Abstract
Glucocorticoids elicit a variety of biological responses in skeletal muscle, including inhibiting protein synthesis and insulin-stimulated glucose uptake and promoting proteolysis. Thus, excess or chronic glucocorticoid exposure leads to muscle atrophy and insulin resistance. Glucocorticoids propagate their signal mainly through glucocorticoid receptors (GR), which, upon binding to ligands, translocate to the nucleus and bind to genomic glucocorticoid response elements to regulate the transcription of nearby genes. Using a combination of chromatin immunoprecipitation sequencing and microarray analysis, we identified 173 genes in mouse C2C12 myotubes. The mouse genome contains GR-binding regions in or near these genes, and gene expression is regulated by glucocorticoids. Eight of these genes encode proteins known to regulate distinct signaling events in insulin/insulin-like growth factor 1 pathways. We found that overexpression of p85α, one of these eight genes, caused a decrease in C2C12 myotube diameters, mimicking the effect of glucocorticoids. Moreover, reducing p85α expression by RNA interference in C2C12 myotubes significantly compromised the ability of glucocorticoids to inhibit Akt and p70 S6 kinase activity and reduced glucocorticoid induction of insulin receptor substrate 1 phosphorylation at serine 307. This phosphorylation is associated with insulin resistance. Furthermore, decreasing p85α expression abolished glucocorticoid inhibition of protein synthesis and compromised glucocorticoid-induced reduction of cell diameters in C2C12 myotubes. Finally, a glucocorticoid response element was identified in the p85α GR-binding regions. In summary, our studies identified GR-regulated transcriptional networks in myotubes and showed that p85α plays a critical role in glucocorticoid-induced insulin resistance and muscle atrophy in C2C12 myotubes.
Glucocorticoids perform vital metabolic functions in skeletal muscle: inhibiting protein synthesis and insulin-stimulated glucose uptake and promoting protein degradation. These effects are critical during stress, producing amino acid precursors for gluconeogenesis, which provides glucose for the brain. Initially, muscle insulin resistance maintains adequate circulating glucose to fuel the brain; however, these effects are deleterious if chronic. Treating animals with glucocorticoids causes a decrease in skeletal muscle size (1–4). Mice treated with glucocorticoids have reduced insulin-stimulated glucose uptake and GLUT4 translocation in myotubes (5–7). Circulating glucocorticoid levels are higher in obese ob/ob and db/db mice than in normal mice. Adrenalectomy in these obese mice improves insulin-stimulated muscle glucose disposal (8). These changes are in part due to the direct effect of glucocorticoids on myotubes, as glucocorticoid treatment in cultured myotubes reduces cell diameters (9–11) and inhibits insulin-stimulated glucose utilization (5, 12).
Although the metabolic effects of glucocorticoids in skeletal muscle are well known, the underlying mechanisms are not fully understood. One way in which glucocorticoids affect glucose and protein metabolism is to antagonize the insulin/insulin-like growth factor 1 (IGF1) pathway (5, 13), which promotes protein synthesis and glucose utilization. Insulin/IGF1 acts by binding to membrane receptors, tyrosine kinases that autophosphorylate and phosphorylate insulin receptor substrates (IRS) (14). Tyrosine-phosphorylated IRS associate with insulin receptors (IR) and activate signaling pathways (14). Mice treated with glucocorticoids have reduced levels of tyrosine-phosphorylated IR and total IRS-1 in skeletal muscle (5). The activity of phosphatidylinositol 3-kinase (PI3K) and Akt, two signaling molecules downstream of IR and IRS-1, is also decreased (5). Glucocorticoids also reduce the activity of mammalian target of rapamycin (mTOR), a protein kinase downstream of Akt and upstream of p70 S6 kinase (p70S6K) (15). Furthermore, glucocorticoid treatment increases the phosphorylation of serine 307 of IRS-1 (pSer307-IRS-1) (5), which disrupts the association of IR and IRS-1, reducing the insulin/IGF1 response (16). However, the mechanism by which glucocorticoids inhibit the insulin/IGF1 pathway is unclear.
Although certain glucocorticoid effects are independent of intracellular glucocorticoid receptors (GR) (17), the majority are mediated by GR, which, upon binding to its ligand, moves to the nucleus and interacts with genomic GR response elements (GRE). It is critical to identify genes directly regulated by GR to learn the physiological mechanisms of glucocorticoids. Here, we used a combination of chromatin immunoprecipitation sequencing (ChIPseq) and microarray analysis to identify direct targets of GR in mouse C2C12 myotubes. We identified potential GR primary targets previously shown to modulate distinct steps of insulin/IGF1 signaling. We focused on one potential GR primary target, p85α. Experiments were conducted to identify its GRE and its role in the suppressive effects of glucocorticoids on myotube diameters, protein synthesis, and the insulin/IGF1-signaling pathway.
Results
Identification of Potential GR Primary Target Genes in C2C12 Myotubes.
