Abstract
Purpose
Lynch syndrome accounts for 2–5% of endometrial cancer cases. Lynch syndrome prediction models have not been evaluated among endometrial cancer cases.
Methods
Area under the receiver operating curve (AUC), sensitivity and specificity of PREMM1,2,6, MMRpredict, and MMRpro scores were assessed among 563 population-based and 129 clinic-based endometrial cancer cases.
Results
A total of 14 (3%) population-based and 80 (62%) clinic-based subjects had pathogenic mutations. PREMM1,2,6, MMRpredict, and MMRpro were able to distinguish mutation carriers from noncarriers (AUC of 0.77, 0.76, and 0.77, respectively), among population-based cases. All three models had lower discrimination for the clinic-based cohort, with AUCs of 0.67, 0.64, and 0.54, respectively. Using a 5% cutoff, sensitivity and specificity were as follows: PREMM1,2,6, 93% and 5% among population-based cases and 99% and 2% among clinic-based cases; MMRpredict, 71% and 64% for the population-based cohort and 91% and 0% for the clinic-based cohort; and MMRpro, 57% and 85% among population-based cases and 95% and 10% among clinic-based cases.
Conclusion
Currently available prediction models have limited clinical utility in determining which patients with endometrial cancer should undergo genetic testing for Lynch syndrome. Immunohistochemical analysis and microsatellite instability testing may be the best currently available tools to screen for Lynch syndrome in endometrial cancer patients.
Keywords: endometrial cancer, genetic screening, genetic testing, Lynch syndrome, prediction models
INTRODUCTION
Lynch syndrome (LS), an inherited syndrome caused by mutations in the mismatch repair (MMR) genes MLH1, MSH2, MSH6, and PMS2,1 is characterized by an increased susceptibility to colorectal cancer (CRC), endometrial cancer (EC), and other malignancies.2 Gene mutation carriers can present with EC as their sentinel malignancy.3 Among women with EC, it is important to identify those who may have LS because these women have a high likelihood of developing a second cancer.4,5 Predictive germline testing is being used clinically to identify mutation carriers who would benefit from increased cancer surveillance as well as to allow presymptomatic genetic testing for family members.6
Given the 2–5% prevalence of LS among EC cases, germline testing of all EC cases is not practical because of its high cost.7,8 Preselecting those who should undergo germline testing is more appropriate and cost-effective.3 Molecular tumor testing, including immunohistochemistry (IHC) or microsatellite instability (MSI) analysis, has been proposed as a practical first step in evaluating women suspected to be at risk for LS.7 However, this approach is not routinely performed for EC. Until molecular testing becomes routine for all EC cases, the utility of predictive tools that utilize personal and family cancer history must be explored.
Clinical prediction rules were recently developed to aid clinicians in identifying patients who should undergo genetic testing for LS. These include PREMM,9,10 MMRpredict,11 and MMRpro,12 which all provide a quantitative risk estimate of having a mutation and may inform clinicians on how to proceed with genetic evaluation. Although the performance of these models has been validated in a number of CRC cohorts13–18 and revealed excellent discriminative ability with high sensitivity and specificity in both population- and clinic-based CRC cases, their performance has not been assessed among EC cases. The purpose of this study was to evaluate the performance of the PREMM1,2,6, MMRpredict, and MMRpro models in distinguishing MMR gene mutation carriers from noncarriers among population- and clinic-based EC cases.
