Abstract
In cyanobacteria, respiratory electron transport takes place in close proximity to photosynthetic electron transport, because the complexes required for both processes are located within the thylakoid membranes. The balance of electron transport routes is crucial for cell physiology, yet the factors that control the predominance of particular pathways are poorly understood. Here we use a combination of tagging with green fluorescent protein and confocal fluorescence microscopy in live cells of the cyanobacterium Synechococcus elongatus PCC 7942 to investigate the distribution on submicron scales of two key respiratory electron donors, type-I NAD(P)H dehydrogenase (NDH-1) and succinate dehydrogenase (SDH). When cells are grown under low light, both complexes are concentrated in discrete patches in the thylakoid membranes, about 100–300 nm in diameter and containing tens to hundreds of complexes. Exposure to moderate light leads to redistribution of both NDH-1 and SDH such that they become evenly distributed within the thylakoid membranes. The effects of electron transport inhibitors indicate that redistribution of respiratory complexes is triggered by changes in the redox state of an electron carrier close to plastoquinone. Redistribution does not depend on de novo protein synthesis, and it is accompanied by a major increase in the probability that respiratory electrons are transferred to photosystem I rather than to a terminal oxidase. These results indicate that the distribution of complexes on the scale of 100–300 nm controls the partitioning of reducing power and that redistribution of electron transport complexes on these scales is a physiological mechanism to regulate the pathways of electron flow.
Keywords: bioenergetics, respiratory complex I, complex II
Both respiratory and photosynthetic electron transport are essential for energy conversion in plants and many microbes. The regulation of electron transport routes is crucial for cell physiology (1). Cyanobacteria make an interesting paradigm for regulation of electron transport, because respiration takes place in close proximity to photosynthesis, with the complexes required for both processes located within the intracytoplasmic thylakoid membranes (2, 3). This colocalization raises the possibility of “unorthodox” electron transport pathways in which electrons cross over from respiratory donors to photosynthetic acceptors, or vice versa. Indeed such unorthodox electron transport pathways are observed and can predominate in cyanobacterial mutants lacking photosystems I (PSI) or photosystem II (PSII) (4, 5) and also in wild-type cyanobacterial cells under certain conditions (5). The factors that might control the predominance of particular electron transport pathways under particular conditions are poorly understood.
Cyanobacterial thylakoid membranes house the photosynthetic reaction centers PSI and PSII, respiratory electron transport complexes including type-I NAD(P)H dehydrogenase (NDH-1), succinate dehydrogenase (SDH), cytochrome (cyt) oxidase, and alternative oxidases, as well as the cyt b6f complex, which functions in both respiratory and photosynthetic electron transport (6). The mobile electron carriers, plastoquinone and plastocyanin (and/or cyt c), are also common to both photosynthetic and respiratory electron transport. NDH-1 and SDH have both been proposed as principal respiratory electron donor complex in cyanobacteria (7, 8). Electrons from respiratory substrates enter the electron transport chain via plastoquinone reduction by NDH-1 or SDH and are passed on through the cyt b6f complex and plastocyanin or cyt c. They then have two possible fates: they could be transferred to a terminal oxidase or they could be transferred to PSI (3). The former case corresponds to conventional respiratory electron transport with net oxidation of the metabolite pool, whereas the latter case is one of several possible routes for cyclic photosynthetic electron transport around PSI, with generation of a proton gradient but no net loss or gain of reducing power (9–11). There is an additional possibility of electron transfer from PSI to oxygen via the flavodiiron proteins Flv1 and Flv3 (12).
Here, we explore the factors that might control the prevalence of respiratory electron transfer to oxidases vs. PSI by using enhanced green fluorescent protein (eGFP) tagging and confocal fluorescence microscopy to investigate the distribution on submicron scales of the two key respiratory electron donors NDH-1 and SDH in the cyanobacterium Synechococcus elongatus PCC 7942. We find that the distribution of the complexes within the thylakoid membranes is under redox-regulated physiological control. Redistribution of the complexes correlates with a major change in the relative probability of the two electron transport pathways.
Results
We chose the unicellular cyanobacterium S. elongatus PCC 7942 (hereafter Synechococcus) as our model organism because it is sequenced, genetically tractable, and has a rather regular arrangement of thylakoid membranes as concentric cylinders aligned along the long axis of the cell, which greatly facilitates studies of protein distribution by fluorescence microscopy (13, 14).
NDH-1 and SDH are the two principal respiratory electron donor complexes in cyanobacteria (7, 8). We tagged both complexes individually, in both cases using expression of the eGFP fusion protein from the native chromosomal locus and under the control of the native promoter, to ensure that the proteins were expressed in context and at physiological levels. Cyanobacterial NDH-1 is partially homologous to mitochondrial complex I (15). Seventeen different protein subunits have been identified as cyanobacterial NDH-1 components, although not all are present in all forms of the complex (8, 16). Recently, another subunit designated NdhS has been identified, possibly involved in ferredoxin binding (17). Biochemical analysis indicates that there are at least two variant forms of the complex with different subunit compositions and, possibly, different physiological roles (18–20). To give a view of the complete distribution of NDH-1, eGFP was fused to the C terminus of the NdhM subunit, which is common to all of the NDH-1 variants (21) (Fig. S1). eGFP was similarly fused to the C terminus of the SdhA subunit of SDH (22, 23) (Fig. S1). In both cases, the transformants were fully segregated (i.e., the alteration was introduced into all copies of the chromosome) and Western blots with anti-GFP antibodies demonstrated that eGFP was fused to a protein of the expected size (Fig. S1). Blue-native gel electrophoresis further showed that NdhM:eGFP was incorporated into fully assembled NDH-1 complexes, and cell growth rates were unaffected by the GFP fusions (Fig. S1), in contrast to mutants with defective SDH or NDH-1 (7, 24). The content of NDH-1 complexes also appears unaffected by the GFP tag on NdhM (Fig. S1). We attempted GFP tagging of some further thylakoid membrane electron carriers. However, GFP-tagging of PSI (on the C terminus of the PsaI subunit) was unsatisfactory as the mutant did not segregate and PsaI:eGFP was not fully incorporated into PSI complexes. GFP tagging of cytochrome oxidase (on the C terminus of the CtaC subunit) did not lead to a detectable GFP fluorescence signal in the cells, probably indicating that levels of cytochrome oxidase in the cell are too low for GFP fluorescence to be detectable above background.