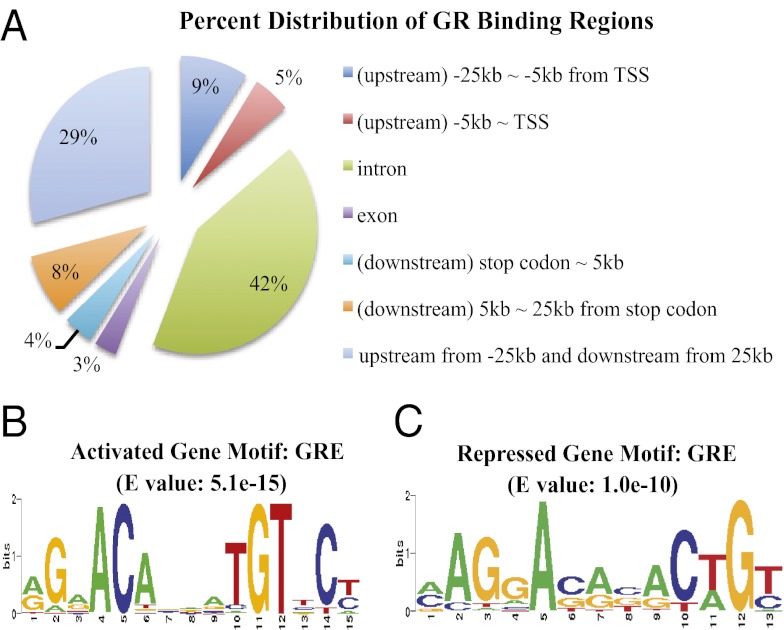
Microarray analyses were conducted in C2C12 myotubes treated for 6 or 24 h with dexamethasone (Dex), a synthetic glucocorticoid, or ethanol (EtOH), a vehicle control. Combining data from both time points, we found that Dex induced the expression of 363 genes by more than 1.5-fold and inhibited the expression of 218 genes by more than 1.5-fold (Dataset S1). To learn which of these genes contains GR-binding regions (GBR) in or near their genomic regions, ChIPseq was performed with C2C12 myotubes treated with Dex or EtOH for 1 h. We located 2,251 genomic positions with sequencing reads significantly enriched in Dex-treated samples compared with EtOH-treated ones, with a P value threshold of 10−5. We used PinkThing to assign these genomic GBR to mouse genes on the basis of proximity and grouped target sites on the basis of their relative position to the nearest gene (GBR are listed in Dataset S2). We found 42% of GBR in the intron regions and 29% either 25 kb upstream of transcription start site (TSS) or downstream of stop codons (Fig. 1A). In contrast, only 5% of GBR are located within 5 kb upstream from TSS (Fig. 1A). Overall, we identified 147 Dex-activated genes and 26 Dex-repressed genes containing GBR in or near their genomic regions (Table S1, Dataset S3).
Fig. 1.
Genomic distribution of GBR and GRE motifs within GBR. (A) Percentage distribution of all identified GBR in different genomic regions. (B) For GBR of glucocorticoid-induced genes, BioProspector analysis identified a motif that is similar to the consensus GRE in STAMP. (C) For GBR of glucocorticoid-repressed genes, BioProspector analysis also identified a motif that is similar to the consensus GRE in STAMP. (B and C) The E value is a parameter that describes the number of hits that one can expect to see by random chance when searching a database of a particular DNA element.
The top categories of gene annotation from gene ontology analysis of 173 GR-regulated genes include those encoding proteins involved in the receptor tyrosine kinase-signaling pathway, blood vessel development, apoptosis, muscle organ development, and cytoskeletal organization (Dataset S4). We performed a combination of BioProspector and STAMP analyses to search for consensus motifs within GBR located in or near genes that were regulated by glucocorticoids. For glucocorticoid-activated genes, the classical GRE was the most represented motif in these analyses on the basis of E values (Fig. 1B). Binding motifs for HSF, AP-1, NF-E2, MAF, NRF-2, core binding, AML, PEBP, FOXP3, HNF3α, T3R, and RREB-1 were all significantly represented (Table S2). For glucocorticoid-repressed genes, the classical GRE motif was also highly represented (Fig. 1C). Moreover, HSF, SMAD3, T3R, XBP-1, ELF-1, HAC-1, ID-1, FOXP3, AbaA, XPF-1, Bach1, and AP-1 were all high on the list of binding motifs (Table S3).
Glucocorticoid-Controlled Genes Involved in Insulin/IGF1 Signaling.
Gene ontology analysis revealed that many glucocorticoid-regulated genes affect receptor tyrosine kinase signaling. We analyzed the potential GR primary targets that can specifically inhibit the insulin/IGF1 pathway, which propagates through receptor tyrosine kinases. As shown in Fig. 2A, at least eight glucocorticoid-regulated genes encode proteins that can modulate the activity of this pathway. One of these genes, Cblb, encodes a ubiquitin E3 ligase involved in IRS-1 degradation (18). Pid-1 inhibits the tyrosine phosphorylation of IRS-1 (19). Grb10 inhibits the physical interaction between IR and IRS-1 (20). Overexpression of p85α induces insulin resistance in vitro and in vivo (21–23). Sesn 1 (24), Ddit4 (15), and Depdc6 (25) inhibit mTOR. Finally, mice with bone-marrow-specific deletion of Sorbs1 are protected from high-fat-diet–induced insulin resistance (26), indicating that overexpression of Sorbs1 could induce insulin resistance. To learn whether these genes were regulated by glucocorticoids in vivo, mice were injected with Dex or PBS control for 24 or 96 h. We found that mice injected with Dex for 24 h had significantly higher expression of Cblb and Pid1 than those injected with PBS in gastrocnemius muscles (Fig. 2B). Furthermore, the expression of Cblb, Pid1, p85α, Sesn1, Ddit4, and Sorbs1 was increased in mice treated with Dex for 96 h compared with the control (Fig. 2B). To determine the effect of chronic glucocorticoid exposure, we used transgenic mice overexpressing corticotropin-releasing hormone (CRH-Tg). CRH causes an increased secretion of adrenocorticotropic hormone, which further stimulates the secretion of adrenal corticosterone. In CRH-Tg mice, the expression of Cblb, Pid1, p85α, Ddit4, Sorbs1, and Sesn1 was markedly elevated in gastrocnemius muscles (Fig. 2C). In summation, at least six of the potential GR primary targets can inhibit insulin/IGF1 signaling in vivo.
Fig. 2.
Gene expression analyses of glucocorticoid-regulated genes in animal models. (A) Eight of the GR primary target genes and their roles in modulating the insulin/IGF1 signaling. (B) Fold induction of gene expression in gastrocnemius muscles of six to eight mice, injected with Dex or PBS for 1 or 4 d. (C) Fold induction of gene expression in gastrocnemius muscles of 12–14 CRH-Tg or WT mice. Error bars represent the SE. *P < 0.01.
