Abstract
Measurement of the naturally occurring hydrogen exchange (HX) behavior of proteins can in principle provide highly resolved thermodynamic and kinetic information on protein structure, dynamics, and interactions. The HX fragment separation-mass spectrometry method (HX-MS) is able to measure hydrogen exchange in biologically important protein systems that are not accessible to NMR methods. In order to achieve high structural resolution in HX-MS experiments, it will be necessary to obtain many sequentially overlapping peptide fragments and be able to identify and analyze them efficiently and accurately by mass spectrometry. This paper describes operations which, when applied to four different proteins ranging in size from 140 to 908 residues, routinely provides hundreds of useful unique peptides, covering the entire protein length many times over. Coverage in terms of the average number of peptide fragments that span each amino acid exceeds 10. The ability to achieve these results required the integrated application of experimental methods that are described here and a computer analysis program, called ExMS, described in a following paper.
Keywords: Hydrogen exchange, Protein, HX-MS, HDX
Introduction
The main chain amide NH hydrogen, one on every amino acid (except proline) in every protein molecule, engages in continual exchange with the hydrogens in water. Hydrogen exchange (HX) rates depend on pH, temperature, immediately neighboring side chains, and isotope effects in ways that are well understood [1–3], so that in the absence of protecting structure, HX rates can be accurately predicted. In structured proteins, the blocking of normal HX chemistry (H-bonding, sterics) can slow HX rates over five to 10 orders of magnitude. The exchange of structurally protected hydrogens is governed by dynamic structural fluctuations that transiently remove the structural barriers and expose the hydrogens to solvent where normal HX chemistry can proceed [4–6]. An understanding of these relationships together with well-designed HX measurements can provide a powerful experimental approach for the study of protein structure, structure change, stability, dynamics, and interactions, resolved to the level of individual amino acids [7–19].
Early HX methods were designed to measure the unresolved H-D exchange kinetics of whole proteins, initially by a mass/density method [20], then IR spectroscopy [21], and then by an accurate and efficient tritium labeling/gel filtration method [22, 23]. In efforts to achieve greater structural resolution, the tritium labeling method was extended to include a proteolytic fragmentation—HPLC separation stage [24–26] that could provide resolution to the level of protein subfragments, typically ranging from five to perhaps 30 residues in length. This method was limited by the inability to rapidly separate many proteolytic fragments by HPLC alone. The rise of two-dimensional (2D) NMR capabilities [27–29] led to the ability to measure HX directly at amino acid resolution [30, 31] and, for a time, eclipsed the further development of the fragmentation separation method. However, routine NMR analysis is limited to relatively small (< ~250 residues) highly soluble proteins that are available in quantity. It has been shown that HX investigations of larger and more demanding protein systems, reaching up even to massively aggregated systems like amyloid, can be achieved by adding a stage of mass spectrometric (MS) separation and analysis to the fragmentation method [10, 32–39]. As in the earlier H-T exchange method [26], proteins are exposed to H-D exchange labeling, samples are taken in time and quenched into a low pH, low temperature condition where exchange is largely halted. The protein is proteolyzed into fragments, they are partially resolved by fast HPLC, and the LC eluant is injected into a mass spectrometer for a second dimension of peptide separation and mass analysis.
A number of problems continue to limit the HX-MS method. Complete online analysis systems have been developed but they have proven to be surprisingly problematic. The measurement provides resolution only to the level of individual fragments, which may wind over some sizeable region in the native protein. The peptide fragments obtained have generally represented something less than the whole length of the protein and, within any given peptide fragment, one generally measures the exchange of only a fraction of the residues because the different amides typically span many orders of magnitude on the HX time axis [40]. Given these problems, one cannot distinguish where in the fragment or precisely where in the protein the hydrogens that are measured are placed. When a change in HX rate is detected, one cannot distinguish which residues in any given fragment change and which do not, or whether the observation detects some small change shared by many sites or a larger change due to some specific subset. Therefore, one is unable to infer the size, identity, energetics, or exact placement of the structure change.
Recent progress shows that the sequence resolution problem can be overcome by a top-down analysis in which the exchange-labeled protein is fragmented while in flight in the mass spectrometer. This approach has been plagued by the scrambling problem in which H and D atoms are redistributed due to the extreme bond vibrational energies that accompany collision induced dissociation (CID). The application of non-ergodic electron capture dissociation and electron transfer dissociation methods now accomplish near amino acid resolution [41–44] for relatively small proteins.
Alternatively, high structural coverage and resolution might be obtained by a bottom up fragmentation-separation approach if one could compare HX data for many sequentially overlapping fragments [45]. However, the ability to obtain very large numbers of peptide fragments and to efficiently analyze them in HX-MS experimentation has not been achieved. Here, we describe a system that can routinely provide many useful peptides, covering the protein length many times over. Also, we have written a computer program called ExMS that can analyze these many peptide fragments under conditions that are useful for HX-MS experiments.
This paper describes the methods used to obtain and validate large numbers of peptide fragments with four different proteins. A following paper describes the ExMS data analysis program [46]. These advances extend the capabilities of modern HX-MS experimentation and move toward ultimate amino acid resolution.