Localization of NDH-1 and SDH Under Low and Moderate Light.
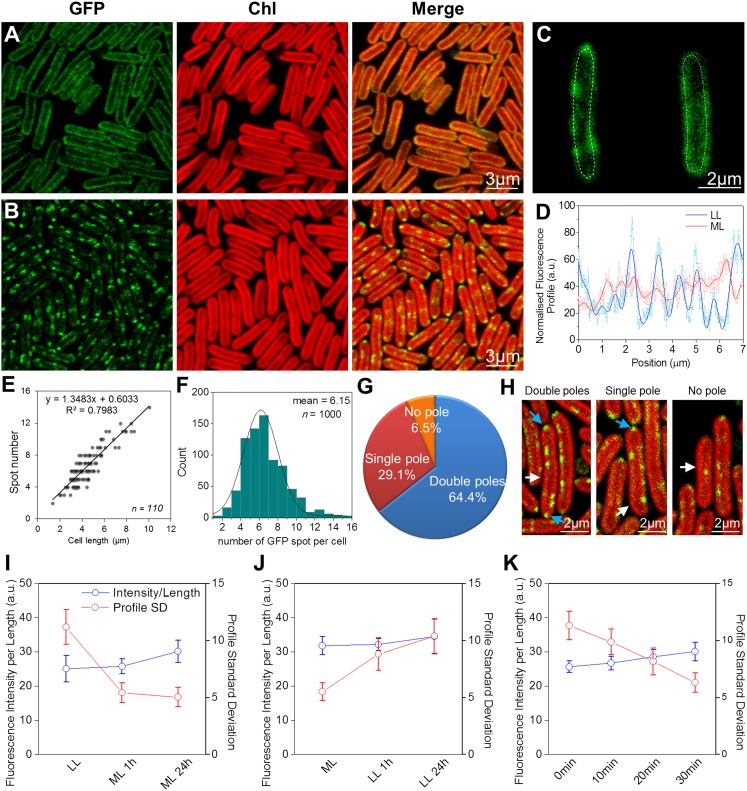
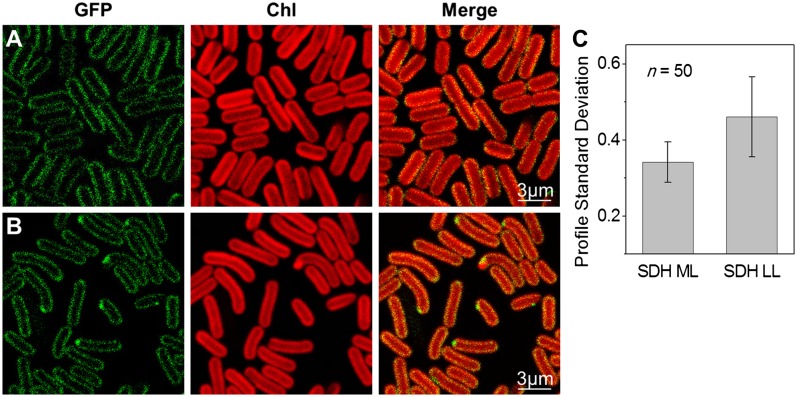
Figs. 1 and 2 show confocal fluorescence micrographs of Synechococcus cells with eGFP tags on NDH-1 and SDH, respectively. The native chlorophyll fluorescence (shown in red) indicates the distribution of the photosynthetic complexes in the thylakoid membranes (13). Consistent with proteomic studies in the cyanobacterium Synechocystis sp. PCC 6803 (25), we could detect NDH-1 and SDH only in the thylakoid membranes. Fluorescently stained cytoplasmic membranes showed up as a distinct halo outside the chlorophyll (Fig. S2), in contrast to what we observed with NDH-1 and SDH where the GFP fluorescence clearly overlapped with the chlorophyll fluorescence (Figs. 1 and 2). In cells grown in moderate light (60 μE⋅m−2⋅s−1) both NDH-1 (Fig. 1A) and SDH (Fig. 2A) were quite evenly distributed in the thylakoids, similarly to the photosynthetic complexes. However, under very low light (6 μE⋅m−2⋅s−1) both complexes were concentrated into discrete patches within the thylakoid membranes (Figs. 1B and 2B). The inhomogeneity of GFP fluorescence was quantified by taking line profiles of fluorescence intensity around the thylakoid membranes (Fig. 1C), smoothing to remove high-frequency noise (Fig. 1D) and then computing the SD (Figs. 1 I and J and 2C). For NDH-1:eGFP the SD of fluorescence intensity was significantly higher under low light conditions (two-tailed Student t test, P = 10−7). For SDH:eGFP the overall fluorescence signal was lower but the difference in distribution under the two conditions remained highly significant (two-tailed Student t test, P = 10−7). The NDH-1 and SDH patches seen under low-light conditions were further characterized in terms of their number, their distribution within the cell, their size, and the approximate number of complexes in each patch. On average there were about six patches of NDH-1:eGFP per cell (Fig. 1F), the number in a particular cell being roughly proportional to its length (Fig. 1E). Cell poles were frequently occupied by NDH-1 patches (Fig. 1G and H). The probability of a patch being located at a cell pole was significantly higher than would be expected on the basis of random distribution (two-tailed Student t test, P = 10−3). The diameter of the NDH-1:eGFP patches was estimated, taking into account the point-spread function of the microscope, and found to range from 60 to 350 nm, with a mean of 160 ± 30 nm (Fig. S3).
