, Top; 0.5% in control vs. 0.4% in DEC1-KD cells), whereas upon treatment with CPT and ETP, the sub-G1 population was increased by CPT (0.5% in control vs. 7.9% in CPT treated) and ETP (0.5% in control vs. 13.4% in ETP treated). However, upon DEC1-KD, the percentage of sub-G1 population was further increased to 18.4% by CPT and to 25% by ETP (Fig. 1D). Similar results were observed with clone 10 upon CPT treatment (Fig. S1D). In addition, colony formation assay showed that upon DEC1-KD in MCF7 cells (Fig. S1E, compare lanes 1 and 3 with 2 and 4, respectively), the sensitivity of MCF7 cells to CPT was markedly increased, whereas doxycycline had no effect (Fig. S1 F–H).
Because both p53 and DEC1 are responsive to multiple extracellular stimuli, we examined whether DEC1 inhibits cell death induced by other stress signals. Interestingly, we found that DEC1 was induced by actinomycin D (ActD, 5 nM; Fig. S2A) and hypoxia (Fig. S2C). Unlike CPT and ETP, 5 nM of ActD initiated ribosomal stress without altering histone γH2AX phosphorylation (Fig. S2A), consistent with a previous report (28). We also found that DEC1-KD enhanced ribosomal stress- and hypoxia-induced cell death (Fig. S2 B and D). Together, these data indicate that DEC1 is capable of suppressing cell death in response to several types of stress signals.
Antiapoptotic Function of DEC1 Is p53-Dependent.
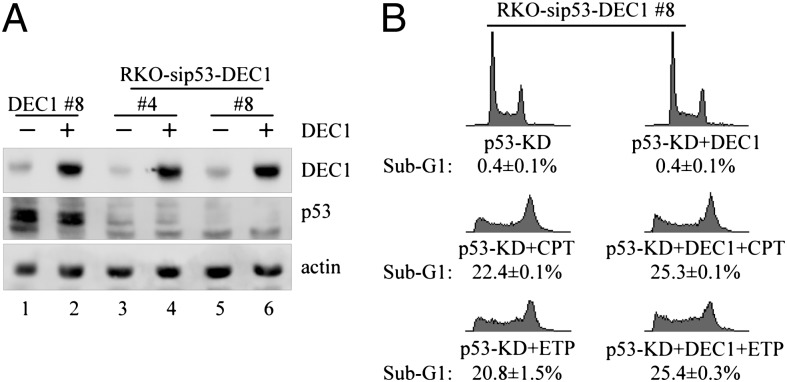
To examine whether p53 is involved in the anti–cell-death activity of DEC1 upon DNA damage, we generated RKO cell lines in which p53 is stably knocked down and DEC1 is inducibly expressed. Western blot analysis showed that a comparable level of DEC1 protein was induced, whereas p53 was undetectable, in clones 4 and 8 compared with that in DEC1-expressing RKO cells (clone 8; Fig. 2A). Interestingly, we showed that ectopic expression of DEC1 did not inhibit DNA damage-induced apoptosis in p53-deficient cells (Fig. 2B and Fig. S3A). To confirm this, we generated MCF7 cell lines in which p53 is stably knocked down and DEC1 is inducibly expressed (Fig. S3B). Similarly, we found that ectopic expression of DEC1 had little if any effect on cell survival upon CPT treatment in p53-deficient cells (Fig. S3C). Thus, loss of p53 abrogates the effect of DEC1 on DNA damage-induced apoptosis.
Fig. 2.
The anti–cell-death activity of DEC1 is p53 dependent. (A) Western blots were prepared with extracts from p53-WT and p53-KD RKO cells uninduced (−) or induced (+) to express DEC1 for 24 h. p53, DEC1, and actin were detected by their respective antibodies. (B) DNA histogram analysis was performed with p53-KD RKO cells uninduced or induced to express DEC1 for 12 h along with mock treatment or treatment with CPT (250 nM) or ETP (20 μg/mL) for 48 h.
DEC1 regulates expression of several genes involved in cell cycle control, including DEC2 (29), ID1 (17), and ΔNp63 (30). However, we found that DEC1 had no obvious effect on the level of p53 protein in RKO cells regardless of DNA damage treatment (Fig. S3D). Consistently, we found that the level of p53 mRNA was not altered in MCF7 cells upon DEC1 induction (Fig. S3E).
DEC1 Inhibits p53-Dependent Expression of MIC-1.