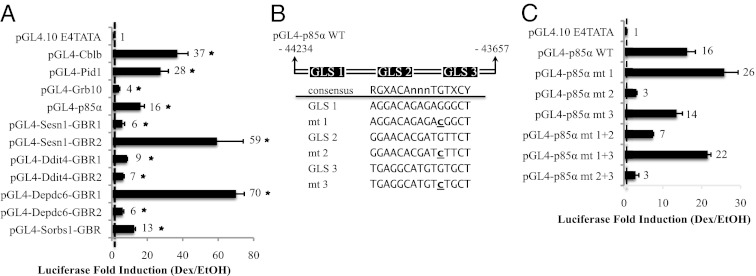
A critical criterion for determining GR primary targets is that GR directly regulates their transcription. Thus, these GBR should be able to mediate the glucocorticoid response. To validate the GBR identified near these eight genes, each GBR was inserted upstream of a TATA box of a heterologous reporter plasmid, pGL4.10-E4TATA, which drives a firefly luciferase gene. A collection of plasmids used is listed in Dataset S5. We found that all GBR tested could mediate glucocorticoid responses (Fig. 3A), suggesting these eight glucocorticoid-regulated genes are primary targets of GR.
Fig. 3.
Glucocorticoid response of GBR. (A) Reporter plasmids harboring different GBR were cotransfected with pcDNA3-hGR and pRL Renilla into C2C12 myoblasts (n > 6/group). Renilla luciferase expression was used to normalize for transfection efficiency. After 24 h of transfection, cells were treated with Dex or EtOH for 16–20 h and assayed for firefly and Renilla luciferase activities. Data show fold induction of normalized luciferase activity (Dex/EtOH-treated samples) from at least six experiments. *P < 0.05. The error bars represent the SE for the fold induction, and the dashed line marks a one-fold induction. (B) pGL4-p85α WT harbors nucleotide (nt) −44234 to −43657 of the p85α genomic region. On the basis of the consensus GRE shown, three GLS were identified. The locations of the GLS are the following: nt −44003 to −43989 (GLS-1), nt −43938 to −43924 (GLS-2), and nt −43913 to +43899 (GLS-3). Point mutation, indicated by underlining, is made in each GLS to generate a mutant (mt). Mt 1 has nt −43993 mutated from G to C; mt 2, nt −43928 from G to C; and mt 3, nt −43903 from G to C. (C) Reporter assay, as described in A, was carried with WT and mt p85α GBR plasmid. pGL4-p85α mt 1+2 represents a double mutant of GLS-1 and -2; pGL4-p85α mt 1+3, a double mutant of GLS-1 and -3; and pGL4-p85α mt 2+3, double mutant of GLS-2 and -3.
Role of p85α in Glucocorticoid-Regulated Insulin Signaling.
The induction of p85α by glucocorticoids in vivo (Fig. 2 B and C) prompted further analysis into its role in mediating glucocorticoid responses in muscle. First, we identified the GRE in the p85α GBR. We searched for DNA sequences matching at least seven nucleotides to the consensus GRE motif identified from BioProspector/STAMP (RGXACAnnnTGTXCY) (Fig. 1C) in the p85α GBR. Three potential GRE-like sequences (GLS) were found, and for each GLS, we mutated the nucleotide from G to C at position 11, which makes direct contact with GR (27) (Fig. 3B). With the reporter assay, we found that GLS-2 mutants had an 80% reduction in the Dex response. Although the GLS-3 mutation had no significant effect, the GLS-1 mutation moderately potentiated the Dex response (Fig. 3C). Double mutations of GLS-2 and GLS-3 had similar effects as GLS-2 single mutations, and double mutations of GLS-1 and GLS-2 resulted in a slight increase in Dex response compared with GLS-2 single mutations (Fig. 3C). These results indicated that, although GLS-1 may be suppressive, GLS-2 plays a central role in mediating the glucocorticoid response. Furthermore, we used TFSEARCH to identify transcription factor-binding motifs in the p85α GBR. We found SRY-, GATA-, USF-, and Ikaros-binding sites located in the vicinity of GLS-2 (Fig. S1).
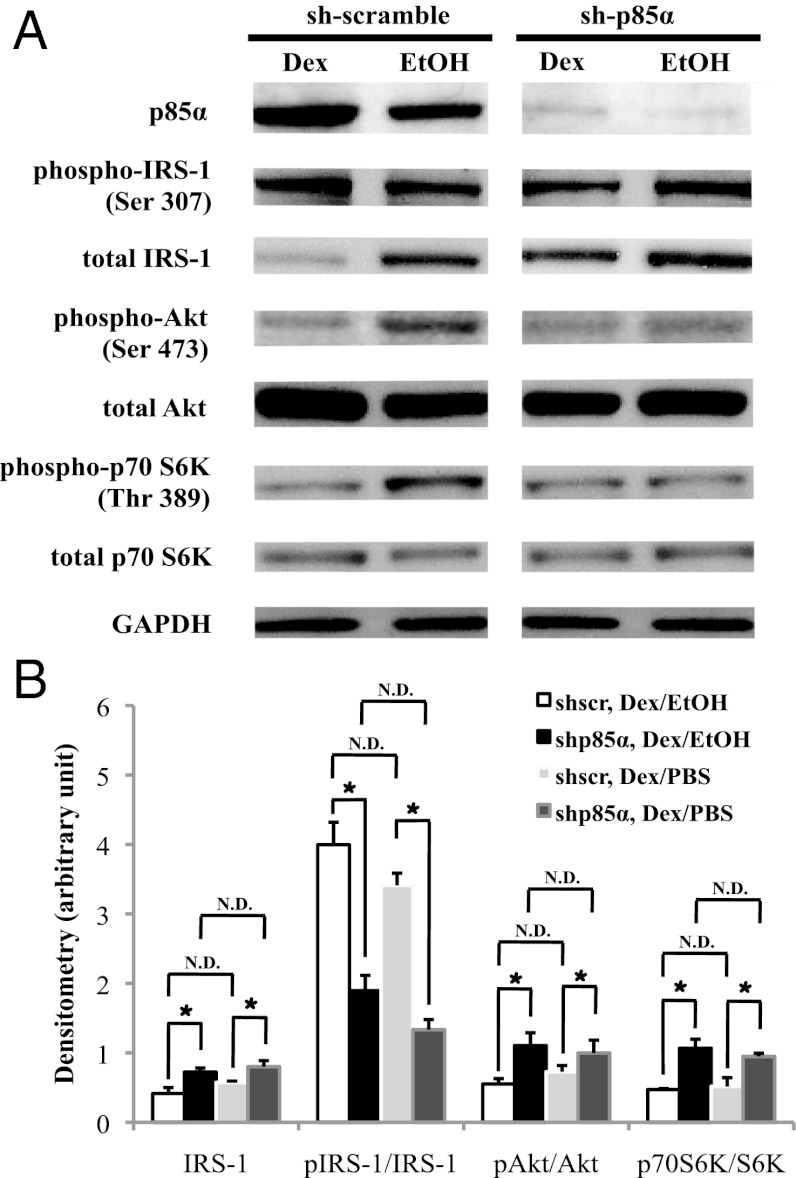
Next, we examined the role of p85α on glucocorticoid-induced inhibition of signaling molecules participating in the insulin/IGF1 pathway. We used lentiviral-mediated small hairpin RNA (shRNA) to decrease p85α expression (sh-p85α) in C2C12 cells. Control cells were infected with lentiviruses expressing shRNA with scramble sequences (sh-scr). Cells were then treated with Dex or EtOH for 72 h. In sh-scr cells, p85α protein levels were increased upon Dex treatment (Fig. 4A). In sh-p85α cells, p85α levels were reduced ∼90% in both Dex- and EtOH-treated cells (Fig. 4A).