Fig. 1.
Localization of NDH-1:eGFP complexes in Synechococcus. (A) NDH-1:eGFP distribution in cells adapted to ML. (B) NDH-1:eGFP distribution in cells under LL. (C) Examples of GFP fluorescence profiles along the thylakoid membranes. (D) Raw and smoothed normalized fluorescence profiles of the tracing lines in C. (E) NDH-1 spot number per cell is proportional to the cell length (n = 110). (F) NDH-1:eGFP spot numbers per cell (n = 1,000). (G) Frequency of occurrence of NDH-1:eGFP spots at the cell poles (n = 800). (H) Examples of polar localization. White and cyan arrows indicate the cell NDH-1:eGFP spots, respectively. (I) Fluorescence profile SDs (red) and normalized fluorescence intensities (blue) of NDH-1:eGFP cells under LL, and 1 h, 24 h after transferring to ML (n = 50, P = 10−7). (J) Fluorescence profile SDs (red) and normalized fluorescence intensities (blue) of NDH-1:eGFP cells adapted to ML, and 1 h, 24 h after transferring to LL (n = 50, P = 10−7). (K) Fluorescence profile SDs and normalized fluorescence intensities for a set of low-light–grown NDH-1:eGFP cells exposed to intense light (500 μE⋅m−2⋅s−1) for 10, 20, and 30 min (n = 50). The corresponding confocal images are shown in Fig. S6.
Fig. 2.
Localization of SDH:eGFP complexes in Synechococcus cells. (A) Confocal images of SDH:eGFP cells under ML. (B) SDH:eGFP cells under LL. (C) SDs of SDH:eGFP fluorescence profiles under moderate and LL (n = 50, P = 10−7).
The number of GFP molecules per patch was estimated by comparing the brightness of the GFP fluorescence to that from the previously characterized patches of cyt bd:GFP in Escherichia coli (26) imaged with the same microscope settings. This indicates a mean of 91 ± 30 NDH-1:eGFP complexes per patch. A similar analysis on SDH:eGFP indicates that individual patches are somewhat larger than NDH-1 patches (mean diameter 280 ± 190 nm) but contain fewer complexes (37 ± 12) and are fewer in number. Around one or two large SDH patches were visible per cell (Fig. 2B). The NDH-1 complexes must be densely packed within their patches. The membrane area occupied by a Thermus thermophilus or E .coli NDH-1 complex can be estimated at 230 nm2 from its crystal structure (27, 28). Thus, assuming the same membrane domain size, 90 NDH-1 complexes would occupy a membrane area of 20,700 nm2, which is similar to the estimated mean area of the patches. The packing of SDH appears to be much less dense than that of NDH-1.
Mobility and Redistribution of NDH-1.
The patches of NDH-1 are not mobile on short timescales. Imaging and fluorescence recovery after photobleaching (FRAP) sequences recorded over 3 min showed no detectable movement (Fig. S4). Cyt bd-I complexes in the E. coli plasma membrane congregate in patches of comparable dimensions, but these patches are highly mobile (26), in striking contrast to the immobility of the NDH-1 patches in Synechococcus. However, this difference is not unexpected, given that the diffusion of complexes in cyanobacterial thylakoids is generally slow and restricted, and PSII is almost immobile (29). On longer timescales, there can clearly be redistribution of the respiratory complexes according to the light environment (Figs. 1 and 2). Redistribution of NDH-1 is largely complete within 1 h (Fig. 1 I and J and Fig. S5). To study shorter timescales, we performed an experiment tracking redistribution within a single set of cells during illumination with more intense light on the microscope stage. This indicates a half-time of 10–20 min for the redistribution (Fig. 1K and Fig. S6). Acclimation to moderate light was accompanied by an increase in GFP fluorescence per cell (Fig. 1I), suggesting synthesis of new NDH-1 complexes. To check whether the NDH-1 redistribution was dependent on de novo protein synthesis, we carried out the same experiment in the presence of the protein synthesis inhibitor lincomycin (30). Lincomycin suppressed the increase in total GFP fluorescence, but the redistribution of fluorescence occurred as before (Fig. S7). Thus, the change in NDH-1 distribution must result from the redistribution of the existing pool of NDH-1 complexes.
Effects of Electron Transport Inhibitors and Bicarbonate on NDH-1 Redistribution.
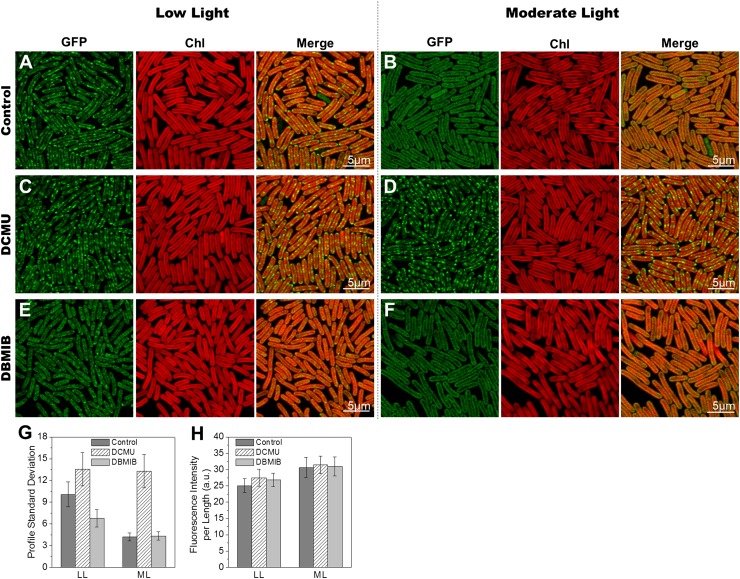
Many light-induced responses in cyanobacteria are triggered by changes in the redox state of electron carriers in the photosynthetic electron transport chain (31). To test whether NDH-1 redistribution is triggered in this fashion, we examined the effects of the electron transport inhibitors 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) and 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone (DBMIB) (Fig. 3). DCMU blocks electron transport from PSII to plastoquinone, whereas DBMIB blocks electron transport from plastoquinol to the cyt b6f complex. Opposite effects of DCMU and DBMIB therefore indicate control by the redox state of plastoquinone (32). Cells expressing NDH-1:eGFP were adapted to either low light or moderate light before addition of the inhibitor and adaptation for 1 h to the opposite light regime. Images of NDH-1:eGFP distribution were recorded and distributions quantified as described above. DCMU completely repressed the redistribution of NDH-1:eGFP in moderate light (Fig. 3 D and G), and induced an even more patchy distribution of NDH-1:eGFP in low light (Fig. 3 C and G). By contrast, DBMIB suppressed the formation of NDH-1:eGFP patches in low light (Fig. 3 E and G). This indicates that redistribution of NDH-1 is triggered by changes in the redox state of plastoquinone, similarly to state transitions (32), the resetting of the cyanobacterial circadian clock (33, 34), and changes in the expression of photosynthetic complexes (35). The patchy distribution of respiratory complexes is triggered by an oxidized plastoquinone pool, whereas the even distribution is triggered by a reduced plastoquinone pool.