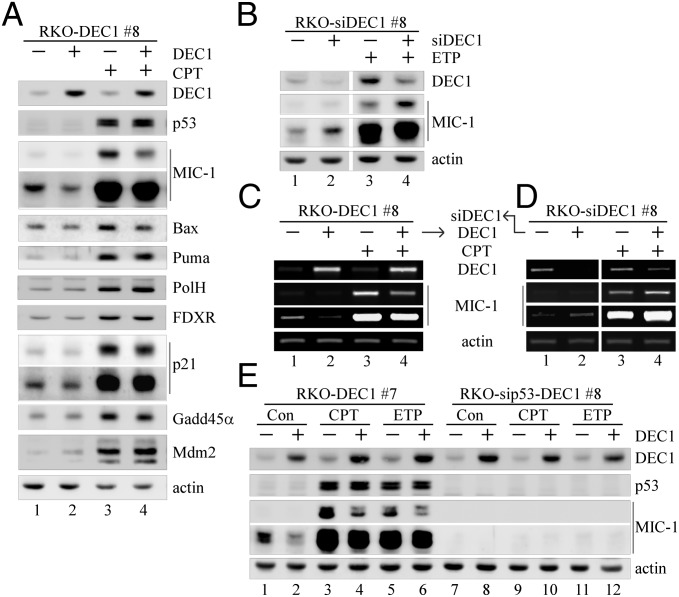
p53 activity in tumor suppression is mediated by its target genes. Thus, it is possible that DEC1 controls the response of a cell to p53-dependent cell survival vs. cell death by altering the expression pattern of p53 target genes. To test this, we examined p53 target genes involved in cell survival and death, including five proapoptotic target genes (MIC-1, Puma, Bax, PolH, and FDXR), two cell-cycle regulators (p21 and GADD45α), and Mdm2. We showed that in RKO cells, DEC1 induction markedly inhibited both the basal and DNA damage-induced expression of MIC-1 and to a much lesser extent Bax, but not Puma, PolH, FDXR, GADD45α, p21, and Mdm2 (Fig. 3A). This observation was confirmed in another RKO cell clone, 7 (Fig. S4A), and two MCF7 cell clones (6 and 16; Fig. S4 B and C), all of which can inducibly express DEC1 under the control of a tetracycline-regulated promoter (16). Conversely, we showed that upon DEC1-KD, both the basal and DNA damage-induced levels of MIC-1 were markedly increased in RKO and MCF7 cells (Fig. 3B, compare lanes 1 and 3 with 2 and 4, respectively; Fig. S4D).
Fig. 3.
MIC-1 expression is repressed by DEC1. (A) Western blots were prepared with extracts from RKO cells uninduced (−) or induced (+) to express DEC1 for 12 h along with mock treatment or treatment with CPT (250 nM) for 18 h. DEC1, p53, MIC-1, Bax, Puma, PolH, FDXR, p21, GADD45α, Mdm2, and actin were detected by their respective antibodies. (B) Western blots were prepared with extracts from RKO cells uninduced or induced to express DEC1 shRNA for 72 h along with mock treatment or treatment with ETP (5 μg/mL) for 9 h. (C) The level of transcripts for DEC1, MIC-1, and actin was measured by RT-PCR with RNAs purified from RKO cells uninduced or induced to express DEC1 for 12 h along with mock treatment or treatment with CPT (250 nM) for 6 h. (D) The experiment was performed as in C except that RKO cells were uninduced or induced to knock down DEC1 for 72 h. (E) Western blots were prepared with extracts from p53-WT and p53-KD RKO cells uninduced or induced to express DEC1 for 12 h along with mock treatment or treatment with CPT (250 nM) or ETP (5 μg/mL) for 12 h.
As a transcription repressor, DEC1 may transcriptionally repress MIC-1 expression. To test this, the level of MIC-1 transcript was measured in the presence or absence of DEC1. We found that upon DEC1 induction, both the basal and DNA damage-induced levels of MIC-1 transcript in RKO cells were reduced (Fig. 3C). Consistent with this, Northern blot analysis showed that the level of MIC-1 transcript was decreased by DEC1 in two individual MCF7 clones (6 and 16; Fig. S4E). Conversely, we found that upon DEC1-KD, both the basal and DNA damage-induced levels of MIC-1 transcripts were increased in RKO cells (Fig. 3D, compare lanes 1 and 3 with lanes 2 and 4, respectively).
To test whether p53 plays a role in DEC1 repression of MIC-1, we examined MIC-1 expression in RKO and MCF7 cells in that DEC1 can be inducibly expressed along with stable knockdown of p53. We showed that upon p53-KD, the level of MIC-1 was low or undetectable regardless of treatment with CPT and ETP (Fig. 3E and Fig. S4F, lanes 7, 9, and 11), which suggests that MIC-1 expression is primarily controlled by p53. As a result, ectopic expression of DEC1 had little discernible effect (Fig. 3E and Fig. S4F, lanes 7–12). As a control, the level of MIC-1 protein in p53-competent cells was substantially induced upon treatment with CPT and ETP, and that both the basal and DNA damage-induced levels of MIC-1 were markedly decreased by DEC1 (Fig. 3E and Fig. S4F, compare lanes 1, 3, and 5 with 2, 4, and 6, respectively).
Interaction of DEC1 and p53 on the MIC-1 Promoter Weakens the DNA-Binding Activity of p53, Leading to Decreased Expression of MIC-1.