MATERIALS AND METHODS
Population-based cases
We analyzed data from 563 unselected subjects with EC enrolled through the Ohio State University Columbus-area LS study from January 1999 to December 2003.6 All subjects provided detailed personal and family cancer history data, and available endometrial tumor specimens were evaluated for MSI (N = 560). IHC staining for loss of protein expression of the MMR genes (MLH1, MSH2, MHS6, and PMS2) was conducted for tumors displaying MSI or when subjects met one of the following criteria: (i) diagnosis at age <50 years, (ii) synchronous or metachronous CRC and EC primaries, and (iii) presence of a first-degree relative with EC or CRC diagnosed at any age. A subset of microsatellite stable tumors were also evaluated by IHC (N = 223). Subjects whose tumors demonstrated MSI and/or abnormal IHC staining underwent germline genetic testing. Molecular tumor testing and mutation analysis were performed using the methods described previously by Hampel et al.6
High-risk clinic-based cases
Data from 129 families affected with EC and recruited through the familial cancer (high-risk) clinics participating in the Colon Cancer Family Registry were analyzed. High-risk clinic-based probands enrolled into the Colon Cancer Family Registry fulfilled one or more of the following eligibility criteria: two or more relatives with a personal history of CRC or LS cancer, a proband diagnosed with CRC at a young age, or a proband presenting at a cancer clinic with LS or Lynch-like syndrome. Cancer-affected relatives and selected unaffected relatives up to at least the second degree were subsequently recruited. A detailed overview of the design and methods pertaining to the Colon Cancer Family Registry was published by Newcomb et al.19 and is available at http://epi.grants.cancer.gov/CFR/.
High-risk families were recruited from three of the six sites participating in the Colon Cancer Family Registry: Mayo Clinic, University of Southern California Consortium, and University of Melbourne (Australasia). Of the 129 unrelated EC cases analyzed, 24 and 74 had MSI and IHC results, respectively, and 70 cases had undergone germline testing. Of the 59 EC cases who did not have germline testing, those who had a first-degree relative with a known deleterious MMR gene mutation were assumed to have an MMR gene mutation, whereas those who had a first-degree relative with a negative germline testing result were assumed not to have an MMR gene mutation. Molecular tumor testing and mutation analysis were performed using the methods described previously.19–21
Statistical analysis
We calculated the risk scores using the PREMM1,2,6, MMRpredict, and MMRpro models. The PREMM1,2,6 score was generated for all subjects using proband-specific variables: gender, history and ages of CRC and EC, and history of other LS-associated cancers (ovary, stomach, small intestine, urinary tract/kidney, bile ducts, glioblastoma multiforme, sebaceous gland tumors, and pancreas). Family history data included the number of first- and second-degree relatives with a history of CRC, EC, and other LS-associated tumors, as well as the youngest ages of diagnosis of CRC and EC.
The variables in the MMRpredict model include age at diagnosis of CRC, gender, location of tumor (proximal versus distal), synchronous or metachronous tumors, history and youngest age of diagnosis of CRC in first-degree relatives, and history of EC in any first-degree relative. Because the MMRpredict model does not account for extracolonic tumors, we used age at diagnosis of EC in place of CRC age. Because location of tumor does not apply to EC, we calculated two sets of MMRpredict scores: one score considered EC a proximal tumor, and the second score considered EC a distal tumor. We generated MMRpredict scores, without considering MSI or IHC, for 156 population-based and 99 clinic-based subjects with EC diagnosis age <55 years to replicate the CRC cohort in which MMRpredict was developed.
The MMRpro model uses the following variables for the proband and first- and second-degree relatives: ages at diagnosis of colorectal and ECs and current age or age at last follow-up for those unaffected by CRC or EC. For relatives who were diagnosed with cancer but did not provide an age at diagnosis, we used the mean age at diagnosis of all patients in each cohort (for CRC, 63 years for population-based cases and 50 years for clinic-based cases; for EC, 62 years for population-based cases and 49 years for clinic-based cases). For those unaffected by cancer without a current age or age at last follow-up, we estimated their date of birth (DOB) as follows: (i) for siblings, the proband’s DOB was used; (ii) for children, nieces, and nephews, a date 30 years later than the proband’s DOB was used; (iii) for parents, aunts, and uncles, a date 25 years before the proband’s DOB was used, and (iv) for grandparents, a date 50 years before the proband’s DOB was used. MMRpro scores, with and without tumor testing results, were generated using the BayesMendel R package for MMRpro22,23 for all clinic-based subjects and for 232 population-based subjects who had a complete family history up to the second-degree relatives.
For each of the three prediction models, discrimination between gene mutation carriers and noncarriers was quantified using the area under the receiver operating curve (AUC) with 95% confidence intervals (CIs). Calibration was assessed by comparing the average predictions from each model with the observed prevalence of mutations. We also obtained the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) using the cut-off levels in the original model development for PREMM1,2,6: ≥5%, ≥10%, ≥20%, and ≥40%, and for MMRpredict: ≥0.5%, ≥5%, ≥20%, and ≥45%. Cut-off levels for MMRpro were chosen arbitrarily to match those of PREMM1,2,6. These results were compared with the sensitivity and specificity of MSI and IHC. All calculations were performed using SAS software, version 9.1 (SAS Institute, Cary, NC).