Experimental
Proteins
Three proteins—α-synuclein (140 residues), maltose binding protein (370 residues), and heat shock protein 104 (908 residues)—were expressed by recombinant methods in E. coli. Apolipoprotein A-1 (243 residues) was isolated from human blood. Purification followed previously described methods [47–50]. Proteases used for proteolytic fragmentation were porcine pepsin, fungal protease XIII, and the two in tandem (Sigma-Aldrich, St. Louis, MO, USA).
Hardware and System Details
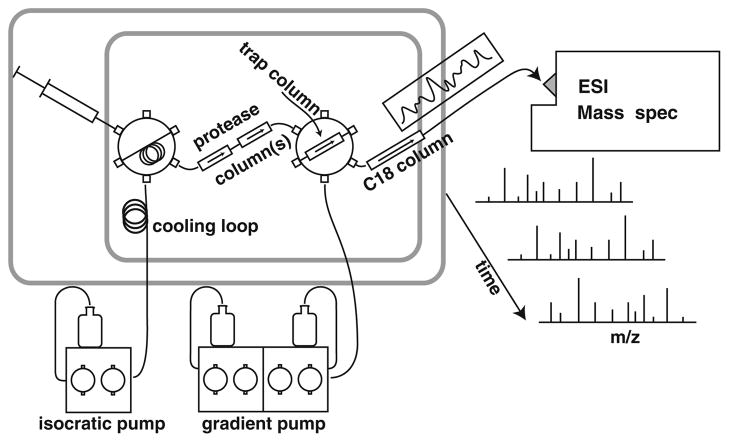
The online flow system used and system details and parameters are illustrated in Figure 1 and described below.
Figure 1.
On-line HX-MS analysis system. The entire flow system is contained within a Peltier cooled chamber (21× 15×25 cm; from an automobile accessory supplier) that maintains 0±1 °C, monitored with a thermocouple thermometer. An internal fan circulates air across the Peltier element and through the chamber. Liquid flow is precooled in a large loop positioned in the airflow. Switching valves are mounted on a ridged board backed with foam insulation with valve handles outside easily accessible for manual operation. The wetted parts of the valves, the injection loop, and columns are contained in the cold chamber. Liquid lines and the thermometer leads are threaded through small holes in the insulation and mounting board. A short length of outflow tubing from the C18 analytical column to the MS electrospray source is packed with ice-filled plastic bags
Experimental samples (~50 μL of ~1 μM protein in 0.1% formic acid at pH 2.5) are injected into the on-line flow system diagrammed in Figure 1. The flow system carries the protein solution at 50–120 μL/min through an immobilized acid protease column for proteolysis (15 or 60 μL total volume each). Digested peptides are directed through a second valve to a small C8 or C18 trap column (1×5 mm, 5 μm beads). After sufficient flow (~3 min at 50 μL/min) to transport the peptides into the trap column and wash away buffer salts, the second valve is switched, placing the trap column in the flow of a low volume HPLC pump. A water/acetonitrile gradient (6 μL/min, 10%–50% AcCN over 10–15 min) elutes peptides from the trap column through an analytical C18 column (0.3×50 mm, 3 μm beads) for rough peptide separation, and then to the electrospray needle for further separation of the peptides by mass. Narrow tubing (25 or 65 μm i.d.) is used in the slow flow chromatography stage. To minimize eluant peptide overlap, we use an acetonitrile gradient shaped to elute constant numbers of peptides per unit time. Typical peptide elution peak widths are 20 sec wide at baseline. We have not found peak widths to be limiting in the MS data even for hundreds of peptides, due especially to the ability of the ExMS analysis program [46] to recognize peptides in MS data even in the presence of significant spectral overlap.
Cleaning steps between serial experiments (several up–down gradients) elute very large peptides not useful for HX-MS experiments, helping to avoid peptide carryover from earlier MS runs and to maintain low column back pressure. If some peptides present a particularly difficult carryover problem [51], the availability of many other peptides makes it feasible to simply remove them from the experimental list. Overall, each experimental cycle takes 20–25 min followed by a 10–15 min cleaning cycle, resulting in a 40 min total time for each run. This allows for as many as 10 experimental HX runs each day in addition to an all-H run to calibrate chromatographic retention time for each peptide and an all-D run to calibrate their back exchange.
The flow rates and column sizes noted were chosen as a compromise between the competing demands of transit time, resolution, and back pressure. Particular care must be taken with flow system connections to assure against leaks under significant back pressure and to minimize unswept dead volume, which can lead to peak trailing and problems with peptide identification, although the ExMS analysis program operates to minimize this problem. Tubing is carefully cut, inspected, and connected, and ports are sprayed out with clean pressurized air on assembly. Common problems include slippage of connectors due to under-tightening, blockage due to over-tightening, and the trapping of particles at connections.
Proteolysis
The quenched experimental protein (pH~2.5, 0 °C) can be fragmented by adding an acid protease in a batch mode but passage through an immobilized protease column is more effective by far [52, 53]. When necessary, protein unfolding can be promoted and proteolysis improved by the inclusion of low concentrations of denaturant (urea, GdmCl, GdmSCN; ~0.5 M) or TCEP to promote disulfide reduction [54]. The protease columns used here are 1 or 2 mm×20 mm guard columns packed with POROS AL to which either pepsin or fungal protease XIII was coupled using Na2SO4 for salting out the protein during the coupling reaction at room temperature. It may be useful to gel-filter protease preparations to remove any contaminating amines such as Tris buffer, which will compete for coupling sites on the beads. Protease columns can be stably stored at 4 °C in pH 2.5 formic acid for months.