To explore the mechanism by which DEC1 regulates p53 activity, we examined whether DEC1 physically interacts with p53. To test this, endogenous DEC1 and p53 were immunoprecipitated by anti-DEC1 and anti-p53, respectively. Western blot analysis showed that endogenous p53 was detected in the DEC1 immunocomplex (Fig. S5A, Left). Previously, we showed that DEC1 and histone deacetylase 1 (HDAC1) physically interact (30). Indeed, HDAC1 was also detected in this complex (Fig. S5A, Left). Conversely, endogenous DEC1 was detected in the p53 immunocomplex (Fig. S5A, Right). In addition, we showed that ectopically expressed HA-tagged p53 was found to interact with ectopically expressed Flag-tagged DEC1 (Fig. S5B). Next, we mapped the domain in DEC1 and p53 necessary for their interaction. To test this, five DEC1 mutants and four p53 mutants were generated (Fig. S5 C and E) and coexpressed in cells (Fig. S5 D and F). We found that the HLH domain (amino acids 66–109) in DEC1 was required because p53 interacted with DEC1(1–412) and DEC1(1–309), but not DEC1(110–412), DEC1(Δ66–109), DEC1(Δ53–65), and DEC1-R58P (Fig. S5D). Similarly, we found that the tetramerization domain (TD; amino acids 325–356) in p53 is required because DEC1 interacted with p53(1–393) and p53(94–393), but not p53(1–324) and p53(Δ325–356; Fig. S5F).
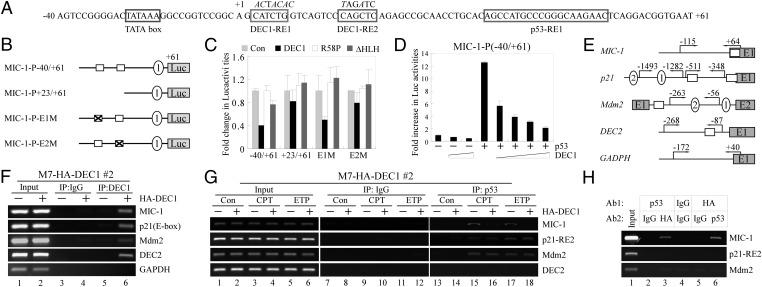
Previous reports (25, 27) and our own study (Fig. 3) suggest that p53 is the primary regulator of MIC-1 transcription at both the basal and stress conditions. Indeed, three p53-response elements (REs) along with four Sp1-REs have been identified in the MIC-1 promoter (23, 25, 27) (Fig. S6A). Because DEC1 is capable of repressing gene expression through class B E-boxes (29), we searched the MIC-1 promoter and found one potential E-box adjacent to the p53-RE3 (Fig. S6A). Thus, a luciferase reporter under the control of the MIC-1 promoter (nt −976 to +61), which contains three p53-REs, one E-box, and Sp1-REs, was generated and designated as MIC-1-P-976/+61 (Fig. S6A). We also generated four other luciferase reporters under the control of the MIC-1 promoter, which lacks the E-Box, p53-RE3 (MIC-1-P-585/+61), or a combination of the E-box, p53-REs, and Sp1-REs (Fig. S6A). We showed that p53, but not mutant (R249S and ΔTD), increased the luciferase activity for each of the luciferase reporters as long as the p53-RE1 is present (Fig. S6B). Thus, to test whether p53-RE1 is sufficient and necessary for the MIC-1 promoter to be regulated by p53, we generated two luciferase reporters under the control of the MIC-1 promoter in which p53-RE1 was disrupted (Fig. S6C). We found that in the absence of p53-RE1, the MIC-1 promoter with p53-RE2/-RE3 (MIC-1-P-976/+41) was weakly responsive to p53, whereas the one with only p53-RE2 (MIC-1-P-585/+41) was inert (Fig. S6D). This observation suggests that p53 regulates the MIC-1 promoter primarily via p53-RE1. Surprisingly, we showed that the MIC-1 promoter was suppressed by DEC1 but not mutant DEC1 (R58P and ΔHLH) regardless of the presence of the E-Box, p53-REs, and Sp1 sites (Fig. S6E). The suppression of the DEC2 promoter by DEC1 was measured as a control (Fig. S6E). Therefore, to identify DEC1-REs in the MIC-1 promoter, we further searched the MIC-1 promoter (nt −40 to +61) and found two imperfect E-boxes, designated as DEC1-RE1/-RE2, which are located downstream of the MIC-1 transcription start site (Fig. 4A). It should be noted that p53-RE1 is also located downstream of the MIC-1 transcription start site (Fig. 4A). To test this, we generated four luciferase reporters under the control of the MIC-1 promoter, which carries both or neither DEC1-RE1/-RE2 (MIC-1-P-40/+61; MIC-1-P+23/+61), or a mutation in one of the DEC1-REs (MIC-1-P-E1M; MIC-1-P-E2M; Fig. 4B). We found that DEC1 had no effect on the MIC-1 promoter that does not carry DEC1-RE2 (CAGCTC element; Fig. 4C). Consistent with above observations, p53 activates the MIC-1 promoter regardless of the presence of DEC1-RE1/-RE2 (Fig. S6F). Nevertheless, we emphasize that in the absence of p53, the basal level of MIC-1 is extremely low (Fig. 3E and Fig. S4F), and as a result, the significance of DEC1 inhibition of MIC-1 expression in the absence of p53 would be limited. Thus, we wanted to investigate whether DEC1 is capable of suppressing the ability of p53 to activate the MIC-1 promoter. Indeed, we found that the ability of p53 to activate the MIC-1 promoter was markedly inhibited by DEC1 in a dose-dependent manner (Fig. 4D).