This study was approved by the Dana-Farber/Harvard Cancer Center institutional review board.
RESULTS
Patient characteristics
A total of 692 EC cases were included in this study: 563 (81%) were population-based cases and 129 (19%) were ascertained through high-risk familial cancer clinics. Among the 563 population-based EC cases, 14 (2.5%) had pathogenic mutations: 2 (14%) in MLH1, 3 (21%) in MSH2, and 9 (64%) in MSH6. In the clinic-based cohort, 80/129 (62%) subjects had pathogenic mutations: 31 (39%) in MLH1, 40 (50%) in MSH2, and 9 (11%) in MSH6. Table 1 presents the subject characteristics stratified by mutation carrier status.
Table 1.
Clinical characteristics of probands, total and by mutation status, in the population- and clinic-based cohorts
| Characteristics | Population-based cohort (N = 563) | Clinic-based cohort (N = 129) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | No mutation | With mutation | P value | Total | No mutation | With mutation | P value | |||||||
| N | (%) | N | (%) | N | (%) | N | (%) | N | (%) | N | (%) | |||
| Total | 563 | (100) | 549 | (97) | 14 | (3) | 129 | (100) | 49 | (38) | 80 | (62) | — | |
| Age at last follow-up, median (range) | 68 | (22–96) | 68 | 61 | 0.03 | 63 | (38–89) | 66 | 61 | 0.11 | ||||
| Living | 463 | (82) | 451 | (82) | 12 | (86) | 1.00 | 42 | (51) | 16 | (46) | 24 | (51) | 0.63 |
| Race | 0.21 | |||||||||||||
| Caucasian | 534 | (95) | 522 | (95) | 12 | (86) | 94 | (73) | 29 | (59) | 65 | (81) | 0.01 | |
| African American | 19 | (3) | 17 | (3) | 2 | (14) | 0 | 0 | — | — | — | — | — | |
| Hispanic | 3 | (0.5) | 3 | (0.6) | 0 | (0) | 1 | (1) | 0 | (0) | 1 | (1) | 0.43 | |
| Asian | 6 | (1) | 6 | (1) | 0 | (0) | 1 | (1) | 1 | (2) | 0 | (0) | 0.20 | |
| Other | 1 | (0.2) | 1 | (0.2) | 0 | (0) | 2 | (2) | 0 | (0) | 2 | (3) | 0.43 | |
| clinical criteria | ||||||||||||||
| Amsterdam II | 17 | (3) | 13 | (2) | 4 | (29) | <.001 | 90 | (70) | 31 | (63) | 59 | (74) | 0.21 |
| Revised Bethesda | 19 | (3) | 16 | (3) | 3 | (21) | 0.01 | 84 | (65) | 25 | (51) | 59 | (74) | 0.01 |
| Proband cancer history | ||||||||||||||
| cRc | ||||||||||||||
| 0 | 555 | (99) | 543 | (99) | 12 | (86) | 0.01 | 95 | (74) | 39 | (80) | 56 | (70) | 0.35 |
| 1 | 7 | (1) | 5 | (1.0) | 2 | (14) | 20 | (16) | 7 | (14) | 13 | (16) | ||
| ≥2 | 1 | (0.2) | 1 | (0.2) | 0 | (0) | 14 | (11) | 3 | (6) | 11 | (14) | ||
| Adenoma | 2 | (0.4) | 2 | (0.4) | 0 | (0) | 1.00 | 6 | (5) | 3 | (6) | 3 | (4) | 0.53 |
| Other LS cancers | 11 | (2) | 10 | (2) | 1 | (7) | 0.24 | 33 | (26) | 7 | (14) | 26 | (33) | 0.02 |
| Multiple LS cancers | 19 | (3) | 16 | (3) | 3 | (21) | 0.01 | 37 | (29) | 10 | (20) | 27 | (34) | 0.10 |
| Mean youngest ages of diagnosis (±SD) | ||||||||||||||
| CRC | 56 | (±15) | 60 | (±15) | 43 | (±4) | 0.19 | 47 | (±12) | 49 | (±14) | 47 | (±11) | 0.38 |
| Endometrial | 61 | (±12) | 61 | (±12) | 53 | (±10) | 0.01 | 48 | (±9) | 49 | (±12) | 48 | (±7) | 0.51 |