To maximize the number of overlapping peptides obtained, we use these two proteases, with broad but different specificities, separately and in tandem. Additional peptides can be produced by additional acid proteases with somewhat different specificity [18, 26]. At the flow rates noted, the protease column transit time, from 3 to 35 s, does not produce a final limit digest. Therefore, larger and smaller columns and slower and faster flow rates can be used to bias toward populations of shorter and longer peptides, respectively. These and other operations can increase the peptide numbers listed here.
Mass Spectrometer
The mass spectrometer used was a ThermoScientific LTQ Orbitrap XL (Thermo Fisher Scientific, Waltham, MA, USA) operated at a 60,000 resolution (~1 s/scan). The MS fragment separation capability is illustrated in Figure S1. Data were collected in profile mode with an AGC target of 106. The mass calibration was checked daily and recalibrated when necessary (<2 ppm rms deviation over 9 masses/100 scans in the calibration mix). Source parameters were: spray voltage 3.5 kV; capillary voltage 40 V; tube lens 170 V; capillary temperature 150 °C. MS/MS CID fragment ions were usually detected in the LTQ stage of the instrument (see main text) in centroid mode at normal scan rate with an AGC target value of 104. CID fragmentation was at 35% energy for 30 ms at Q of 0.25. HCD fragmentation energy was 35.
Results and Discussion
The proteolytic fragments described here were identified initially by data-dependent MS/MS experiments. Peptides in the MS/MS list were evaluated by a quality score (Ppep) that we independently calibrated to reduce the probability of false assignments to 1/1000. Large numbers of high scoring peptide fragments were found. We tested the ability of the ExMS analysis program [46] to find and definitively validate these many peptides in MS spectra under conditions that mimic MS-HX experiments with partially deuterated samples. The accuracy of the peptide fragment assignments was validated in a variety of ways. These steps are detailed below.
Peptide Identification
Protein fragments were prepared for MS/MS analysis by passage through the same online system designed for HX-MS experiments (Figure 1). Samples unfolded at the low pH quench condition used in HX-MS experiments were manually injected into the cold online flow system, where the protein was proteolyzed in immobilized acid protease columns, the peptide fragments were caught on a trap column, washed, and then gradient eluted through a small HPLC column directly into the mass spectrometer by electrospray for further mass analysis.
Starting with all-H protein samples, high resolution parent ion data were collected in the Orbitrap detector, selected peptide ions were CID-fragmented, and then detected in either the LTQ or Orbitrap stage. The four most intense peptide ions in each scan were selected for MS/MS analysis using a dynamic exclusion list to deselect already identified peptides. Three separate MS/MS runs were done for each proteolysis condition (pepsin, fungal protease XIII, the two in tandem), using an exclusion list for the second and third runs to avoid previously processed peptides with excellent Ppep scores (see below).
SEQUEST (ThermoScientific Bioworks 3.3.1) was used to analyze the MS/MS results against a large database of protein sequences including the four experimental proteins studied here and all potential contaminants, namely the acid proteases used, the list of proteins used in this lab, human and dog keratin, and the entire E. coli proteome because the experimental proteins were expressed in E. coli. An enlarged decoy set containing also the human proteome was used with apoA-1 because it was isolated from human blood. Search tolerance was 4 ppm for parent ions and 0.1 u for MS/MS fragment ions.
Ppep Calibration and Use
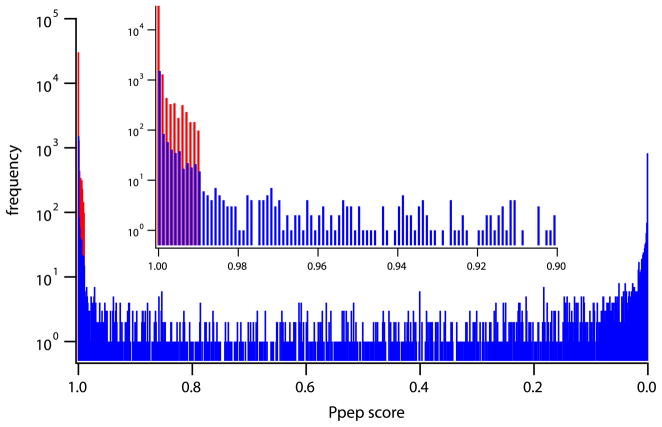
We used the Bioworks Ppep score to grade the quality of peptide MS/MS assignments. Ppep is a proprietary, relatively poorly documented statistical goodness of fit parameter provided in Bioworks. Although this quality parameter was designed for a different purpose, it proved to be useful here. We calibrated the ability of the Ppep score to distinguish false peptide identifications in the present system. MS/MS data were analyzed against a decoy sequence database with the amino acid sequence of all proteins reversed [55]. In this case, matches obtained clearly represent false identifications. Figure 2 shows in red the number distribution of false identifications plotted against their Ppep score. (Higher Ppep score indicates lower identification quality.) The red distribution in Figure 2 shows that in order to reduce the false identification probability to <1/1000, the Ppep cutoff score would have to be set at 0.990. We adopted this cutoff value.
Figure 2.