Fig. 3.
Effects of electron transport inhibitors on redistribution of NDH-1 complexes. (A) NDH-1:eGFP cells under LL. (B) LL-adapted NDH-1:eGFP cells acclimated to ML for 1 h. (C) NDH-1:eGFP cells acclimated to LL for 1 h in the presence of DCMU. (D) NDH-1:eGFP cells acclimated to ML for 1 h in the presence of DCMU. (E) NDH-1:eGFP cells acclimated to LL for 1 h in the presence of DBMIB. (F) NDH-1:eGFP cells acclimated to ML for 1 h in the presence of DBMIB. (G) Fluorescence profile SD for NDH-1:GFP cells under LL and 1 h after transferring to ML with or without electron transport inhibitors (n = 50; P = 10−8, LL ± DCMU; P = 10−12, LL ± DBMIB; P = 10−30, ML ± DCMU). (H) Normalized fluorescence intensities of NDH-1:eGFP cells under LL and 1 h after transferring to ML (n = 50).
Because NDH-1 is important for CO2/HCO3− assimilation in cyanobacteria (16), we also tested the effect of addition of 10 mM NaHCO3 to the medium (cells were otherwise grown aerobically without supplementation of CO2/HCO3−). However, supplementation with NaHCO3 had no detectable effect on NDH-1:eGFP distribution in either low light or moderate light.
Electron Transport Pathways Under Low and Moderate Light.
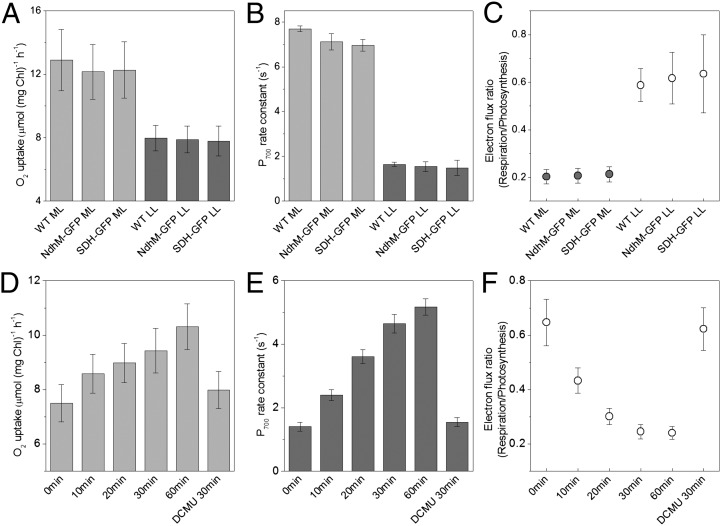
The functional significance of the changes in NDH-1 and SDH distribution induced by changes in light environment was explored by electron transport measurements on cells adapted to the two conditions. Respiratory electron transport (i.e., transfer of electrons to a terminal oxidase) was assessed by measuring oxygen uptake in the dark (Fig. 4A). Electron transfer to PSI was assessed using microsecond flash photolysis at 703 nm (36) to determine the kinetics of rereduction of P700 following a train of flashes sufficient to oxidase P700 and the plastoquinone pool (Fig. S8). The measurements were carried out in the presence of DCMU, so that the electrons required to rereduce P700+ would have to come from respiratory donors or by cyclic electron transfer (37). Measurements were carried out on wild-type Synechococcus cells as well as the two GFP-tagged strains. All three strains showed similar rates of electron transport under both conditions (Fig. 4 A and B), confirming that the eGFP-tagged NDH-1 and SDH complexes are fully functional. Low-light–adapted cells show somewhat lower rates of respiratory oxygen uptake (Fig. 4A), which may be due to lower levels of photosynthetically produced respiratory substrates. There is a much larger difference in the rate constants for P700+ rereduction, which are about four times higher in moderate-light cells compared with low-light cells (Fig. 4B). For comparison of the data in Fig. 4 A and B, the rate constants for P700+ rereduction were converted into predicted steady-state fluxes of electrons to PSI at saturating light intensity, using the concentration of PSI reaction centers obtained from the maximum 703 nm absorbance change. Respiratory oxygen uptake rates were converted into electron flux to the oxidases by multiplying by 4 (because four electrons are required to reduce O2 to 2H2O). The ratio of the two fluxes (respiration/photosynthesis) then gives an indication of the probability that an electron is transferred to a terminal oxidase rather than to PSI. This ratio is about three times higher in low light compared with moderate light (Fig. 4C). A time course for the change in electron flux ratio during acclimation to moderate light (Fig. 4F) shows comparable kinetics to the change in distribution of NDH-1:eGFP (Fig. 1 I and K). Preincubation with DCMU during a 30-min moderate light acclimation completely suppresses the change in electron flux ratio (Fig. 4 D–F). This provides a further demonstration that electron transport activity correlates with NDH-1 distribution, because DCMU also suppresses the redistribution of NDH-1 during moderate light acclimation (Fig. 3).