| Other LS cancers | 55 | (±10) | 54 | (±10) | 66 | (—) | 0.26 | 57 | (±10) | 52 | (±9) | 58 | (±10) | 0.22 |
| Family cancer history | ||||||||||||||
| CRC | ||||||||||||||
| Any FDR/SDR | 0.09 | 0.85 | ||||||||||||
| No | 441 | (78) | 433 | (79) | 8 | (57) | 14 | (11) | 5 | (10) | 9 | (11) | ||
| Yes | 122 | (22) | 116 | (21) | 6 | (43) | 115 | (89) | 44 | (90) | 71 | (89) | ||
| Number of FDR | 0.05 | 0.29 | ||||||||||||
| 0 | 470 | (84) | 461 | (84) | 9 | (64) | 20 | (16) | 6 | (12) | 14 | (18) | ||
| 1 | 83 | (15) | 79 | (14) | 4 | (29) | 42 | (33) | 20 | (41) | 22 | (28) | ||
| ≥2 | 10 | (2) | 9 | (2) | 1 | (7) | 66 | (52) | 23 | (47) | 43 | (54) | ||
| Number of SDR | 0.02 | 0.01 | ||||||||||||
| 0 | 523 | (93) | 512 | (93) | 11 | (79) | 78 | (61) | 36 | (74) | 42 | (53) | ||
| 1 | 31 | (5) | 30 | (6) | 1 | (7) | 25 | (19) | 10 | (20) | 15 | (19) | ||
| ≥2 | 9 | (2) | 7 | (1) | 2 | (14) | 25 | (19) | 3 | (6) | 22 | (28) | ||
| Endometrial cancer | ||||||||||||||
| Any FDR/SDR | 0.01 | 0.93 | ||||||||||||
| No | 497 | (88) | 489 | (89) | 8 | (57) | 81 | (63) | 31 | (63) | 50 | (62) | ||
| Yes | 66 | (12) | 60 | (11) | 6 | (43) | 48 | (37) | 18 | (37) | 30 | (38) | ||
| Number of FDR | 0.01 | 0.55 | ||||||||||||
| 0 | 521 | (92) | 511 | (93.1) | 10 | (72) | 90 | (70) | 37 | (76) | 53 | (67) | ||
| 1 | 39 | (7) | 36 | (6.6) | 3 | (21) | 33 | (26) | 10 | (20) | 23 | (29) | ||
| ≥2 | 3 | (1) | 2 | (0.4) | 1 | (7) | 5 | (4) | 2 | (4) | 3 | (4) | ||
| Number of SDR | 0.17 | 0.63 | ||||||||||||
| 0 | 534 | (95) | 522 | (95) | 12 | (86) | 114 | (89) | 42 | (86) | 72 | (91) | ||
| 1 | 28 | (5) | 26 | (5) | 2 | (14) | 12 | (9) | 6 | (12) | 6 | (8) | ||
| ≥2 | 1 | (0.2) | 1 | (0.2) | 0 | (0) | 2 | (2) | 1 | (2) | 1 | (1) | ||
| Other LS cancer | ||||||||||||||
| Any FDR/SDR | 0.01 | 0.87 | ||||||||||||
| No | 434 | (77) | 428 | (78) | 6 | (43) | 62 | (48) | 24 | (49) | 38 | (47) | ||
| Yes | 129 | (23) | 121 | (22) | 8 | (57) | 67 | (52) | 25 | (51) | 42 | (53) | ||
| Number of FDR | <0.001 | 0.88 | ||||||||||||
| 0 | 481 | (85) | 470 | (86) | 11 | (79) | 78 | (60) | 31 | (63) | 47 | (59) | ||
| 1 | 72 | (13) | 72 | (13) | 0 | (0) | 37 | (29) | 13 | (27) | 24 | (30) | ||
| ≥2 | 10 | (2) | 7 | (1) | 3 | (21) | 14 | (11) | 5 | (10) | 9 | (11) | ||
| Number of SDR | 0.003 | 0.64 | ||||||||||||
| 0 | 505 | (90) | 497 | (90) | 8 | (57) | 103 | (80) | 39 | (80) | 64 | (80) | ||
| 1 | 48 | (8) | 43 | (8) | 5 | (36) | 18 | (14) | 8 | (16) | 10 | (13) | ||
| ≥2 | 10 | (12) | 9 | (2) | 1 | (7) | 8 | (6) | 2 | (4) | 6 | (7) | ||
| Mean youngest ages of diagnosis in FDR and SDR (±SD) | ||||||||||||||
| CRC | 63 | (±15) | 63 | (±15) | 51 | (±16) | 0.06 | 45 | (±13) | 48 | (±12) | 43 | (±13) | 0.09 |
| Endometrial | 59 | (±15) | 60 | (±15) | 56 | (±13) | 0.58 | 51 | (±12) | 50 | (±15) | 52 | (±11) | 0.72 |
| Other LS cancers | 59 | (±18) | 60 | (±17) | 43 | (±17) | 0.02 | 51 | (±18) | 51 | (±19) | 51 | (±18) | 0.95 |
CRC, colorectal cancer; FDR, first-degree relative; LS, Lynch syndrome; SDR, second-degree relative.