Distributions of Bioworks Ppep scores. Known false identifications, in red, represent SEQUEST hits for MS/MS data run against a large database with reversed sequences. Data in blue represent hits on the experimental protein for MS/MS data run against the large database with all proteins in their forward sequences. Results for the four proteins studied are merged to provide a large statistical sampling. The inset focuses on the poorest scoring matches. These results show that the cutoff Ppep value necessary to reduce false identifications to <0.1% is 0.990. (Other measures of assignment quality could be calibrated in the same way.)
The MS/MS data were also analyzed with the experimental protein and all other proteins in the large sequence database in their correct sequence. Figure 2 shows in blue the distribution of hits on the experimental protein versus Ppep score. A list of potentially useful peptides was assembled from the MS/MS results by rejecting apparent peptide matches with Ppep scores >0.990, which amounted to 40% of hits on the experimental proteins.
Contaminant peptides with scores below (better than) the cutoff score were found to originate only from the pepsin used (two overlapping C-terminal peptides). No E. coli protein had any peptide with a good score whether the experimental protein itself was included or omitted in the search database. In the case of apo A-I, hits were found on some other lipoproteins included in the human data base and seen at very low levels in PAGE analysis.
To confirm the usefulness of these many peptides for HX-MS experiments, another requirement is that the peptides must be found by the ExMS program [46] in MS spectra under HX-MS conditions. For each experimental protein, we did MS runs with fully protonated samples and with samples that had been equilibrated in 50% D2O solvent, just as for HX-MS experimentation. The ExMS analysis searches through the MS spectra for each peptide in turn and subjects each found peptide to a series of validation tests. The all-H MS runs provided an additional test for the correctness of the MS/MS identification of the peptides since their exact theoretical isotopic peak positions and envelope shape are known. ExMS directly found close to 100% of the listed peptides. Peptide identification is most challenging in the 50% D condition where isotopic envelope widths and therefore spectral overlap are maximized and m/z values to be expected for individual peaks are less well defined. These experiments provided the opportunity to test and optimize the various settable parameters in the ExMS program under the most difficult conditions. The optimized version of ExMS automatically found 50% to 75% of the peptide list, and most of the rest were easily determined by using the manual check function of the program. Peptides not definitively found (~20%) were removed from the peptide list.
Peptide Numbers
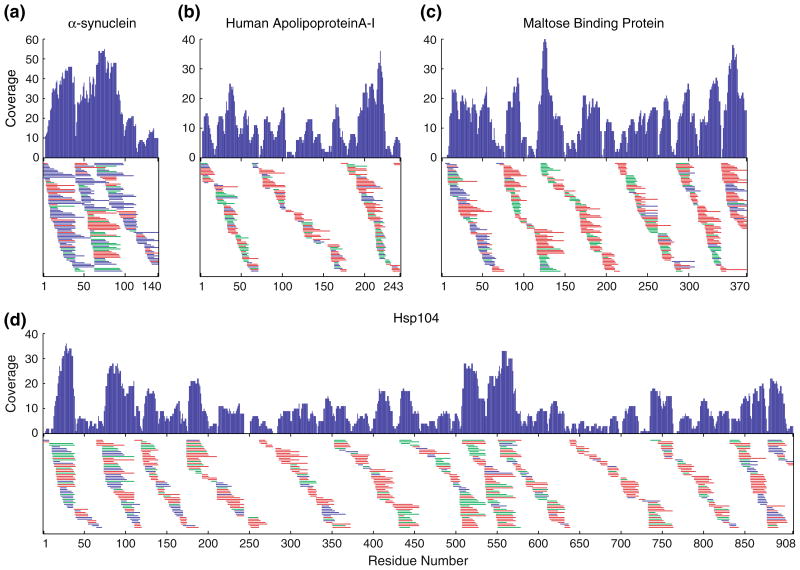
For the four proteins studied here, Figure 3 shows the unique peptides (many with more than one charge state, not shown) that were initially identified by MS/MS with Ppep<0.990 and were also found by ExMS in both all-H and 50% D MS runs. The number of useful peptides approach or exceed the number of amino acids in each protein, namely 222 from α-synuclein (140 amino acids), 239 for human apolipoprotein A-I (243 amino acids), 443 for maltose binding protein (370 amino acids), and 664 for heat shock protein 104 (908 amino acids). Figure 3 also shows the coverage per amino acid that is provided by the multiple overlapping peptides. The amino acid coverage obtained is variable, sometimes even falling to zero when high probability cut sites are closely spaced [56]. More importantly, the usual coverage per residue is large, averaging over 10 peptides per amino acid position.
Figure 3.
Unique peptides and amino acid coverage obtained for four proteins by the methods described here. Peptides are represented by horizontal bars. The different colors distinguish peptides obtained by proteolysis with porcine pepsin (red), fungal protease XIII (blue), and the two proteases in tandem (green). The coverage graphs show the number of peptides that span each amino acid residue position. For each protein, the list of peptides identified by MS/MS was culled to reduce potentially false identifications to one per thousand, and to eliminate those not definitively found by the ExMS program in MS spectra from runs with all-H protein and 50% D-labeled protein
Table S1 reports the survival history of peptides for each protein through the stages of identification and validation. The record indicates that these numbers could be substantially increased by the use of additional acid proteases and MS/MS runs, as well as manipulation of the degree of proteolysis (flow rate, protease column size, pH).