Fig. 4.
Electron transport activities in Synechococcus cells (wild-type and GFP-tagged transformants) adapted to LL or ML. (A) Rates of respiratory oxygen uptake in the dark (n = 5). (B) Rate constants for P700+ rereduction in the presence of DCMU (n = 5). (C) Ratio of respiratory electron flux to oxidases vs. PSI (under saturating light) calculated from the data in A and B. It gives an indication of the probability that respiratory electrons are transferred a terminal oxidase rather than to PSI. (D–F) Time-course for the changes in electron transport activity during acclimation from LL to ML, plus effect of DCMU on acclimation to ML for 30 min (n = 15, pooled data from the three strains). Two-tailed Student t tests for changes in electron flux ratio in F are: P = 2 × 10−10 (LL vs. 1 h in ML). P = 4 × 10−9 (30 min in ML ± DCMU).
Discussion
NDH-1 and SDH complexes cluster into 100- to 300-nm patches in the thylakoid membranes in low light (Figs. 1 and 2). Exposure to moderate light induces the dispersal of these clusters of respiratory complexes within 30 min (Figs. 1 and 2), and this change in distribution correlates with a major increase in the probability that respiratory electrons are transferred to PSI rather than to a terminal oxidase (Fig. 4). The distribution of respiratory complexes under moderate light matches much more closely the distribution of the photosynthetic complexes, at least as far as can be assessed at optical resolution (Figs. 1 and 2). This matching distribution suggests intermingling of the photosynthetic and respiratory complexes, leading to a greater prevalence of electron exchange between the two chains. Both respiratory electron transport and cyclic photosynthetic electron transport via PSI generate a proton gradient for ATP production. In physiological terms, the distinction is that respiratory electron transport leads to a net oxidation of the metabolite pool, whereas cyclic photosynthetic electron transport does not lead to any net change in redox status (10, 11). Therefore, the switch in electron transport modes under different light regimes is likely to reflect the need to control the redox balance of the cell, as does the switch between photosynthetic linear and cyclic electron transport in the green alga Chlamydomonas, for example (11). Redistribution of the respiratory complexes in Synechococcus is triggered by changes in the redox state of an electron carrier close to the plastoquinone pool (Fig. 3), which is consistent with the idea that this is a mechanism to control the redox balance of the cell. State 1–state 2 transitions (a rapid modulation of excitation energy distribution between PSII and PSI) are also triggered by changes in the redox state of plastoquinone in cyanobacteria (32). Therefore it is possible that the same signal transduction pathway leads both to the redistribution of respiratory complexes that we describe here and to reorganization of photosynthetic light-harvesting complexes during state transition. A distinction is that the 10- to 30-min timescale of redistribution of the respiratory complexes (Fig. 1) is considerably slower than the 1- to 5-min timescale of state transitions in cyanobacteria (38). This may be explained by the slow timescale of protein diffusion in the thylakoid membrane (29), which contrasts with the rapid movement or local conformational changes of the light-harvesting phycobilisomes on the membrane surface (29, 38–40). As with state transitions in cyanobacteria, the physical factors that lead to the redistribution of the respiratory complexes remain to be determined. Covalent modification, either of the complexes themselves or their interaction partners, is a strong possibility.
Other recent studies have indicated clustering, on similar length scales, of oxidative phosphorylation complexes in the plasma membranes of E. coli (26) and Bacillus subtilis (41) and the clustering of both photosynthetic and respiratory complexes in the plasma membrane of the atypical cyanobacterium Gloeobacter violaceus, which lacks thylakoid membranes (42). Taken together, these studies suggest that electron transport typically occurs within clusters of complexes colocalized on submicron scales, and our present study shows that redistribution of electron transport on these scales is accompanied by major changes in electron transport activity. Therefore, a typical length scale for electron transport (defined by the distances that mobile electron carriers such as quinones and plastocyanin typically diffuse between donor and acceptor complexes) might be on the order of 100 nm.
The search for mechanistic explanations for the “streaming” of electrons between specific pairs of donor and acceptor complexes has focused on the search for “supercomplexes” with strong associations on the nanometer scale (43–48). Biochemical approaches and electron microscopy have provided support for the presence of supercomplexes in some bioenergetic membranes, including respiratory supercomplexes (43–45, 47, 48), and respiratory–photosynthetic supercomplexes (46); however, direct evidence that electron transport is confined to such supercomplexes is lacking. Whereas supercomplexes could well play a role in controlling some pathways of electron flow (49), our study indicates that the larger-scale distribution of electron transport complexes within heterogeneous bioenergetic membranes is also an important factor. This could be an important consideration for the future reengineering of organisms for enhanced biofuel production, for example.
The factors that physically control the distribution of respiratory complexes within cyanobacterial thylakoid membranes are subjects for future study. It will also be important to get a more complete picture of the protein composition of the membrane patches defined here by the clustering of NDH-1 and SDH complexes under low light.
Materials and Methods
Strains and Growth Conditions.
S. elongatus PCC 7942 was grown in BG-11 medium (50) at 30 °C under 60 μE⋅m−2⋅s−1 (moderate light, ML) or 6 μE⋅m−2⋅s−1 (low light, LL) white light in culture flasks with constant shaking or on BG-11 plates containing 1.5% (wt/vol) agar. Cultures were grown in air without any additional CO2 source, except for the experiment where we supplemented the liquid medium with 10 mM NaHCO3 (SI Materials and Methods). The electron transport inhibitors DCMU and DBMIB were added to 20 μM and 10 μM, respectively, where indicated. Cells were adapted for 1 h under LL or ML in the presence of DCMU or DBMIB before microscopy. For electron transport measurements in cells preadapted with DCMU, LL-grown cells were adapted for 30 min in ML in the presence of DCMU before measurement.