Molecular tumor testing in the population-based cohort
MSI
Abnormal MSI results were observed in 131 (23%) population-based EC cases, with 23 (4%) having MSI-low tumors and 108 (19%) MSI-high tumors. Ten of the 13 (77%) mutation carriers who took MSI testing had MSI-high tumors, 2 (15%) had MSI-low tumors, and 1 (8%) had a microsatellite stable tumor. One mutation carrier did not have MSI because of insufficient tumor sample. All MLH1 and MSH2 mutation carriers had MSI-high tumors. Among the MSH6 mutation carriers, 1 (12%) had a microsatellite stable tumor, 2 (25%) had MSI-low tumors, 5 (63%) had MSI-high tumors, and 1 did not have MSI testing because of insufficient tumor sample. Abnormal MSI results had 92% sensitivity, 78% specificity, a PPV of 9%, and an NPV of 99.8% for identifying cases with germline mutations. In terms of identifying cases with abnormal IHC, MSI yielded a sensitivity and specificity of 93%, a PPV of 88%, and an NPV of 96%. Conversely, the sensitivity and specificity of IHC staining to detect MSI-high/low tumors were 88 and 96%, respectively.
IHC
Abnormal results were seen in 93 (27%) of 348 cases who had IHC testing for MLH1, of which 2 had a germline MLH1 mutation. Abnormal results were seen in 19 (5%) of 352 cases who underwent IHC testing for MSH2. Of these, 2 were MSH2 mutation carriers. One MSH2 mutation carrier had a normal IHC for MSH2. Among 337 cases tested for IHC for MSH6, 28 (8%) had abnormal results. Of these, 2 had germline MSH2 mutations and 8 had germline MSH6 mutations. Loss of PMS2 was seen in 74 (25%) of 293 cases tested for IHC for PMS2. Only 1 MLH1 mutation carrier had loss of PMS2, which also had associated loss of MLH1 on IHC. Overall, abnormal IHC results consistent with the gene mutation were observed in the tumors of 12 (86%) mutation carriers, yielding a sensitivity of 86%, specificity of 67%, PPV of 10%, and NPV of 99%.
Molecular tumor testing in the high-risk clinic-based cohort
MSI
Only 24 of the high-risk clinic-based cases underwent MSI testing; of these, abnormal MSI results were observed in 21 (87%) cases, with 1 (4%) having MSI-low tumor and 20 (83%) with MSI-high tumors. Among the 16 mutation carriers who had MSI testing, 15 (94%) had MSI-high tumors and 1 (6%) had an MSI-low tumor. Of the 9 MLH1 mutation carriers who had MSI testing, 1 (11%) had MSI-low tumor and 8 (89%) had MSI-high tumors. All MSH2 mutation carriers (6/6) who had MSI testing had MSI-high tumors. Only 1 MSH6 mutation carrier had MSI testing that was MSI-high. Abnormal MSI results had 100% sensitivity, 38% specificity, PPV of 76%, and NPV of 100% for identifying cases with germline mutations. In terms of identifying cases with abnormal IHC, MSI yielded a sensitivity of 100%, specificity of 60%, a PPV of 90%, and an NPV of 100%. Conversely, the sensitivity and specificity of IHC staining to detect MSI-high/low tumors were 90 and 100%, respectively.