The Validity of MS/MS Peptide Identification
We empirically calibrated in our system a quality score (Ppep) against known incorrect MS/MS identifications and accepted only those assignments with a score that reduces the false positive probability to <1/1000. These peptides were further tested by ExMS to ensure that they exhibit their theoretically expected isotopic peaks and envelope shapes. The unusually large number of useful peptide fragments found raises the possibility that they may include incorrectly identified peptides or correctly identified peptides that in fact represent contaminant proteins. Accordingly, further tests were done.
To control for peptide fragments that might come from contaminants in our protein preparations, the SEQUEST analysis of MS/MS data was run against a large data base containing all possible contaminant proteins. Individual hits with good Ppep scores on any contaminant protein were rare, as expected from the analysis in Figure 2, with the exceptions noted before (two C-terminal pepsin fragments; known low level apolipoprotein contaminants). This was true whether the experimental protein was included in the comparison data base or not. Another indicator is that the number of peptides found for our different proteins increases in proportion to the size of the experimental protein (Figure 3), whereas the same contaminant decoy set was used for the different proteins except for the greatly expanded apoA-I set (the human proteome), which did not produce more hits.
The possibility of peptide production by in-source fragmentation was checked by increasing the source fragmentation voltage, and in other experiments the capillary and tube lens voltage. The number of peptides found did not change except at high source fragmentation voltage (>60 V) where the number fell precipitously.
Tests were done to check the quality of the MS/MS identifications. Figure S2 shows the number of b and y daughter ions found in MS/MS analyses of α-synuclein peptides with good Ppep score. The number averages about 1.5 times the number of residues in each peptide. Of the 543 peptides found (all three protease combinations), only one has a suspiciously low number of observed daughter ions.
In the MS/MS experiments described before, parent peptides selected based on Orbitrap scans were fragmented by CID and daughter ions were analyzed in the LTQ instrument. In tests done with α-synuclein, we fragmented by both HCD and CID and detected daughter ions in the Orbitrap. Of 129 previously identified peptic peptides derived from CID/LTQ analysis, 107 were validated by CID/Orbitrap analysis and 103 by HCD/Orbitrap analysis.
A large fraction (~70%) of our peptides were found in more than one charge state. The MS/MS analysis reached the same identification from these independent CID analyses. Almost all of the peptides paired in this way show very similar chromatographic retention time (ΔRT<5 s). For the very few retention time violations found, one peptide of the pair had poor Ppep score and appears to be due to low level chromatographic peak tailing.
MS/MS analysis is more difficult if two or more different peptides are co-fragmented by CID. The situation was checked for our largest protein where this problem will be maximized. For heat shock protein 104 cleaved by pepsin + fungal protease, CID/SEQUEST identified 564 peptides. Of these, 70 had co-eluting ions that fell within Δm/z=±0.5 of the maximum intensity peak and so were co-selected for fragmentation in the LTQ. Of these, 66 had excellent scores in the MS/MS analysis and four were simply good.
The fact that many groupings of peptides are found with common termini (Figure 3), due to high probability cut points, speaks for their accurate identification. We checked our pepsin cut sites against the list of prohibited sites found by Hamuro et al. [56] (no Arg, His, Lys, or Pro at the P1 site). One violation was found, at an Arg Phe site.
A severe test is offered by checking for consistency in HX-MS experiments. In H-D exchange experiments so far done, identical but differently charged peptides yielded results in excellent quantitative agreement, indicating that they were correctly identified. Among sequentially overlapping peptides, no inconsistencies have been found.
In summary, many independent measures document the reality and accurate identification of the large numbers of peptide fragments found in this work.
Peptide Inventory
It is commonly assumed that the number of useful peptide fragments in HX-MS experiments is limited by the small number of fragments produced by proteolysis. We inventoried the peptide fragments in a pepsin plus fungal protease digest of α-synuclein. A home-written program was used to search the MS spectra for “features” that appear to represent peptide fragments. There are thousands, spread over orders of magnitude in intensity. Analogous observations have been made before. We culled the list down to about 700 distinctly peptide-like features with non-redundant mono-isotopic mass. All were found to match some theoretical α-synuclein peptide.
The large number of peptides produced and observed that appear to represent true α-synuclein peptide fragments compares with the much smaller number identified in this work, namely 97 unique peptides in three MS/MS runs for α-synuclein (pepsin + fungal protease, Table S1). One assumes that a similar analysis would find similar numbers for other proteins. These results tend to change one’s perspective. The question is not how the many peptides that we find could possibly be present but rather why so few are commonly found of the many that are there. A partial answer can be found in the record of peptide identification in Table S1, which suggests that the numbers found might be easily increased.
Conclusions
To gain access to the large amount of structural, biophysical, and functional information that is in principle available from HX-MS experimentation, and ultimately to extend experimental HX-MS resolution to the amino acid level, it is most important to maximize the number of peptide fragments studied. Analysis shows that acid proteolysis produces many more peptides than have usually been identified (see inventory above). This paper describes methods that access and validate a number of peptide fragments that approximate the number of amino acids in each of four different proteins ranging in size from 140 to 908 residues. At the level of initial MS/MS identification, important determinants are the use of multiple independent proteases, multiple non-overlapping MS/MS runs, the reliability of the physical set up, and the resolution and sensitivity of the mass spectrometer. At the level of MS analysis, this work also helped to develop the ExMS computer analysis program and used it to efficiently and definitively find, validate, and characterize these many peptides under conditions pertinent for HX-MS experiments. These same approaches should be widely applicable in other laboratories. The ExMS program is described in a following paper [46].