Fig. 4.
Effect of p85α RNAi in glucocorticoid response. (A) Sh-p85α and sh-scr C2C12 myotubes treated with Dex or EtOH for 72 h. Immunoblots were used to detect the indicated proteins. (B) Quantification of the densitometry of total IRS-1 and the ratio of pSer307-IRS-1, pSer473-Akt, and pThr389-S6K bands to that of IRS-1, Akt, and p70S6K bands, respectively, with Dex/EtOH or Dex/PBS. Data were normalized to GAPDH optical density. These results were averaged from at least three immunoblots. *P < 0.05 and N.D. specifies “no difference.” The error bars represent the SE for the fold induction and relative abundance.
We monitored the phosphorylation status of serine 473 of Akt (pSer473-Akt) and threonine 389 of p70S6K (pThr389-S6K). These phosphorylations are required to potentiate their kinase activity. In contrast, pSer307-IRS-1 disrupts its interaction with the insulin/IGF1 receptor. In sh-scr cells, pSer473-Akt and pThr389-S6K levels were reduced upon Dex treatment (Fig. 4A). Although pSer307-IRS-1 levels were comparable between Dex- and EtOH-treated cells, IRS-1 protein expression was decreased upon Dex treatment in sh-scr cells (Fig. 4A). After normalizing to the IRS-1 protein present, there was a significant increase of pSer307-IRS-1 in Dex-treated sh-scr cells. In sh-p85α cells, Dex treatment did not suppress the levels of pSer473-Akt and pThr389-S6K (Fig. 4 A and B). Although total IRS-1 protein levels decreased upon Dex treatment, this reduction was weaker than that of Dex-treated sh-scr cells (Fig. 4 A and B). Considering the similarity of pSer307-IRS-1 levels and the difference in total IRS-1 levels between Dex- and EtOH-treated sh-scr and sh-p85α cells, the ability of Dex to induce pSer307-IRS-1 was significantly compromised by knocking down p85α (Fig. 4B). Notably, EtOH treatment resembles PBS-treated sh-scr and sh-p85α cells (Fig. 4B). Overall, our data indicated that reducing the expression of p85α compromised the ability of glucocorticoids to inhibit the activity of Akt and p70S6K, to reduce IRS-1 protein levels, and to induce phosphorylation at serine 307 of IRS-1.
Role of p85α in Glucocorticoid-Induced Muscle Atrophy.
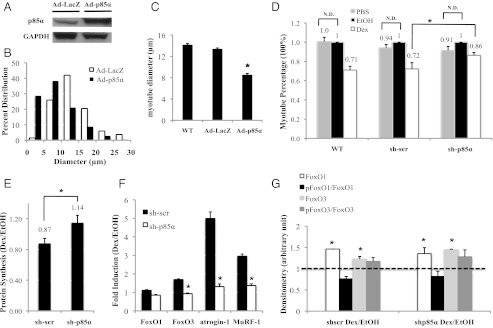
We investigated whether p85α is involved in glucocorticoid-induced muscle atrophy. C2C12 myotubes were infected with adenoviruses expressing p85α (Ad-p85α) or a control of LacZ (Ad-LacZ). Cell diameters were then measured 72 h after infection. p85α protein levels are approximately fourfold higher in Ad-p85α than in Ad-LacZ cells (Fig. 5A). We found that significantly more Ad-p85α myotubes had smaller cell diameters than Ad-LacZ ones (Fig. 5B). The average cell diameter of Ad-p85α myotubes was about 30% smaller than Ad-LacZ ones (Fig. 5C). These results demonstrate that overexpressing p85α mimics the effect of glucocorticoids in reducing C2C12 myotube diameters.
Fig. 5.
Effect of p85α overexpression and knock-down on C2C12 myotube cell diameters. (A) Immunoblot was performed in Ad-LacZ and Ad-p85α myotubes to detect p85α protein levels. (B) The distribution of cell diameters in Ad-LacZ and Ad-p85α myotubes. (C) The average cell diameter of WT, Ad-LacZ, and Ad-p85α myotubes. (D) WT, sh-scr, and sh-p85α myotubes were treated with Dex, EtOH, or PBS for 72 h; EtOH-treated WT, sh-scr, and sh-p85α myotube diameters were set as 1 (100%) for Dex-treated or PBS-treated WT, sh-scr, and sh-p85α myotubes, respectively. N.D. specifies “no difference.” (E) Protein synthesis level was measured for sh-scr and sh-p85α myotubes, with the EtOH-treated sh-scr and sh-p85α myotube protein synthesis level set as 1 for Dex-treated sh-scr and sh-p85α myotubes, respectively. (F) FoxO1, FoxO3, atrogin-1, and MuRF-1 gene expressions in sh-scr and sh-p85α myotubes. Rpl19 primer was the internal control. (G) Protein levels of phosphorylated and total FoxO1 and FoxO3 are shown for sh-scr and sh-p85α myotubes. (C–G) *P < 0.05. The error bars represent the SE for diameters. These results are averaged from at least three independent experiments.