Fig. 4.
DEC1 inhibits p53 to transactivate the MIC-1 promoter. (A) Sequence of the MIC-1 promoter (nt −40 to +61) with the locations of TATA box, DEC1-RE1/RE2 (with mutations shown in italics), and p53-RE1. (B) Schematic presentation of luciferase constructs under the control of the MIC-1 promoter with intact, truncated, or mutated E-boxes. (C) The luciferase activity was measured in the presence of WT or mutant DEC1. (D) The luciferase assay was measured in the presence of p53 along with an increasing dose of DEC1. (E) Schematic presentation of the MIC-1, p21, Mdm2, DEC2, and GAPDH promoters with the locations of primers used for ChIP assays. (F) MCF7 cells uninduced (−) or induced (+) to express HA-tagged DEC1 for 18 h were cross-linked with formaldehyde followed by sonication. Chromatin was immunoprecipitated (IP) with anti-HA to precipitate HA-DEC1, or a control IgG. The binding of DEC1 to target promoters was quantified by PCR. (G) The experiment was performed as in F except that anti-p53 was used to precipitate p53–DNA complexes, and MCF7 cells were mock treated or treated with CPT (250 nM) or ETP (5 μg/mL) for 12 h. (H) MCF7 cells were induced to express HA-DEC1 along with treatment with CPT (250 nM) for 16 h. The first ChIP performed with anti-p53 was re-ChIPed with anti-HA and control IgG. Conversely, the first ChIP performed with anti-HA was re-ChIPed with anti-p53 and control IgG. The first ChIP performed with control IgG was only re-ChIPed with control IgG.
Next, we measured whether DEC1 directly binds to the MIC-1 and other p53 target gene promoters (Fig. 4E). To test this, a ChIP assay was performed with DEC1-producing MCF7 cells. As shown in Fig. S6G, HA-DEC1 was expressed in MCF7 cells upon induction with doxycycline. DEC1 binding to the DEC2 promoter was measured as a positive control. We found that DEC1 bound to the MIC-1, p21, Mdm2, and DEC2 promoters (Fig. 4F), which is not surprising because several putative E-boxes are located on the p21 promoter and an E-box is located on the Mdm2 promoter (Table S1). These observations lead us to examine whether DEC1 modulates the ability of p53 to recognize these promoters. As previously reported (25, 31), we found that endogenous p53 in MCF7 cells bound to p53-REs on the MIC-1, p21, and Mdm2 promoters upon treatment with CPT and ETP (Fig. 4G, compare lanes 15 and 17 with lane 13). Interestingly, upon DEC1 induction, the extent of p53 bound to the MIC-1 promoter was markedly inhibited (Fig. 4G, compare lanes 16 and 18 with lanes 15 and 17, respectively). However, DEC1 had no effect on the extent of p53 bound to the p21 and Mdm2 promoters (Fig. 4G, compare lanes 16 and 18 with lanes 15 and 17, respectively). p53 binding to the DEC2 promoter was measured as a negative control (Fig. 4G).
To further investigate why DEC1 selectively inhibits MIC-1 but not p21 and Mdm2, we analyzed the distance between DEC1-RE and p53-RE on these promoters. We found that the distance between the DEC1-RE and p53-RE is only 17 bp on the MIC-1 promoter, but more than 280 bp on other promoters (Table S1). Therefore, we hypothesize that both DEC1 and p53 have to bind to the same promoter in close proximity, which then facilitates DEC1-p53 interaction on the MIC-1 promoter. To test this, a ChIP-reChIP assay was performed and showed that DEC1 and p53 were present on the same chromatin fragment on the MIC-1 but not p21 and Mdm2 promoters (Fig. 4H).
As shown above, Bax expression was also inhibited by DEC1 (Fig. 3A). Surprisingly, we found that DEC1 bound to the Bax promoter (Fig. S7A), but had no effect on p53 binding to the Bax promoter (Fig. S7B); this suggests that DEC1 inhibits Bax expression through another mechanism. Previous reports showed that the Bax promoter contains four E-boxes downstream of four p53-half sites (32) and a p53-RE in intron 1 (33) (Fig. S7A). To test whether Bax is transcriptionally inhibited by DEC1, we generated two luciferase reporters under the control of the Bax promoter: Bax-P-433/+388, which contains p53 half-sites, E-boxes, and the p53-RE; and Bax-P-433/+8, which lacks the p53-RE (Fig. S7C). We showed that the intronic p53-RE was responsive to p53 (Fig. S7D). In addition, we showed that DEC1 inhibited the Bax promoter regardless of the p53-RE (Fig. S7E).