Supplementary Material
Acknowledgments
The authors thank John Engen, Yoshitomu Hamuro, and Virgil Woods Jr. for exceptionally helpful comments on the work and on the manuscript. They gratefully acknowledge support by research grants from the NIH (GM031847), NSF (MCB1020649), and the Mathers Foundation.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s13361-011-0235-4) contains supplementary material, which is available to authorized users.
References
- 1.Bai Y, Milne JS, Mayne L, Englander SW. Primary Structure Effects on Peptide Group Hydrogen Exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Connelly GP, Bai Y, Jeng MF, Englander SW. Isotope Effects in Peptide Group Hydrogen Exchange. Proteins. 1993;17:87–92. doi: 10.1002/prot.340170111. [DOI] [PubMed] [Google Scholar]
- 3.Molday RS, Englander SW, Kallen RG. Primary Structure Effects on Peptide Group Hydrogen Exchange. Biochemistry. 1972;11:150–158. doi: 10.1021/bi00752a003. [DOI] [PubMed] [Google Scholar]
- 4.Hvidt A, Nielsen SO. Hydrogen Exchange in Proteins. Adv Protein Chem. 1966;21:287–386. doi: 10.1016/s0065-3233(08)60129-1. [DOI] [PubMed] [Google Scholar]
- 5.Linderstrøm-Lang K. Symposium on Protein Structure. Methuen; London: 1958. Deuterium Exchange and Protein structure. [Google Scholar]
- 6.Englander SW, Kallenbach NR. Hydrogen Exchange and Structural Dynamics of Proteins and Nucleicacids. Q Rev Biophys. 1983;16:521–655. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]
- 7.Kang S, Prevelige P. Mass Spectrometry Analysis for Protein–Protein Interactions and Dynamics. John Wiley and Sons; Hoboken, NJ: 2008. Hydrogen/deuterium exchange studies in viruses; pp. 105–122. [Google Scholar]
- 8.Gross ML, Zhou M, Hambly D. Mass SApectrometry analysis for Protein–Protein Interactions and Dynamics. John Wiley and Sons; Hoboken, NJ: 2008. Two Approaches to Mass Spectrometric Protein Footprinting: PLIMSTEX and FPOP; pp. 243–270. [Google Scholar]
- 9.Hamuro Y, Coales S, Hamuro L, Woods VL., Jr . Mass Spectrometry Analysis for Protein–Protein Interactions and Dynamics. John Wiley and Sons; Hoboken, NJ: 2008. Use of Enhanced Peptide Amide Hydrogen/Deuterium Exchange-Mass Spectrometry (DXMS) in the Examination of Protein–Protein Interactions; pp. 123–156. [Google Scholar]
- 10.Komives EA. Mass Spectrometry Analysis for Protein–Protein Interactions and Dynamics. John Wiley and Sons; Hoboken, NJ: 2008. Deuterium Exchange Approaches for Examining Protein Interactions: Case Studies of Complex Formation; pp. 91–104. [Google Scholar]
- 11.Tang L, Hopper E, Tong Y, Sadowsky J, Peterson K, Gellman S, Fitzgerald M. H/D Exchange- and Mass Sectrometry-Based Strategy for the Thermodynamic Analysis of Protein–Ligand Interactions. Anal Chem. 2007;79:5869–5877. doi: 10.1021/ac0700777. [DOI] [PubMed] [Google Scholar]
- 12.Dai S, Fitzgerald M. A mass spectrometry-based probe of equililibrium intermediates in protein folding reactions. Biochemistry. 2006;45:12890–12897. doi: 10.1021/bi061295h. [DOI] [PubMed] [Google Scholar]
- 13.Engen JR. Analysis of Protein Conformation and Dynamics by Hydrogen/Deuterium Exchange MS. Anal Chem. 2009;81:7870–7875. doi: 10.1021/ac901154s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaveti S, Engen JR. Protein Interactions Probed with Mass Spectrometry. Methods Mol Biol. 2006;316:179–197. doi: 10.1385/1-59259-964-8:179. [DOI] [PubMed] [Google Scholar]
- 15.Engen JR. Analysis of Protein Complexes with Hydrogen Exchange and Mass Spectrometry. Analyst. 2003;128:623–628. doi: 10.1039/b212800b. [DOI] [PubMed] [Google Scholar]
- 16.Garcia RA, Pantazatos D, Villarreal FJ. Hydrogen/Deuterium Exchange Mass Spectrometry for Investigating Protein–Ligand Interactions. Assay Drug Devel Tech. 2004;2:81–91. doi: 10.1089/154065804322966342. [DOI] [PubMed] [Google Scholar]
- 17.Tsutsui Y, Wintrode PL. Hydrogen/deuterium, exchange-mass spectrometry: A powerful tool for probing protein structure, dynamics and interactions. Current Med Chem. 2007;14:2344–2358. doi: 10.2174/092986707781745596. [DOI] [PubMed] [Google Scholar]
- 18.Wales TE, Engen JR. Hydrogen Exchange Mass Spectrometry for the Analysis of Protein Dynamics. Mass Spectrom Rev. 2006;25:158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 19.Kaltashov IA, Eyles SJ. Studies of Biomolecular Conformations and Conformational Dynamics by Mass Spectrometry. Mass Spectrom Rev. 2002;21:37–71. doi: 10.1002/mas.10017. [DOI] [PubMed] [Google Scholar]