Following the treatment of sh-scr and sh-p85α myotubes with Dex, EtOH, or PBS for 72 h, we measured cell diameters. The average diameter of Dex-treated sh-scr myotubes was 28% smaller than those of EtOH-treated sh-scr myotubes (Fig. 5D). A similar effect was observed in wild-type (WT) myotubes, with no lentiviral infection, treated with Dex and EtOH (Fig. 5D). In contrast, the average diameter of Dex-treated sh-p85α myotubes was 14% smaller than that of EtOH-treated sh-p85α myotubes (Fig. 5D). Although glucocorticoids still decreased C2C12 myotube diameter, reduced p85α expression significantly compromised this decrease. Notably, the average myotube diameters of EtOH-treated WT, sh-scr, and sh-p85α myotubes were comparable.
In addition, we investigated the effect of glucocorticoids on protein synthesis in sh-scr and sh-p85α myotubes. Myotubes were treated with Dex or EtOH for 72 h, and protein synthesis was measured using a fluorescence assay. We found that Dex-treated sh-scr myotubes had 13% lower nascent protein synthesis than EtOH-treated sh-scr myotubes (Fig. 5E). In contrast, Dex-treated sh-p85α myotubes had 14% higher nascent protein synthesis than EtOH-treated sh-p85α myotubes (Fig. 5E). Notably, the protein synthesis rates in EtOH-treated sh-scr and sh-p85α myotubes were similar. Our data suggest that p85α mediates glucocorticoid-reduced protein synthesis.
Previous studies have shown that, in myotubes, glucocorticoids stimulate the expression of atrogenes such as FoxO1, FoxO3, atrogin-1 (a.k.a. MAFbx), and MuRF-1, which contribute to glucocorticoid-induced muscle atrophy (28, 29). We found that 72 h of Dex treatment induced FoxO3, atrogin-1, and MuRF-1 gene expression in sh-scr myotubes (Fig. 5F). However, this induction was diminished in sh-p85α myotubes (Fig. 5F). These results indicate that p85α is involved in glucocorticoid-induced FoxO3, atrogin-1, and MuRF-1 gene expression. Immunoblotting showed that Dex treatment up-regulated total FoxO1 and FoxO3 proteins, whereas the levels of phosphorylated-FoxO1 (pFoxO1) and FoxO3 (pFoxO3) were unchanged (Fig. 5G). Thus, the ratio of pFoxO1 to total FoxO1 and the ratio of pFoxO3 to total FoxO3 were reduced. However, these ratios were similar in sh-scr and sh-p85α myotubes.
Discussion
In this report, we present several findings. First, we identified potential GR primary target genes in C2C12 myotubes. Identification will facilitate future studies into the mechanisms underlying glucocorticoid actions in skeletal muscle physiology. Second, through ChIPseq, we localized genome-wide GBR in C2C12 myotubes, which is the first step in understanding the mechanisms governing GR-regulated gene transcription. Finally, we found eight potential GR primary targets that can modulate distinct steps in insulin/IGF1 signaling. Specifically, we have shown that p85α induction plays a key role in mediating glucocorticoid inhibition of the insulin/IGF1 response.
We previously identified GBR in another mouse cell type, 3T3-L1 adipocytes (30). The distribution of GBR in genomic regions is similar in adipocytes and myotubes. Whereas only 5% of GBR lie within 5 kb upstream of TSS, many are localized in introns greater than 25 kb upstream of TSS or greater than 25 kb downstream of stop codons. In motif analyses of GBR from glucocorticoid-activated genes, classical GRE sequences were highly represented. AP-1 and HNF3α, two binding motifs that have been shown to act with GR for maximal activation of transcription (31, 32), also scored highly. The binding site of HNF3α is similar to that of FoxO1 and FoxO3, which perform similar metabolic functions to GR in myotubes, as they promote proteolysis, reduce protein synthesis, and reduce glucose utilization (33). Therefore, GR and FoxO may act together to transcriptionally regulate genes involved in these physiological processes. The classical GRE sequence is also highly represented in GBR of genes repressed by glucocorticoids, but the mechanism is unclear. The AP-1 element, which mediates glucocorticoid repression (34), was highly represented in the GBR of glucocorticoid-repressed genes. Most motifs identified in GBR of glucocorticoid-regulated genes have not been linked to transcriptional repression by GR.
Gene ontology analysis recognized some GR primary targets involved in the regulation of apoptosis. In myotubes, glucocorticoids were shown to potentiate apoptosis (35). This analysis also identified gene groups involved in muscle organ development and cytoskeletal organization, suggesting that glucocorticoids may modulate mechanical properties of muscle, a concept that has not been extensively studied. Genes involved in blood vessel development were also highly represented. Many of these genes regulate angiogenesis, which plays an important role in modulating skeletal muscle health (36). The role of glucocorticoids in angiogenesis has been described (37), but the impact of this function on skeletal muscle is not entirely clear.