IHC
Abnormal results were seen in 26 (37%) of 70 cases who had IHC testing for MLH1, of which 22 had a germline MLH1 mutation. Abnormal results were seen in 28 (38%) of 74 cases who underwent IHC testing for MSH2. Of these, 21 were MSH2 mutation carriers. Among 69 cases who had IHC testing for MSH6, 31 (45%) had abnormal results. Of these, 19 had germline MSH2 mutations, and 5 had germline MSH6 mutations. Loss of PMS2 was seen in 22 (42%) of 52 cases tested for IHC for PMS2. A total of 18 MLH1 mutation carriers had loss of PMS2, which also had associated loss of MLH1 on IHC. Fifty-one of the 80 mutation carriers had IHC testing, of which 48 (94%) had abnormal results consistent with the gene mutation. In general, having an abnormal IHC result in any of the four MMR genes yielded a sensitivity of 94%, specificity of 48%, PPV of 80%, and NPV of 79% for identifying germline mutation carriers.
Prediction scores in the population-based cohort
Discriminative ability
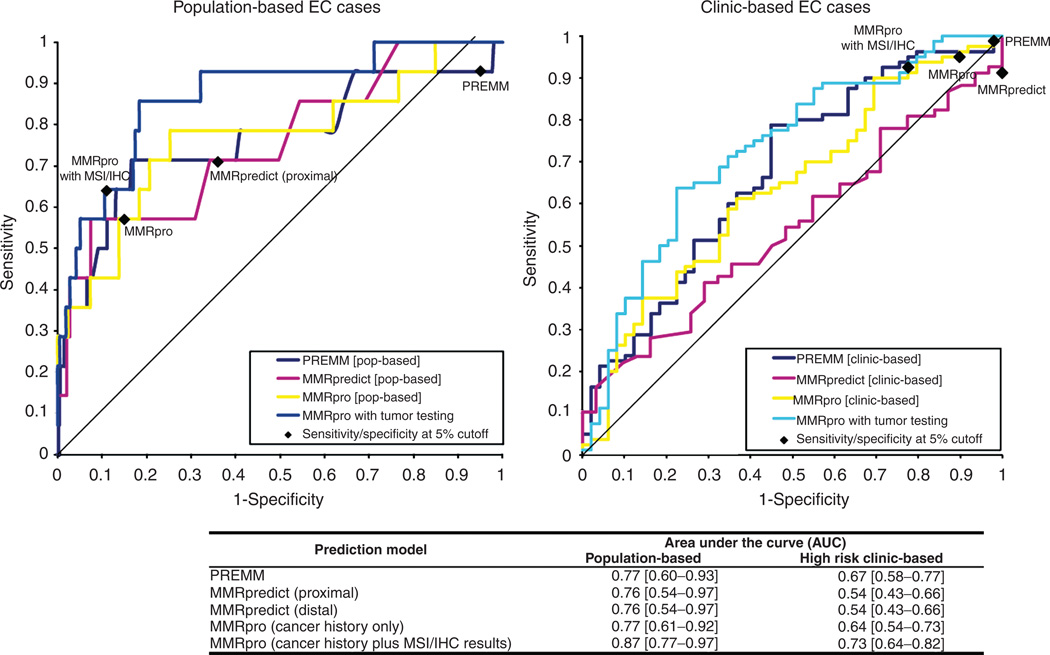
PREMM1,2,6, MMRpredict, and MMRpro were able to distinguish mutation carriers from noncarriers with an AUC of 0.77 (CI: 0.60–0.93), 0.76 (CI: 0.54–0.97), and 0.77 (CI: 0.61–0.92), respectively, for the population-based cohort (Figure 1). For a fair comparison of discrimination, we also ran the analysis in the 56 EC cases that had a complete set of three scores, which yielded comparable results, with an AUC of 0.74, 0.79, and 0.87 for PREMM1,2,6, MMRpredict, and MMRpro, respectively. All predictive models tended to overestimate the risk of finding a mutation in this cohort. Compared with an observed mutation prevalence of 3% in this cohort, average prediction scores for having a mutation were as follows—PREMM1,2,6, 8%; MMRpredict (proximal), 11%; and MMRpro, 6%.