- 20.Linderstrom-Lang KU. Deuterium Exchange Between Peptides and Water. Chem Soc Spec Publ. 1955;2:1–20. [Google Scholar]
- 21.Hvidt A, Kagi JHR, Ottesen M. Effect of Oxidized Nicotinamide-Adenine Dinucleotide on Hydrogen-Deuterium Exchange of Yeast Alcohol Dehydrogenase as Measured by Infrared Spectrophotometry. Biochim Biophys Acta. 1963;75:290–296. doi: 10.1016/0006-3002(63)90614-0. [DOI] [PubMed] [Google Scholar]
- 22.Englander SWA. Hydrogen Exchange Method Using Tritium and Sephadex. Application to Ribonuclease. Biochemistry. 1963;2:798–807. doi: 10.1021/bi00904a030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Englander SW, Downer NW, Teitelbaum H. Hydrogen Exchange. Ann Rev Biochem. 1972;41:903–924. doi: 10.1146/annurev.bi.41.070172.004351. [DOI] [PubMed] [Google Scholar]
- 24.Rosa JJ, Richards FM. An Experimental Procedure for Increasing the Structural Resolution of Chemical Hydrogen Exchange Measurements on Proteins: Application to Ribonuclease S Peptide. J Mol Biol. 1979;133:399–416. doi: 10.1016/0022-2836(79)90400-5. [DOI] [PubMed] [Google Scholar]
- 25.Englander SW, Calhoun DB, Englander JJ, Kallenbach NR, Liem RKH, Malin EL, Mandal C, Rogero JR. Individual Breathing Reactions Measured in Hemoglobin by Hydrogen Exchange Methods. Biophys J. 1980;32:577–590. doi: 10.1016/S0006-3495(80)84991-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Englander JJ, Rogero JR, Englander SW. Protein Hydrogen Exchange Studied by the Fragment Separation Method. Anal Biochem. 1985;147:234–244. doi: 10.1016/0003-2697(85)90033-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagayama K, Kumar A, Wüthrich K, Ernst RR. Experimental Techniques of Two-Dimensional Correlated Spectroscopy. J Magn Res. 1980;40:321–334. [Google Scholar]
- 28.Sorensen OW, Eich GW, Levitt MH, Bodenhausen G, Ernst RR. Product Operator Formalism for the Description of NMR Pulse Experiments. Prog NMR Spectrom. 1984;16:163–192. [Google Scholar]
- 29.Bax A, Lerner L. Two-Dimensional NMR Spectroscopy. Science. 1986;232:960–967. doi: 10.1126/science.3518060. [DOI] [PubMed] [Google Scholar]
- 30.Wagner G, Wuthrich K. Amide Proton Exchange and Surface Conformation of BPTI in Solution: Studies with 2D NMR. J Mol Biol. 1982;160:343–361. doi: 10.1016/0022-2836(82)90180-2. [DOI] [PubMed] [Google Scholar]
- 31.Wand AJ, Englander SW. Two-Dimensional Proton NMR Studies of Cytochrome c. Biochemistry. 1985;24:5290–5294. doi: 10.1021/bi00341a002. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Z, Smith DL. Determination of Amide Hydrogen Exchange by Mass Spectrometry: A New Tool for Protein Structure Elucidation. Protein Sci. 1993;2:522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoofnagle AN, Resing KA, Ahn NG. Protein Analysis by Hydrogen Exchange Mass Spectrometry. Ann Rev Biophys Biomol Struct. 2003;18:18. doi: 10.1146/annurev.biophys.32.110601.142417. [DOI] [PubMed] [Google Scholar]
- 34.Kaltashov IA, Eyles SJ. Mass Spectrometry in Biophysics: Conformation and Dynamics of Biomolecules. John Wiley; Hoboken, NJ: 2005. p. 458. [Google Scholar]
- 35.Kaltashov IA, Engen JR, Gross ML. Hydrogen Exchange and Covalent Modification: Focus on Biomolecular Structure, Dynamics, and Function. J Am Soc Mass Spectrom. 2006;17:I–II. [Google Scholar]
- 36.Konermann L, Tong X, Pan Y. Protein Structure and Dynamics Studied by Mass Spectrometry: H/D Exchange, Hydroxyl Radical Labeling, and Related Approaches. J Mass Spectrom. 2008;43:1021–1036. doi: 10.1002/jms.1435. [DOI] [PubMed] [Google Scholar]
- 37.Yan X, Maier CS. Hydrogen/Deuterium Exchange Mass Spectrometry. Methods Mol Biol. 2009;492:255–271. doi: 10.1007/978-1-59745-493-3_15. [DOI] [PubMed] [Google Scholar]
- 38.Marcsisin SR, Engen JR. Hydrogen Exchange Mass Spectrometry: What Is It and What Can It Tell Us? Analyt Bioanalyt Chem. 2010;397:967–972. doi: 10.1007/s00216-010-3556-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Del Mar C, Greenbaum EA, Mayne L, Englander SW, Woods VL. Structure and Properties of α-Synuclein and Other Amyloids Determined at the Amino Acid Level. Proc Natl Acad Sci USA. 2005;102:15477–15482. doi: 10.1073/pnas.0507405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coales SJ, Sook Yen E, Lee JE, Ma A, Morrow JA, Hamuro Y. Expansion of Time Window for Mass Spectrometric Measurement of Amide Hydrogen/Deuterium Exchange Reactions. Rapid Commun Mass Spectrom. 2010;24:3585–3592. doi: 10.1002/rcm.4814. [DOI] [PubMed] [Google Scholar]