We focused on gene clusters that modulate insulin/IGF1 receptor tyrosine kinase signaling because glucocorticoids decrease insulin-stimulated glucose utilization and protein synthesis and increase proteolysis. In skeletal muscle, glucocorticoids affect multiple steps in the insulin/IGF1-signaling pathway. Therefore, it is conceivable that glucocorticoids induce a group of genes to mediate these effects. We focused on elucidating the role of p85α, the regulatory subunit of PI3K, in glucocorticoid-inhibited insulin signaling. Excess p85α can compete with PI3K (a heterodimer of p85α and the catalytic subunit of PI3K) to interact with IRS-1 (23, 38), resulting in a decrease in insulin response. In contrast, reducing the expression of p85α improves insulin sensitivity (39). Moreover, the induction of p85α gene expression by glucocorticoids and the functional interaction between GR and the p85α /PI3K pathway were previously described (40–42). We demonstrated that p85α is a GR primary target by identifying a GRE in the p85α GBR. We used RNAi to decrease p85α expression in C2C12 cells to analyze its role in glucocorticoid responses. The levels of pSer473-Akt and pThr389-S6K were a little lower in EtOH-treated sh-p85α cells compared with EtOH-treated sh-scr cells (Fig. 4B), suggesting that the reduction of p85α expression decreased the activity of Akt and p70S6K. However, previous studies indicated that Pik3r2, another regulatory subunit of PI3K, can compensate for p85α function in skeletal muscle-specific p85α knockout mice (43). In our sh-p85α myotubes, Pik3r2 and the remaining p85α should have been enough to support downstream activities. Therefore, the reduced suppressive effect of glucocorticoids was unlikely due to the lack of activity in insulin/IGF1 signaling.
Our results from p85α RNAi experiments raise several interesting points. First, p85α reduction limited the ability of Dex to decrease IRS-1 protein expression; however, IRS-1 gene expression was unchanged after 6 or 24 h of Dex treatment according to microarray analysis. Therefore, Dex likely reduced IRS-1 protein stability, but the mechanism of p85α in glucocorticoid-promoted IRS-1 degradation is unclear. Second, p85α reduction decreased Dex-induced phosphorylation of IRS-1 at serine 307. Several kinases, including ΙκB kinase β (IKKβ), p70S6K, c-Jun N-terminal kinases (JNK), and PKCθ, have been shown to phosphorylate IRS-1 at serine 307 (16). To our knowledge, there are no reports that glucocorticoids can increase IKKβ activity. Because glucocorticoids decrease p70S6K activity, p70S6K is unlikely to mediate this event. Interestingly, in obese and type 2 diabetic patients, p85α protein expression correlates with the activity of JNK, PKCθ, and pSer307-IRS-1 (44). Establishing the link between glucocorticoids, p85α, JNK, and PKCθ requires further study.
Our studies have shown that p85α is involved in glucocorticoid-induced atrophy and glucocorticoid-inhibited protein synthesis in C2C12 myotubes. Dex treatment resulted in a 50% smaller reduction of myotube diameter in sh-p85α cells than in control cells. The remaining Dex-induced reduction of diameter in sh-p85α myotubes may be due to residual p85α. Alternatively, other GR-regulated genes might contribute to the Dex effect. Supporting this alternative, a study showed that reducing Ddit4 expression attenuates glucocorticoid-inhibited protein synthesis in L6 myotubes (15). This study suggests that additional genes, such as the remaining six GR primary targets, may also have a nonredundant effect and could mediate glucocorticoid-induced muscle atrophy by suppressing insulin/IGF1 signaling.
The ability of Dex to decrease cell diameters in sh-p85α myotubes may be due to Dex-induced protein degradation, as the inhibitory effect of Dex on protein synthesis was abolished in these cells. Glucocorticoids have been shown to increase the expression of MuRF-1 and atrogin-1, two ubiquitin E3 ligases implicated as causes of muscle atrophy (40). In mice lacking MuRF1, the ability of glucocorticoids to induce muscle atrophy is compromised (3). The previously identified MuRF1 GRE (45) was not found in our ChIPseq; however, it is possible that our GR antibody did not recognize the conformation of GR while bound to the MuRF1 GRE. FoxO1 is required for maximal glucocorticoid-activated MuRF1 gene transcription. Although we did not find any GBR near or within the atrogin-1 gene, its transcription is activated by FoxO1 and FoxO3 (11). In agreement with previous reports, we observed a reduced ratio of pFoxO1 to total FoxO1 upon Dex treatment (29). Also, we found a decreased ratio of pFoxO3 to total FoxO3. However, these ratios were similar between sh-scr and sh-p85α myotubes, despite the reduced ability of Dex to suppress Akt activity in the latter. Because Dex increased total FoxO1 and FoxO3 proteins, it added another layer of complexity for the calculation and comparison between their phosphorylation status in sh-scr and sh-p85α cells. The immunoblotting might not be sensitive enough to consistently detect the difference. Upon 72 h Dex treatment, FoxO3, MuRF-1, and atrogin-1 gene expressions were reduced in sh-p85α myotubes, but levels of total FoxO3 protein in sh-scr and sh-p85α myotubes were similar. A longer Dex treatment could be required to observe a change in FoxO3 protein level. Nonetheless, the fact that p85α is involved in Dex-activated atrogene expression suggests that it has a role in glucocorticoid-induced protein degradation.
In summary, we have identified GR-controlled transcriptional networks in myotubes and focused on one that can modulate insulin/IGF1 signaling. We highlighted the role of p85α in this crosstalk between glucocorticoid and insulin action. Future studies will test other GR primary targets to complete the picture of glucocorticoid-induced insulin resistance and muscle atrophy in skeletal muscle.
Materials and Methods
Cell Culture.
The protocol for cell culture is shown in SI Materials and Methods.
Animals.
The protocol is described in SI Materials and Methods. The Office of Laboratory Animal Care at the University of California, Berkeley (#R306-0111) approved all animal experiments reported in this article.
ChIPseq.
The protocol of ChIP, the preparation of the genomic DNA library, the analyses of sequencing data, annotation of genes, motif research, and gene ontology analysis are described in SI Materials and Methods.
Microarray and Data Analysis.
The protocols are presented in SI Materials and Methods. The microarray data are available at the Gene Expression Omnibus Web site (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE28840.
RNA Isolation and Quantitative PCR.
Protocols are described in SI Materials and Methods. Primer sequences are listed in Dataset S5.
Plasmids, Transfection, and Luciferase Reporter Assay.
Protocols are described in SI Materials and Methods. Primer sequences are listed in Dataset S5.
Lentiviral Infection and Western Blot.
The protocol is presented in SI Materials and Methods.
Muscle Atrophy Assay.
The protocol is shown in SI Materials and Methods.