MIC-1 Is a Downstream Effector of DEC1 to Modulate DNA Damage-Induced Cell Death.
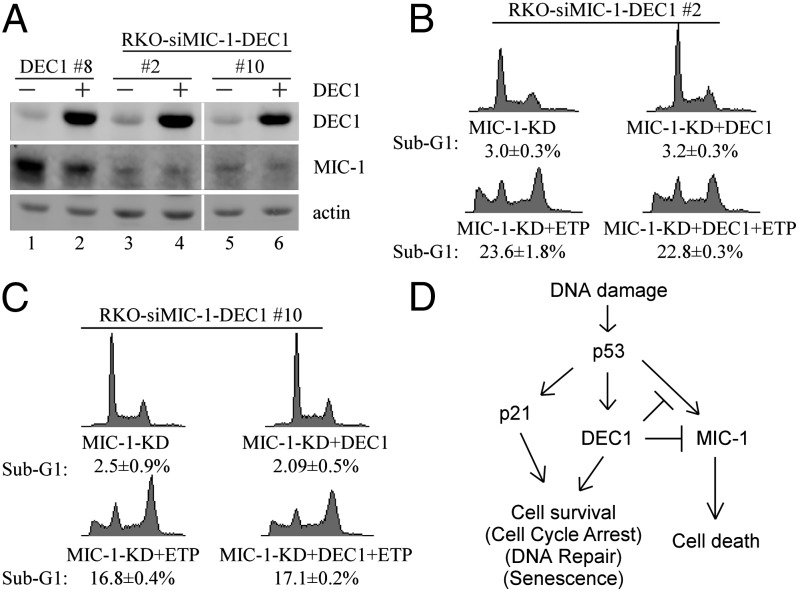
To determine the role of MIC-1 in the antiapoptotic activity of DEC1, we generated multiple RKO and MCF7 cell lines in which MIC-1 is stably knocked down and DEC1 is inducibly expressed. DEC1-expressing RKO clone 8 and MCF7 clone 6 were used as controls. Western blot analysis showed that comparable levels of DEC1 protein were expressed, whereas the level of MIC-1 protein was low in MIC-1-KD RKO cell clones 2 and 10 (Fig. 5A) and MCF7 cell clones 5 and 24 (Fig. S8A). We showed that upon knockdown of MIC-1, ectopic expression of DEC1 in RKO cells had no effect on DNA damage-induced apoptosis (Fig. 5 B and C), which is different from the above observation that ectopic expression of DEC1 prevented RKO cells from undergoing DNA damage-induced apoptosis (Fig. 1B and Fig. S1A). We also showed that upon knockdown of MIC-1, ectopic expression of DEC1 in MCF7 cells had no effect on DNA damage-induced growth suppression (Fig. S8B), which is different from the observation that ectopic expression of DEC1 promoted cell survival in MCF7 cells upon DNA damage (Fig. S1C). These results suggest that MIC-1 is a downstream effector of DEC1 in suppressing DNA damage-induced cell death in a p53-dependent manner.
Fig. 5.
MIC-1 is required for DEC1 to inhibit DNA damage-induced cell death. (A) Western blots were prepared with extracts from MIC-1-WT or MIC-1-KD RKO cells uninduced (−) or induced (+) to express DEC1 for 24 h. (B and C) DNA histogram analysis was performed with MIC-1-KD RKO cells uninduced or induced to express DEC1 for 12 h along with mock treatment or treatment with ETP (20 μg/mL) for 48 h. (D) A model for the role of DEC1 in the p53 pathway in response to DNA damage.
Discussion
We previously showed that DEC1, a target of p53, mediates G1 arrest and premature senescence in p53- and p21-independent manners (16). In this study, we showed that DEC1 inhibits DNA damage-induced apoptosis in a p53-dependent manner. Interestingly, we showed that DEC1 attenuates p53 induction of MIC-1, but not other apoptotic target genes (Puma, FDXR, and PolH), p21, GADD45α, and Mdm2. Moreover, we showed that similar to p53-KD (Fig. 2B and Fig. S3 A–C), depletion of MIC-1 abrogates the ability of DEC1 to suppress DNA damage-induced cell death (Fig. 5 and Fig. S8). It is well established that in response to DNA damage, p53 is activated and then induces an array of target genes, including DEC1 for inducing cell cycle arrest and senescence, and MIC-1 for inducing apoptosis. However, a number of known and unknown factors, including the cellular context and the type of a stress signal, would dictate the response of a cell to choose p53-dependent cell survival vs. cell death. Here, we hypothesize that DEC1 suppresses p53 induction of MIC-1 and thus controls the response of a cell to p53-dependent cell survival vs. cell death to a stress signal (Fig. 5D).