Generation of Constructs.
PCR products of about 3 kb, including ∼1 kb homologous sequence upstream and downstream of synpcc7942_1982 (ndhM) or synpcc7942_0641 (sdhA), were cloned using the pGEM-T Easy cloning system (Promega) as detailed in the Promega manual. Enhanced green fluorescent protein (eGFP) fusions were created by inserting the GFP:apramycin region amplified from the plasmid pIJ786 (PBL Biomedical Laboratories) to the C terminus of ndhM or sdhA using the Redirect strategy (51, 52). Plasmids were verified by PCR and sequencing and used to transform Synechococcus cells following the method described earlier (53). PCR and agarose gel electrophoresis were applied to check genetic segregation of the ndhM or sdhA loci in the transformants. The DNA oligonucleotides used in this work are shown in Table S1.
Confocal Microscopy and Image Analysis.
Synechococcus cells were immobilized by drying a droplet of cell suspension onto BG-11 agar plates. Blocks of agar with the cells adsorbed onto the surface were covered with a coverslip and placed under the microscope. Laser-scanning confocal microscopy used a Leica TCS-SP5 with a 63× oil-immersion objective (NA 1.4) and excitation at 488 nm. Images (12-bit, 512 × 512 pixels) were recorded at 200 Hz, averaging each scan line six times. Emission was recorded simultaneously at 500–520 nm (GFP fluorescence) and 670–720 nm (chlorophyll fluorescence). The confocal pinhole was set to give z axis resolution of about 0.77 μm. To correct for the autofluorescent background from the cells, images were recorded before and after bleaching by scanning repeatedly at 6× laser power. This bleached GFP fluorescence while having no detectable effect on the background. Postbleach images were then subtracted from prebleach to give clean GFP images (54). Imaging time-courses were recorded while illuminating the cells with white light at 500 μE⋅m−2⋅s−1 on the sample stage between image acquisitions. Live-cell images were recorded from at least five different cultures. All images were captured with all pixels below saturation. The inhomogeneity of GFP fluorescence was quantified by taking line profiles of fluorescence intensity around the thylakoid membranes, smoothing to remove high frequency noise and then computing the SD from the mean fluorescence intensity. The point-spread function of the microscope was measured by recording images with the same microscope settings of 170 nm diameter yellow-green fluorescent microspheres (PS-Speck microscope point source kit; Invitrogen), and the information used to estimate the true diameter of GFP patches observed in membranes. Background subtraction and image analysis were carried out using ImageJ software (National Institutes of Health). Significance was assessed using a two-tailed Student t test. Results are presented as mean ± SD.
Respiratory Electron Transport.
The respiratory electron transport measurements are described in SI Materials and Methods.
P700+ Rereduction Measurements.
P700 (the primary electron donor of PSI) rereduction kinetics were determined by analysis of flash-induced absorbance changes at 703 nm in a laboratory-built spectrophotometer at room temperature as described previously (36). See SI Materials and Methods. The kinetics of P700+ rereduction were fitted with a single exponential decay (Fig. S8).
Membrane Preparation, Protein Analysis, and Immunoblotting.
The membrane preparation, protein analysis, and immunoblotting are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
L.-N.L. acknowledges the support of a Marie Curie Fellowship from the European Commission (Contract FP7-PEOPLE-2009-IEF 254575). S.J.B. was supported by Biotechnology and Biological Sciences Research Council Grant BB/G021856/1. P.R.R. acknowledges Biotechnology and Biological Sciences Research Council Grant BB/H000097/1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1120960109/-/DCSupplemental.
References
- 1.Millar AH, Whelan J, Soole KL, Day DA. Organization and regulation of mitochondrial respiration in plants. Annu Rev Plant Biol. 2011;62:79–104. doi: 10.1146/annurev-arplant-042110-103857. [DOI] [PubMed] [Google Scholar]
- 2.Vermaas WF. Photosynthesis and Respiration in Cyanobacteria. Encyclopedia of Life Sciences. London: Nature Publishing Group; 2001. pp. 245–251. [Google Scholar]
- 3.Peschek GA, Obinger C, Paumann M. The respiratory chain of blue-green algae (cyanobacteria) Physiol Plant. 2004;120:358–369. doi: 10.1111/j.1399-3054.2004.00274.x. [DOI] [PubMed] [Google Scholar]
- 4.Vermaas WF. Molecular genetics of the cyanobacterium Synechocystis sp. PCC 6803: Principles and possible biotechnology applications. J Appl Phycol. 1996;8:263–273. [Google Scholar]
- 5.Grossman AR, Mackey KRM, Bailey S. A perspective on photosynthesis in the oligotrophic oceans: Hypotheses concerning alternate routes of electron flow. J Phycol. 2010;46:629–634. [Google Scholar]
- 6.Schmetterer G. Cyanobacterial respiration. In: Bryant DA, editor. The Molecular Biology of Cyanobacteria. Vol 1. Dordrecht: Kluwer; 1994. pp. 409–435. [Google Scholar]
- 7.Cooley JW, Vermaas WF. Succinate dehydrogenase and other respiratory pathways in thylakoid membranes of Synechocystis sp. strain PCC 6803: Capacity comparisons and physiological function. J Bacteriol. 2001;183:4251–4258. doi: 10.1128/JB.183.14.4251-4258.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogawa T, Mi H. Cyanobacterial NADPH dehydrogenase complexes. Photosynth Res. 2007;93:69–77. doi: 10.1007/s11120-006-9128-y. [DOI] [PubMed] [Google Scholar]