Protein Synthesis Assay.
The protocol is described in SI Materials and Methods.
Supplementary Material
Acknowledgments
This work is supported by National Institutes of Health Grant R01DK083591 and Muscular Dystrophy Association Grant 186068. T.K. is supported by a Dissertation Award Fellowship from the University of California Tobacco-Related Diseases Research Program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. G.L.H. is a guest editor invited by the Editorial Board.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, http://www.ncbi.nlm.nih.gov/geo/ (accession no. GSE28840).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1111334109/-/DCSupplemental.
References
- 1.Hu Z, Wang H, Lee IH, Du J, Mitch WE. Endogenous glucocorticoids and impaired insulin signaling are both required to stimulate muscle wasting under pathophysiological conditions in mice. J Clin Invest. 2009;119:3059–3069. doi: 10.1172/JCI38770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auclair D, Garrel DR, Chaouki Zerouala A, Ferland LH. Activation of the ubiquitin pathway in rat skeletal muscle by catabolic doses of glucocorticoids. Am J Physiol. 1997;272:C1007–C1016. doi: 10.1152/ajpcell.1997.272.3.C1007. [DOI] [PubMed] [Google Scholar]
- 3.Baehr LM, Furlow JD, Bodine SC. Muscle sparing in muscle RING finger 1 null mice: Response to synthetic glucocorticoids. J Physiol. 2011;589:4759–4776. doi: 10.1113/jphysiol.2011.212845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shimizu N, et al. Crosstalk between glucocorticoid receptor and nutritional sensor mTOR in skeletal muscle. Cell Metab. 2011;13(2):170–182. doi: 10.1016/j.cmet.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Morgan SA, et al. 11beta-hydroxysteroid dehydrogenase type 1 regulates glucocorticoid-induced insulin resistance in skeletal muscle. Diabetes. 2009;58:2506–2515. doi: 10.2337/db09-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dimitriadis G, et al. Effects of glucocorticoid excess on the sensitivity of glucose transport and metabolism to insulin in rat skeletal muscle. Biochem J. 1997;321:707–712. doi: 10.1042/bj3210707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weinstein SP, Wilson CM, Pritsker A, Cushman SW. Dexamethasone inhibits insulin-stimulated recruitment of GLUT4 to the cell surface in rat skeletal muscle. Metabolism. 1998;47(1):3–6. doi: 10.1016/s0026-0495(98)90184-6. [DOI] [PubMed] [Google Scholar]
- 8.Ohshima K, Shargill NS, Chan TM, Bray GA. Effects of dexamethasone on glucose transport by skeletal muscles of obese (ob/ob) mice. Int J Obes. 1989;13(2):155–163. [PubMed] [Google Scholar]
- 9.Menconi M, Gonnella P, Petkova V, Lecker S, Hasselgren PO. Dexamethasone and corticosterone induce similar, but not identical, muscle wasting responses in cultured L6 and C2C12 myotubes. J Cell Biochem. 2008;105:353–364. doi: 10.1002/jcb.21833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stitt TN, et al. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol Cell. 2004;14:395–403. doi: 10.1016/s1097-2765(04)00211-4. [DOI] [PubMed] [Google Scholar]
- 11.Sandri M, et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gathercole LL, Bujalska IJ, Stewart PM, Tomlinson JW. Glucocorticoid modulation of insulin signaling in human subcutaneous adipose tissue. J Clin Endocrinol Metab. 2007;92:4332–4339. doi: 10.1210/jc.2007-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schakman O, Gilson H, Thissen JP. Mechanisms of glucocorticoid-induced myopathy. J Endocrinol. 2008;197(1):1–10. doi: 10.1677/JOE-07-0606. [DOI] [PubMed] [Google Scholar]
- 14.Lee YH, White MF. Insulin receptor substrate proteins and diabetes. Arch Pharm Res. 2004;27:361–370. doi: 10.1007/BF02980074. [DOI] [PubMed] [Google Scholar]
- 15.Wang H, Kubica N, Ellisen LW, Jefferson LS, Kimball SR. Dexamethasone represses signaling through the mammalian target of rapamycin in muscle cells by enhancing expression of REDD1. J Biol Chem. 2006;281:39128–39134. doi: 10.1074/jbc.M610023200. [DOI] [PubMed] [Google Scholar]
- 16.Gual P, Le Marchand-Brustel Y, Tanti JF. Positive and negative regulation of insulin signaling through IRS-1 phosphorylation. Biochimie. 2005;87(1):99–109. doi: 10.1016/j.biochi.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 17.Song IH, Buttgereit F. Non-genomic glucocorticoid effects to provide the basis for new drug developments. Mol Cell Endocrinol. 2006;246(1-2):142–146. doi: 10.1016/j.mce.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 18.Nakao R, et al. Ubiquitin ligase Cbl-b is a negative regulator for insulin-like growth factor 1 signaling during muscle atrophy caused by unloading. Mol Cell Biol. 2009;29:4798–4811. doi: 10.1128/MCB.01347-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu WL, et al. Over-expression of NYGGF4 (PID1) inhibits glucose transport in skeletal myotubes by blocking the IRS1/PI3K/AKT insulin pathway. Mol Genet Metab. 2011;102:374–377. doi: 10.1016/j.ymgme.2010.11.165. [DOI] [PubMed] [Google Scholar]
- 20.Wick KR, et al. Grb10 inhibits insulin-stimulated insulin receptor substrate (IRS)-phosphatidylinositol 3-kinase/Akt signaling pathway by disrupting the association of IRS-1/IRS-2 with the insulin receptor. J Biol Chem. 2003;278:8460–8467. doi: 10.1074/jbc.M208518200. [DOI] [PubMed] [Google Scholar]
- 21.Barbour LA, et al. Increased P85alpha is a potent negative regulator of skeletal muscle insulin signaling and induces in vivo insulin resistance associated with growth hormone excess. J Biol Chem. 2005;280:37489–37494. doi: 10.1074/jbc.M506967200. [DOI] [PubMed] [Google Scholar]