- 41.Pan JX, Han J, Borchers CH, Konermann L. Hydrogen/Deuterium Exchange Mass Spectrometry with Top-Down Electron Capture Dissociation for characterizing Structural Transitions of a 17 kDa Protein. J Am Chem Soc. 2009;131:12801–12808. doi: 10.1021/ja904379w. [DOI] [PubMed] [Google Scholar]
- 42.Abzalimov RR, Kaplan DA, Easterling ML, Kaltashov IA. Protein Conformations Can be Probed in Top-Down HDX MS Experiments Utilizing Electron Transfer Dissociation of Protein Ions without Hydrogen Scrambling. J Am Soc Mass Spectrom. 2009;20:1514–1517. doi: 10.1016/j.jasms.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rand KD, Zehl M, Jensen ON, Jorgensen TJD. Protein Hydrogen Exchange Measured at Single-Residue Resolution by Electron Transfer Dissociation Mass Spectrometry. Anal Chem. 2009;81:5577–5584. doi: 10.1021/ac9008447. [DOI] [PubMed] [Google Scholar]
- 44.Zehl M, Rand KD, Jensen ON, Jorgensen TJD. Electron Transfer Dissociation Facilitates the Measurement of Deuterium Incorporation into Selectively Labeled Peptide with Single Residue Resolution. J Am Chem Soc. 2008;130:17453–17459. doi: 10.1021/ja805573h. [DOI] [PubMed] [Google Scholar]
- 45.Althaus E, Canzar S, Ehrler C, Emmett M, Karrenbauer A, Marshall A, Meyer-Base A, Tipton J, Zhang H. Computing H-D Exchange Rates of Single Residues from Data of Proteolytic Fragments. BMC Bioinformatics. 2010;11:424. doi: 10.1186/1471-2105-11-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kan ZY, Mayne L, Chetty PS, Englander SW. ExMS: Data Analysis for HX-MS Experiments. J Am Soc Mass Spectrom. 2011 doi: 10.1007/s13361-011-0236-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chun S, Strobel S, Bassford PJ, Randall L. Folding of Maltose Binding Protein. Evidence for the Identity of the Rate-Determining Step In Vivo and In Vitro. J Biol Chem. 1993;268:20855–20862. [PubMed] [Google Scholar]
- 48.Weisweiler P. Isolation and Quantitation of Apolipoproteins A-I and A-II from Human High-Density Lipoproteins by Fast-Protein Liquid Chromatography. Clin Chim Acta. 1987;169:249–254. doi: 10.1016/0009-8981(87)90325-1. [DOI] [PubMed] [Google Scholar]
- 49.Giasson BI, Murray IV, Trojanowski JQ, Lee VM. A Hydrophobic Stretch of 12 Amino Acid Residues in the Middle of α-Synuclein is Essential for Filament Assembly. J Biol Chem. 2001;276:2380–2386. doi: 10.1074/jbc.M008919200. [DOI] [PubMed] [Google Scholar]
- 50.Shorter J, Lindquist S. Hsp104 Catalyzes Formation and Elimination of Self-Relicating Sup35 Prion Conformers. Science. 2004;304:1793–1797. doi: 10.1126/science.1098007. [DOI] [PubMed] [Google Scholar]
- 51.Fang J, Rand KD, Beuning PJ, Engen JR. False EX1 signatures caused by sample carryover during HXMS analyses. Int J Mass Spectrom. 2011;302:19–25. doi: 10.1016/j.ijms.2010.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu Y, Kaveti S, Engen JR. Extensive Deuterium Back-Exchange in Certain Immobilized Pepsin Columns Used for H/D Exchange Mass Spectrometry. Anal Chem. 2006;78:1719–1723. doi: 10.1021/ac0518497. [DOI] [PubMed] [Google Scholar]
- 53.Wang LT, Pan H, Smith DL. Hydrogen Exchange-Mass spectrometry—Optimization of Digestion Conditions. Mol Cell Proteom. 2002;1:132–138. doi: 10.1074/mcp.m100009-mcp200. [DOI] [PubMed] [Google Scholar]
- 54.Hamuro Y, Burns L, Canaves J, Hoffman R, Taylor S, Woods V. Domain Organization of d-akap2 Revealed by Enhanced Deuterium Exchange-Mass Spectrometry (DXMS) J Mol Biol. 2002;321:703–714. doi: 10.1016/s0022-2836(02)00419-9. [DOI] [PubMed] [Google Scholar]
- 55.Hather G, Higdon R, Bauman A, von Haller PD, Kolker E. Estimating False Discovery Rates for Peptide and Protein Identification Using Randomized Databases. Proteomics. 2010;10:2369–2376. doi: 10.1002/pmic.200900619. [DOI] [PubMed] [Google Scholar]
- 56.Hamuro Y, Coales SJ, Molnar KS, Tuske SJ, Morrow JA. Specificity of Immobilized Porcine Pepsin in H/D Exchange Compatible Conditions. Rapid Commun Mass Spectrom. 2008;22:1041–1046. doi: 10.1002/rcm.3467. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.