- 9.Munekage Y, et al. Cyclic electron flow around photosystem I is essential for photosynthesis. Nature. 2004;429:579–582. doi: 10.1038/nature02598. [DOI] [PubMed] [Google Scholar]
- 10.Shikanai T. Cyclic electron transport around photosystem I: Genetic approaches. Annu Rev Plant Biol. 2007;58:199–217. doi: 10.1146/annurev.arplant.58.091406.110525. [DOI] [PubMed] [Google Scholar]
- 11.Rochaix JD. Reprint of: Regulation of photosynthetic electron transport. Biochim Biophys Acta. 2011;1807:878–886. doi: 10.1016/j.bbabio.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 12.Allahverdiyeva Y, et al. Interplay between flavodiiron proteins and photorespiration in Synechocystis sp. PCC 6803. J Biol Chem. 2011;286:24007–24014. doi: 10.1074/jbc.M111.223289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mullineaux CW, Sarcina M. Probing the dynamics of photosynthetic membranes with fluorescence recovery after photobleaching. Trends Plant Sci. 2002;7:237–240. doi: 10.1016/s1360-1385(02)02283-5. [DOI] [PubMed] [Google Scholar]
- 14.Savage DF, Afonso B, Chen AH, Silver PA. Spatially ordered dynamics of the bacterial carbon fixation machinery. Science. 2010;327:1258–1261. doi: 10.1126/science.1186090. [DOI] [PubMed] [Google Scholar]
- 15.Berger S, Ellersiek U, Steinmüller K. Cyanobacteria contain a mitochondrial complex I-homologous NADH-dehydrogenase. FEBS Lett. 1991;286:129–132. doi: 10.1016/0014-5793(91)80957-5. [DOI] [PubMed] [Google Scholar]
- 16.Battchikova N, Eisenhut M, Aro EM. Cyanobacterial NDH-1 complexes: novel insights and remaining puzzles. Biochim Biophys Acta. 2011;1807:935–944. doi: 10.1016/j.bbabio.2010.10.017. [DOI] [PubMed] [Google Scholar]
- 17.Battchikova N, et al. Identification of novel Ssl0352 protein (NdhS), essential for efficient operation of cyclic electron transport around photosystem I, in NADPH:plastoquinone oxidoreductase (NDH-1) complexes of Synechocystis sp. PCC 6803. J Biol Chem. 2011;286:36992–37001. doi: 10.1074/jbc.M111.263780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang P, et al. Expression and functional roles of the two distinct NDH-1 complexes and the carbon acquisition complex NdhD3/NdhF3/CupA/Sll1735 in Synechocystis sp PCC 6803. Plant Cell. 2004;16:3326–3340. doi: 10.1105/tpc.104.026526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernát G, Appel J, Ogawa T, Rögner M. Distinct roles of multiple NDH-1 complexes in the cyanobacterial electron transport network as revealed by kinetic analysis of P700+ reduction in various Ndh-deficient mutants of Synechocystis sp. strain PCC6803. J Bacteriol. 2011;193:292–295. doi: 10.1128/JB.00984-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohkawa H, Pakrasi HB, Ogawa T. Two types of functionally distinct NAD(P)H dehydrogenases in Synechocystis sp. strain PCC6803. J Biol Chem. 2000;275:31630–31634. doi: 10.1074/jbc.M003706200. [DOI] [PubMed] [Google Scholar]
- 21.Prommeenate P, Lennon AM, Markert C, Hippler M, Nixon PJ. Subunit composition of NDH-1 complexes of Synechocystis sp. PCC 6803: Identification of two new ndh gene products with nuclear-encoded homologues in the chloroplast Ndh complex. J Biol Chem. 2004;279:28165–28173. doi: 10.1074/jbc.M401107200. [DOI] [PubMed] [Google Scholar]
- 22.Yankovskaya V, et al. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science. 2003;299:700–704. doi: 10.1126/science.1079605. [DOI] [PubMed] [Google Scholar]
- 23.Sun F, et al. Crystal structure of mitochondrial respiratory membrane protein complex II. Cell. 2005;121:1043–1057. doi: 10.1016/j.cell.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 24.Ogawa T. A gene homologous to the subunit-2 gene of NADH dehydrogenase is essential to inorganic carbon transport of Synechocystis PCC6803. Proc Natl Acad Sci USA. 1991;88:4275–4279. doi: 10.1073/pnas.88.10.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pisareva T, et al. Model for membrane organization and protein sorting in the cyanobacterium Synechocystis sp. PCC 6803 inferred from proteomics and multivariate sequence analyses. J Proteome Res. 2011;10:3617–3631. doi: 10.1021/pr200268r. [DOI] [PubMed] [Google Scholar]
- 26.Lenn T, Leake MC, Mullineaux CW. Clustering and dynamics of cytochrome bd-I complexes in the Escherichia coli plasma membrane in vivo. Mol Microbiol. 2008;70:1397–1407. doi: 10.1111/j.1365-2958.2008.06486.x. [DOI] [PubMed] [Google Scholar]
- 27.Efremov RG, Baradaran R, Sazanov LA. The architecture of respiratory complex I. Nature. 2010;465:441–445. doi: 10.1038/nature09066. [DOI] [PubMed] [Google Scholar]
- 28.Hunte C, Zickermann V, Brandt U. Functional modules and structural basis of conformational coupling in mitochondrial complex I. Science. 2010;329:448–451. doi: 10.1126/science.1191046. [DOI] [PubMed] [Google Scholar]
- 29.Sarcina M, Tobin MJ, Mullineaux CW. Diffusion of phycobilisomes on the thylakoid membranes of the cyanobacterium Synechococcus 7942. Effects of phycobilisome size, temperature, and membrane lipid composition. J Biol Chem. 2001;276:46830–46834. doi: 10.1074/jbc.M107111200. [DOI] [PubMed] [Google Scholar]
- 30.Dalla Chiesa M, et al. Reduced turnover of the D1 polypeptide and photoactivation of electron transfer in novel herbicide resistant mutants of Synechocystis sp. PCC 6803. Eur J Biochem. 1997;248:731–740. doi: 10.1111/j.1432-1033.1997.00731.x. [DOI] [PubMed] [Google Scholar]