p53 regulates its targets by binding to p53-REs as a tetramer. An intact p53 TD is critical for efficient DNA binding (34), protein–protein interaction (35), and posttranslational modifications (36). Here, we showed that DEC1 physically associates with p53 via the TD (Fig. S5) and attenuates p53 binding to its target MIC-1 promoter (Fig. 4). In addition, transactivation of the MIC-1 promoter by p53 is diminished by DEC1. Importantly, we showed that in order for DEC1 to suppress p53 activation of the MIC-1 promoter, both DEC1 and p53 have to bind to the same promoter and interact with each other on the promoter (Fig. 4). However, although both DEC1 and p53 bind to p21 and Mdm2 promoters, DEC1 is unable to inhibit the ability of p53 to bind to p53-REs on these promoters, probably due to lack of interaction between DEC1 and p53 on the promoters (Fig. 4), and, consequently, DEC1 is unable to suppress p53 induction of p21 and Mdm2 (Fig. 3 and Fig. S4). We note that DEC1 slightly inhibits p53 induction of Bax expression (Fig. 3A) but does not inhibit the ability of p53 to bind to the Bax promoter (Fig. S7B). In addition, DEC1 potentially inhibits Bax expression regardless of p53 (Fig. S7E), which suggests that p53 is capable of inducing apoptosis in the absence of Bax, consistent with published studies (37, 38). Together, our data suggest that upon binding to the MIC-1 promoter, DEC1 and p53 form a complex, which weakens the DNA-binding activity of p53 to the MIC-1 promoter. Thus, further study is warranted to examine whether DEC1 disrupts proper formation of p53 tetramers upon binding to the p53 TD.
The activities of transcription factors are often modulated by transcription cofactors, such as HDACs. It has been shown that HDAC1 is recruited by p53 to repress p53 target gene promoters (39) and inhibits p53 activity through p53 deacetylation (40). Here, we showed that HDAC1 is present in the p53–DEC1 complex (Fig. S5A). Interestingly, we previously reported that HDAC2 inhibits p53 transcriptional activity (31), and HDAC2 associates with DEC1 and attenuates DEC1 activation of the ΔNp63 promoter (30). Thus, DEC1 potentially represses p53 transcriptional activity on the MIC-1 promoter via recruiting HDAC1/2 corepressors, which needs to be further explored.
MIC-1, a TGF-β superfamily cytokine, plays a role in cell proliferation, cell mobility, and the response of cancer cells to a therapy. Under physiological conditions, the level of MIC-1 protein is undetectable in most tissues except placenta and brain (41). However, MIC-1 expression is increased during neoplastic transformation, along with an elevated level of secreted mature MIC-1 in sera from cancer patients (41). Thus, the level of serum MIC-1 is explored as a diagnostic tool. Several factors are known to up-regulate MIC-1 expression, including Sp1 (23) and p53 (25, 27). Like other TGF-β family members, MIC-1 acts as a tumor suppressor at the early stage of tumorigenesis, but at the late stage is associated with tumor invasion and metastasis (41). Here, we showed that DEC1 inhibits MIC-1 expression under both the basal and stress conditions. Interestingly, the basal expression of MIC-1 in the absence of p53 is extremely low in multiple cell lines (Fig. 3 and Fig. S4). Thus, we hypothesize that in the early stage of tumorigenesis when p53 is still wild type and can be readily activated, overexpression of DEC1 in some tumors would inhibit p53 induction of MIC-1, and thus promote tumor development.
DEC1/2 along with CLOCK (NPAS2), BMAL1, Cry1/2, and Per1/2 are five clock-gene families regulating the circadian rhythm (14). Importantly, recent observations showed that clock genes are involved in DNA damage-induced apoptosis and tumorigenesis. Loss of Per1 or Per2 impairs DNA damage-induced apoptosis (42, 43), whereas Cry-null cells exhibit increased apoptotic response upon DNA damage (44). These findings suggest that clock genes differentially modulate the extent of DNA damage-induced cell death. Interestingly, rhythmic expression of p53 is synchronized with the expression pattern of Per1 in oral mucosa (45). In addition, impaired p53 activation upon γ-irradiation was found in Per2-null thymocytes (43). However, the mechanism by which the clock genes regulate p53 is not clear. Therefore, our findings suggest that DEC1 links the circadian clock with the p53 pathway, and thus may be explored to improve cancer chronotherapy.
Materials and Methods
Antibodies used in this work are listed in SI Materials and Methods. Detailed information for cell line generation, plasmid construction, DNA histogram analysis, luciferase reporter assay, RT-PCR, ChIP assay, and ChIP-reChIP assay are available in SI Materials and Methods. Primers for RT-PCR, ChIP assay, and ChIP-reChIP assay are summarized in Table S2.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health Grants CA076069, CA123227, and CA102188.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1203185109/-/DCSupplemental.