Figure 1.
Receiver operating characteristic curves and AUC of the LS prediction models. AUC, area under the curve; EC, endometrial cancer; IHC, immunohistochemistry; LS, Lynch syndrome; MSI, microsatellite instability.
Table 2 presents the distribution of subjects for the population-based cohort according to the prespecified risk groups for each prediction model.
Table 2.
Distribution of subject scores according to clinical prediction rules, population-based cohort
| Total | No mutation | With mutation | |||||
|---|---|---|---|---|---|---|---|
| Characteristics | N | (%) | N | (%) | N | (%) | P value |
| PREM1,2,6 | |||||||
| <5% | 30 | (5) | 29 | (5) | 1 | (7) | |
| 5–9% | 443 | (79) | 439 | (80) | 4 | (29) | |
| 10–19% | 63 | (11) | 59 | (11) | 4 | (29) | |
| 20–29% | 13 | (2) | 12 | (2) | 1 | (7) | |
| 30–39% | 6 | (1) | 5 | (1) | 1 | (7) | |
| ≥40% | 8 | (2) | 5 | (1) | 3 | (21) | |
| Total | 563 | (100) | 549 | (100) | 14 | (100) | |
| Mean score (±SD) | 8 | (±8) | 8 | (±7) | 23 | (±24) | 0.03 |
| MMRpredict (proximal) | |||||||
| <0.5% | 0 | (0) | 0 | (0) | 0 | (0) | |
| 0.5–4% | 97 | (62) | 95 | (64) | 2 | (29) | |
| 5–19% | 38 | (24) | 37 | (25) | 1 | (14) | |
| 20–44% | 6 | (4) | 6 | (4) | 0 | (0) | |
| ≥45% | 15 | (10) | 11 | (7) | 4 | (57) | |
| Total | 156 | (100) | 149 | (100) | 7 | (100) | |
| MMRpredict (distal) | |||||||
| <0.5% | 61 | (39) | 60 | (40) | 1 | (14) | |
| 0.5–4% | 73 | (47) | 71 | (48) | 2 | (29) | |
| 5–19% | 11 | (7) | 10 | (7) | 1 | (14) | |
| 20–44% | 7 | (4) | 5 | (3) | 2 | (29) | |
| ≥45% | 4 | (3) | 3 | (2) | 1 | (14) | |
| Total | 156 | (100) | 149 | 100 | 7 | (100) | |
| Mean score (±SD) – proximal | 11 | (±20) | 10 | ±18 | 42 | ±39 | 0.08 |
| Mean score (±SD) – distal | 5 | (±12) | 4 | ±10 | 26 | ±35 | 0.14 |
| MMRpro | |||||||
| <5% | 191 | (82) | 185 | (85) | 6 | (43) | |
| 5–9% | 18 | (8) | 16 | (7) | 2 | (14) | |
| 10–19% | 8 | (3) | 7 | (3) | 1 | (7) | |
| 20–29% | 4 | (2) | 4 | (2) | 0 | (0) | |
| 30–39% | 1 | (1) | 1 | (1) | 0 | (0) | |
| ≥40% | 10 | (4) | 5 | (2) | 5 | (36) | |
| Total | 232 | (100) | 218 | (100) | 14 | (100) | |
| Mean score (±SD) | 6 | (±18) | 4 | (±13) | 34 | (±44) | 0.02 |
PREMM1,2,6 scores
In this cohort, 533 (95%) subjects garnered a PREMM1,2,6 score ≥5%. The mean PREMM1,2,6 score was 8 (SD = 8), with mutation carriers having a higher mean PREMM1,2,6 score than noncarriers (23 vs. 8, respectively, P = 0.03). Using a ≥5% cut-off score, the PREMM1,2,6 model had 93% sensitivity, 5% specificity, a PPV of 2%, and an NPV of 97%.