- 31.Mullineaux CW. How do cyanobacteria sense and respond to light? Mol Microbiol. 2001;41:965–971. doi: 10.1046/j.1365-2958.2001.02569.x. [DOI] [PubMed] [Google Scholar]
- 32.Mullineaux CW, Allen JF. State 1-State 2 transitions in the cyanobacterium Synechococcus 6301 are controlled by the redox state of electron carriers between Photosystems I and II. Photosynth Res. 1990;23:297–311. doi: 10.1007/BF00034860. [DOI] [PubMed] [Google Scholar]
- 33.Ivleva NB, Gao T, LiWang AC, Golden SS. Quinone sensing by the circadian input kinase of the cyanobacterial circadian clock. Proc Natl Acad Sci USA. 2006;103:17468–17473. doi: 10.1073/pnas.0606639103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wood TL, et al. The KaiA protein of the cyanobacterial circadian oscillator is modulated by a redox-active cofactor. Proc Natl Acad Sci USA. 2010;107:5804–5809. doi: 10.1073/pnas.0910141107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fujita Y, Murakami A, Ohki K. Regulation of photosystem composition in the cyanobacterial photosynthetic system: The regulation occurs in response to the redox state of the electron pool located between the two photosystems. Plant Cell Physiol. 1987;28:283–292. [Google Scholar]
- 36.Rich PR, Madgwick SA, Moss DA. The interactions of duroquinol, DBMIB and NQNO with the chloroplast cytochrome bf complex. Biochim Biophys Acta. 1991;1058:312–328. [Google Scholar]
- 37.Nixon PJ, Rich PR. In: Chlororespiratory Pathways and Their Physiological Significance. The Structure and Function of Plastids, Advances in Photosynthesis and Respiration. Wise RR, Hoober JK, editors. Vol 23. The Netherlands: Springer; 2006. pp. 237–251. [Google Scholar]
- 38.Joshua S, Mullineaux CW. Phycobilisome diffusion is required for light-state transitions in cyanobacteria. Plant Physiol. 2004;135:2112–2119. doi: 10.1104/pp.104.046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu LN, Aartsma TJ, Thomas JC, Zhou BC, Zhang YZ. FRAP analysis on red alga reveals the fluorescence recovery is ascribed to intrinsic photoprocesses of phycobilisomes than large-scale diffusion. PLoS ONE. 2009;4:e5295. doi: 10.1371/journal.pone.0005295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu LN, et al. Watching the native supramolecular architecture of photosynthetic membrane in red algae: Topography of phycobilisomes and their crowding, diverse distribution patterns. J Biol Chem. 2008;283:34946–34953. doi: 10.1074/jbc.M805114200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson AS, van Horck S, Lewis PJ. Dynamic localization of membrane proteins in Bacillus subtilis. Microbiology. 2004;150:2815–2824. doi: 10.1099/mic.0.27223-0. [DOI] [PubMed] [Google Scholar]
- 42.Rexroth S, et al. The plasma membrane of the cyanobacterium Gloeobacter violaceus contains segregated bioenergetic domains. Plant Cell. 2011;23:2379–2390. doi: 10.1105/tpc.111.085779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schägger H, Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dudkina NV, Eubel H, Keegstra W, Boekema EJ, Braun HP. Structure of a mitochondrial supercomplex formed by respiratory-chain complexes I and III. Proc Natl Acad Sci USA. 2005;102:3225–3229. doi: 10.1073/pnas.0408870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schäfer E, et al. Architecture of active mammalian respiratory chain supercomplexes. J Biol Chem. 2006;281:15370–15375. doi: 10.1074/jbc.M513525200. [DOI] [PubMed] [Google Scholar]
- 46.Peng L, Shimizu H, Shikanai T. The chloroplast NAD(P)H dehydrogenase complex interacts with photosystem I in Arabidopsis. J Biol Chem. 2008;283:34873–34879. doi: 10.1074/jbc.M803207200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Iwai M, et al. Isolation of the elusive supercomplex that drives cyclic electron flow in photosynthesis. Nature. 2010;464:1210–1213. doi: 10.1038/nature08885. [DOI] [PubMed] [Google Scholar]
- 48.Bultema JB, Braun HP, Boekema EJ, Kouril R. Megacomplex organization of the oxidative phosphorylation system by structural analysis of respiratory supercomplexes from potato. Biochim Biophys Acta. 2009;1787:60–67. doi: 10.1016/j.bbabio.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 49.Ramírez-Aguilar SJ, et al. The composition of plant mitochondrial supercomplexes changes with oxygen availability. J Biol Chem. 2011;286:43045–43053. doi: 10.1074/jbc.M111.252544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;111:1–61. [Google Scholar]
- 51.Gust B, Kieser T, Chater KF. REDIRECT Technology: PCR-Targeting System in Streptomyces coelicolor. Norwich, UK: John Innes Centre; 2002. [Google Scholar]
- 52.Gust B, et al. Lambda red-mediated genetic manipulation of antibiotic-producing Streptomyces. Adv Appl Microbiol. 2004;54:107–128. doi: 10.1016/S0065-2164(04)54004-2. [DOI] [PubMed] [Google Scholar]
- 53.Golden SS. Mutagenesis of cyanobacteria by classical and gene-transfer-based methods. Methods Enzymol. 1988;167:714–727. doi: 10.1016/0076-6879(88)67083-2. [DOI] [PubMed] [Google Scholar]
- 54.Spence E, et al. Membrane-specific targeting of green fluorescent protein by the Tat pathway in the cyanobacterium Synechocystis PCC6803. Mol Microbiol. 2003;48:1481–1489. doi: 10.1046/j.1365-2958.2003.03519.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.