References
- 1.Beckerman R, Prives C. Transcriptional regulation by p53. Cold Spring Harb Perspect Biol. 2010;2:a000935. doi: 10.1101/cshperspect.a000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harms K, Nozell S, Chen X. The common and distinct target genes of the p53 family transcription factors. Cell Mol Life Sci. 2004;61:822–842. doi: 10.1007/s00018-003-3304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu G, Chen X. DNA polymerase eta, the product of the xeroderma pigmentosum variant gene and a target of p53, modulates the DNA damage checkpoint and p53 activation. Mol Cell Biol. 2006;26:1398–1413. doi: 10.1128/MCB.26.4.1398-1413.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen X, Ko LJ, Jayaraman L, Prives C. p53 levels, functional domains, and DNA damage determine the extent of the apoptotic response of tumor cells. Genes Dev. 1996;10:2438–2451. doi: 10.1101/gad.10.19.2438. [DOI] [PubMed] [Google Scholar]
- 5.Walker KK, Levine AJ. Identification of a novel p53 functional domain that is necessary for efficient growth suppression. Proc Natl Acad Sci USA. 1996;93:15335–15340. doi: 10.1073/pnas.93.26.15335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu J, Jiang J, Zhou W, Zhu K, Chen X. Differential regulation of cellular target genes by p53 devoid of the PXXP motifs with impaired apoptotic activity. Oncogene. 1999;18:2149–2155. doi: 10.1038/sj.onc.1202533. [DOI] [PubMed] [Google Scholar]
- 7.Zhu J, Zhang S, Jiang J, Chen X. Definition of the p53 functional domains necessary for inducing apoptosis. J Biol Chem. 2000;275:39927–39934. doi: 10.1074/jbc.M005676200. [DOI] [PubMed] [Google Scholar]
- 8.Samuels-Lev Y, et al. ASPP proteins specifically stimulate the apoptotic function of p53. Mol Cell. 2001;8:781–794. doi: 10.1016/s1097-2765(01)00367-7. [DOI] [PubMed] [Google Scholar]
- 9.Bourdon JC, et al. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005;19:2122–2137. doi: 10.1101/gad.1339905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai C, et al. Differential effects on p53-mediated cell cycle arrest vs. apoptosis by p90. Proc Natl Acad Sci USA. 2011;108:18937–18942. doi: 10.1073/pnas.1110988108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.MacLachlan TK, Takimoto R, El-Deiry WS. BRCA1 directs a selective p53-dependent transcriptional response towards growth arrest and DNA repair targets. Mol Cell Biol. 2002;22:4280–4292. doi: 10.1128/MCB.22.12.4280-4292.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das S, et al. Hzf Determines cell survival upon genotoxic stress by modulating p53 transactivation. Cell. 2007;130:624–637. doi: 10.1016/j.cell.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanaka T, Ohkubo S, Tatsuno I, Prives C. hCAS/CSE1L associates with chromatin and regulates expression of select p53 target genes. Cell. 2007;130:638–650. doi: 10.1016/j.cell.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 14.Honma S, et al. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature. 2002;419:841–844. doi: 10.1038/nature01123. [DOI] [PubMed] [Google Scholar]
- 15.Collado M, et al. Tumour biology: Senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- 16.Qian Y, Zhang J, Yan B, Chen X. DEC1, a basic helix-loop-helix transcription factor and a novel target gene of the p53 family, mediates p53-dependent premature senescence. J Biol Chem. 2008;283:2896–2905. doi: 10.1074/jbc.M708624200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qian Y, Chen X. ID1, inhibitor of differentiation/DNA binding, is an effector of the p53-dependent DNA damage response pathway. J Biol Chem. 2008;283:22410–22416. doi: 10.1074/jbc.M800643200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, et al. Abundant expression of Dec1/stra13/sharp2 in colon carcinoma: Its antagonizing role in serum deprivation-induced apoptosis and selective inhibition of procaspase activation. Biochem J. 2002;367:413–422. doi: 10.1042/BJ20020514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ehata S, et al. Transforming growth factor-beta promotes survival of mammary carcinoma cells through induction of antiapoptotic transcription factor DEC1. Cancer Res. 2007;67:9694–9703. doi: 10.1158/0008-5472.CAN-07-1522. [DOI] [PubMed] [Google Scholar]
- 20.Thin TH, Li L, Chung TK, Sun H, Taneja R. Stra13 is induced by genotoxic stress and regulates ionizing-radiation-induced apoptosis. EMBO Rep. 2007;8:401–407. doi: 10.1038/sj.embor.7400912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bootcov MR, et al. MIC-1, a novel macrophage inhibitory cytokine, is a divergent member of the TGF-beta superfamily. Proc Natl Acad Sci USA. 1997;94:11514–11519. doi: 10.1073/pnas.94.21.11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Böttner M, Suter-Crazzolara C, Schober A, Unsicker K. Expression of a novel member of the TGF-beta superfamily, growth/differentiation factor-15/macrophage-inhibiting cytokine-1 (GDF-15/MIC-1) in adult rat tissues. Cell Tissue Res. 1999;297:103–110. doi: 10.1007/s004410051337. [DOI] [PubMed] [Google Scholar]