- 22.Ueki K, et al. Positive and negative roles of p85 alpha and p85 beta regulatory subunits of phosphoinositide 3-kinase in insulin signaling. J Biol Chem. 2003;278:48453–48466. doi: 10.1074/jbc.M305602200. [DOI] [PubMed] [Google Scholar]
- 23.Draznin B. Molecular mechanisms of insulin resistance: Serine phosphorylation of insulin receptor substrate-1 and increased expression of p85alpha: the two sides of a coin. Diabetes. 2006;55:2392–2397. doi: 10.2337/db06-0391. [DOI] [PubMed] [Google Scholar]
- 24.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peterson TR, et al. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137:873–886. doi: 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lesniewski LA, et al. Bone marrow-specific Cap gene deletion protects against high-fat diet-induced insulin resistance. Nat Med. 2007;13:455–462. doi: 10.1038/nm1550. [DOI] [PubMed] [Google Scholar]
- 27.Luisi BF, et al. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. 1991;352:497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- 28.Bodine SC, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 29.Zhao W, et al. Dependence of dexamethasone-induced Akt/FOXO1 signaling, upregulation of MAFbx, and protein catabolism upon the glucocorticoid receptor. Biochem Biophys Res Commun. 2009;378:668–672. doi: 10.1016/j.bbrc.2008.11.123. [DOI] [PubMed] [Google Scholar]
- 30.Yu CY, et al. Genome-wide analysis of glucocorticoid receptor binding regions in adipocytes reveal gene network involved in triglyceride homeostasis. PLoS ONE. 2010;5:e15188. doi: 10.1371/journal.pone.0015188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang JC, Strömstedt PE, O’Brien RM, Granner DK. Hepatic nuclear factor 3 is an accessory factor required for the stimulation of phosphoenolpyruvate carboxykinase gene transcription by glucocorticoids. Mol Endocrinol. 1996;10:794–800. doi: 10.1210/mend.10.7.8813720. [DOI] [PubMed] [Google Scholar]
- 32.Miner JN, Yamamoto KR. Regulatory crosstalk at composite response elements. Trends Biochem Sci. 1991;16:423–426. doi: 10.1016/0968-0004(91)90168-u. [DOI] [PubMed] [Google Scholar]
- 33.Gross DN, van den Heuvel AP, Birnbaum MJ. The role of FoxO in the regulation of metabolism. Oncogene. 2008;27:2320–2336. doi: 10.1038/onc.2008.25. [DOI] [PubMed] [Google Scholar]
- 34.De Bosscher K, Vanden Berghe W, Haegeman G. The interplay between the glucocorticoid receptor and nuclear factor-kappaB or activator protein-1: Molecular mechanisms for gene repression. Endocr Rev. 2003;24:488–522. doi: 10.1210/er.2002-0006. [DOI] [PubMed] [Google Scholar]
- 35.Lee MC, Wee GR, Kim JH. Apoptosis of skeletal muscle on steroid-induced myopathy in rats. J Nutr. 2005;135:1806S–1808S. doi: 10.1093/jn/135.7.1806S. [DOI] [PubMed] [Google Scholar]
- 36.Hudlicka O, Brown M, Egginton S. Angiogenesis in skeletal and cardiac muscle. Physiol Rev. 1992;72:369–417. doi: 10.1152/physrev.1992.72.2.369. [DOI] [PubMed] [Google Scholar]
- 37.Logie JJ, et al. Glucocorticoid-mediated inhibition of angiogenic changes in human endothelial cells is not caused by reductions in cell proliferation or migration. PLoS ONE. 2010;5:e14476. doi: 10.1371/journal.pone.0014476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barbour LA, et al. Human placental growth hormone increases expression of the p85 regulatory unit of phosphatidylinositol 3-kinase and triggers severe insulin resistance in skeletal muscle. Endocrinology. 2004;145:1144–1150. doi: 10.1210/en.2003-1297. [DOI] [PubMed] [Google Scholar]
- 39.Mauvais-Jarvis F, et al. Reduced expression of the murine p85alpha subunit of phosphoinositide 3-kinase improves insulin signaling and ameliorates diabetes. J Clin Invest. 2002;109(1):141–149. doi: 10.1172/JCI13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng B, Ohkawa S, Li H, Roberts-Wilson TK, Price SR. FOXO3a mediates signaling crosstalk that coordinates ubiquitin and atrogin-1/MAFbx expression during glucocorticoid-induced skeletal muscle atrophy. FASEB J. 2010;24:2660–2669. doi: 10.1096/fj.09-151480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fujita T, Fukuyama R, Enomoto H, Komori T. Dexamethasone inhibits insulin-induced chondrogenesis of ATDC5 cells by preventing PI3K-Akt signaling and DNA binding of Runx2. J Cell Biochem. 2004;93:374–383. doi: 10.1002/jcb.20192. [DOI] [PubMed] [Google Scholar]
- 42.Andrade MV, Hiragun T, Beaven MA. Dexamethasone suppresses antigen-induced activation of phosphatidylinositol 3-kinase and downstream responses in mast cells. J Immunol. 2004;172:7254–7262. doi: 10.4049/jimmunol.172.12.7254. [DOI] [PubMed] [Google Scholar]
- 43.Luo J, et al. Loss of class IA PI3K signaling in muscle leads to impaired muscle growth, insulin response, and hyperlipidemia. Cell Metab. 2006;3:355–366. doi: 10.1016/j.cmet.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 44.Bandyopadhyay GK, Yu JG, Ofrecio J, Olefsky JM. Increased p85/55/50 expression and decreased phosphotidylinositol 3-kinase activity in insulin-resistant human skeletal muscle. Diabetes. 2005;54:2351–2359. doi: 10.2337/diabetes.54.8.2351. [DOI] [PubMed] [Google Scholar]
- 45.Waddell DS, et al. The glucocorticoid receptor and FOXO1 synergistically activate the skeletal muscle atrophy-associated MuRF1 gene. Am J Physiol Endocrinol Metab. 2008;295:E785–E797. doi: 10.1152/ajpendo.00646.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.