- 23.Baek SJ, Horowitz JM, Eling TE. Molecular cloning and characterization of human nonsteroidal anti-inflammatory drug-activated gene promoter. Basal transcription is mediated by Sp1 and Sp3. J Biol Chem. 2001;276:33384–33392. doi: 10.1074/jbc.M101814200. [DOI] [PubMed] [Google Scholar]
- 24.Paralkar VM, et al. Cloning and characterization of a novel member of the transforming growth factor-beta/bone morphogenetic protein family. J Biol Chem. 1998;273:13760–13767. doi: 10.1074/jbc.273.22.13760. [DOI] [PubMed] [Google Scholar]
- 25.Tan M, Wang Y, Guan K, Sun Y. PTGF-beta, a type beta transforming growth factor (TGF-beta) superfamily member, is a p53 target gene that inhibits tumor cell growth via TGF-beta signaling pathway. Proc Natl Acad Sci USA. 2000;97:109–114. doi: 10.1073/pnas.97.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hromas R, et al. PLAB, a novel placental bone morphogenetic protein. Biochim Biophys Acta. 1997;1354:40–44. doi: 10.1016/s0167-4781(97)00122-x. [DOI] [PubMed] [Google Scholar]
- 27.Li PX, et al. Placental transforming growth factor-beta is a downstream mediator of the growth arrest and apoptotic response of tumor cells to DNA damage and p53 overexpression. J Biol Chem. 2000;275:20127–20135. doi: 10.1074/jbc.M909580199. [DOI] [PubMed] [Google Scholar]
- 28.Dai MS, et al. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol Cell Biol. 2004;24:7654–7668. doi: 10.1128/MCB.24.17.7654-7668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, et al. DEC1 negatively regulates the expression of DEC2 through binding to the E-box in the proximal promoter. J Biol Chem. 2003;278:16899–16907. doi: 10.1074/jbc.M300596200. [DOI] [PubMed] [Google Scholar]
- 30.Qian Y, Jung YS, Chen X. DeltaNp63, a target of DEC1 and histone deacetylase 2, modulates the efficacy of histone deacetylase inhibitors in growth suppression and keratinocyte differentiation. J Biol Chem. 2011;286:12033–12041. doi: 10.1074/jbc.M110.207241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harms KL, Chen X. Histone deacetylase 2 modulates p53 transcriptional activities through regulation of p53-DNA binding activity. Cancer Res. 2007;67:3145–3152. doi: 10.1158/0008-5472.CAN-06-4397. [DOI] [PubMed] [Google Scholar]
- 32.Miyashita T, Reed JC. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 33.Thornborrow EC, Patel S, Mastropietro AE, Schwartzfarb EM, Manfredi JJ. A conserved intronic response element mediates direct p53-dependent transcriptional activation of both the human and murine bax genes. Oncogene. 2002;21:990–999. doi: 10.1038/sj.onc.1205069. [DOI] [PubMed] [Google Scholar]
- 34.Friedman PN, Chen X, Bargonetti J, Prives C. The p53 protein is an unusually shaped tetramer that binds directly to DNA. Proc Natl Acad Sci USA. 1993;90:3319–3323. doi: 10.1073/pnas.90.8.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pietsch EC, et al. The tetramerization domain of p53 is required for efficient BAK oligomerization. Cancer Biol Ther. 2007;6:1576–1583. doi: 10.4161/cbt.6.10.4719. [DOI] [PubMed] [Google Scholar]
- 36.Shieh SY, Ahn J, Tamai K, Taya Y, Prives C. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 2000;14:289–300. [PMC free article] [PubMed] [Google Scholar]
- 37.Yakovlev AG, et al. BOK and NOXA are essential mediators of p53-dependent apoptosis. J Biol Chem. 2004;279:28367–28374. doi: 10.1074/jbc.M313526200. [DOI] [PubMed] [Google Scholar]
- 38.Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell. 2001;7:683–694. doi: 10.1016/s1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- 39.Bansal N, et al. Tumor suppressor protein p53 recruits human Sin3B/HDAC1 complex for down-regulation of its target promoters in response to genotoxic stress. PLoS ONE. 2011;6:e26156. doi: 10.1371/journal.pone.0026156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ito A, et al. MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J. 2002;21:6236–6245. doi: 10.1093/emboj/cdf616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Welsh JB, et al. Large-scale delineation of secreted protein biomarkers overexpressed in cancer tissue and serum. Proc Natl Acad Sci USA. 2003;100:3410–3415. doi: 10.1073/pnas.0530278100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gery S, et al. The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol Cell. 2006;22:375–382. doi: 10.1016/j.molcel.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 43.Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 44.Lee JH, Sancar A. Circadian clock disruption improves the efficacy of chemotherapy through p73-mediated apoptosis. Proc Natl Acad Sci USA. 2011;108:10668–10672. doi: 10.1073/pnas.1106284108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bjarnason GA, et al. Circadian expression of clock genes in human oral mucosa and skin: Association with specific cell-cycle phases. Am J Pathol. 2001;158:1793–1801. doi: 10.1016/S0002-9440(10)64135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.