MMRpredict scores
Using the diagnosis of EC in place of a proximal CRC, all 175 cases obtained MMRpredict scores above 0.5%, the cut-off recommended for genetic testing of CRC patients. The mean MMRpredict score was 11 (SD = 20); mean score for mutation carriers was higher than for noncarriers (42 vs. 10, respectively, P = 0.08). MMRpredict had 100% sensitivity using a 0.5% threshold but 0% specificity and a PPV of 5%. Using a 5% cut-off, MMRpredict had a sensitivity of 71%, specificity of 64%, PPV of 9%, and NPV of 98%.
However, when EC was substituted for a distal CRC, an MMRpredict score cut-off of 0.5% missed one (14%) mutation carrier in this cohort with a specificity of 40%, whereas a 5% cut-off produced 57% sensitivity and 88% specificity.
MMRpro scores
Only 41/232 (18%) population-based EC cases for which MMRpro scores were calculated received MMRpro scores ≥5%. The mean MMRpro score for this cohort was 6 (SD = 18), with a higher mean score for mutation carriers compared with noncarriers (34 vs. 4, respectively, P = 0.02). Using a score cut-off of ≥5%, MMRpro missed 6 (43%) mutation carriers, yielding 57% sensitivity, 85% specificity, a PPV of 20%, and an NPV of 97%.
When molecular tumor testing results were incorporated into the MMRpro calculation, an MMRpro score >5% missed 5 (36%) mutation carriers and had a sensitivity of 64%, specificity of 89%, PPV of 27%, and NPV of 98%.
Prediction scores in the clinic-based cohort
Discriminative ability
For the clinic-based cohort, PREMM1,2,6 and MMRpro were able to distinguish mutation carriers from noncarriers with AUCs of 0.67 (CI: 0.58–0.77) and 0.64 (CI: 0.54–0.73). However, MMRpredict did not perform as well, with an AUC of 0.54 (0.43–0.66) (Figure 1). To fairly compare the discriminative ability among the three models, we also ran the analysis in the 99 EC cases that had a complete set of three scores, which yielded comparable results, with AUCs of 0.60, 0.54, and 0.62 for PREMM1,2,6, MMRpredict, and MMRpro, respectively. Compared with an observed mutation prevalence of 62% in this cohort, PREMM1,2,6 and MMRpredict underestimated the risk of finding a mutation, with average prediction scores of 49% for PREMM1,2,6 and 60% for MMRpredict (proximal). In contrast, MMRpro overestimated the risk of finding a mutation with an average prediction score of 74%.
Table 3 presents the distribution of subjects for the clinic-based cohort according to the prespecified risk groups for each prediction model.
Table 3.
Distribution of subject scores according to clinical prediction rules, clinic-based cohort
| Total | No mutation |
With mutation |
|||||
|---|---|---|---|---|---|---|---|
| Characteristics | N | (%) | N | (%) | N | (%) | P value |
| PREM1,2,6 | |||||||
| <5% | 2 | (2) | 1 | (2) | 1 | (1) | |
| 5–9% | 5 | (4) | 3 | (6) | 2 | (2) | |
| 10–19% | 20 | (15) | 13 | (27) | 7 | (9) | |
| 20–29% | 19 | (15) | 10 | (20) | 9 | (11) | |
| 30–39% | 14 | (11) | 3 | (6) | 11 | (14) | |
| ≥40% | 69 | (53) | 19 | (39) | 50 | (63) | |
| Total | 129 | (100) | 49 | (100) | 80 | (100) | |
| Mean score (±SD) | 49 | (±29) | 39 | (±28) | 55 | (±29) | 0.001 |
| MMRpredict (proximal) | |||||||
| <0.5% | 0 | (0) | 0 | (0) | 0 | (0) | |
| 0.5–4% | 6 | (6) | 0 | (0) | 6 | (9) | |
| 5–19% | 18 | (18) | 5 | (16) | 13 | (19) | |
| 20–44% | 11 | (11) | 4 | (13) | 7 | (10) | |
| ≥45% | 64 | (65) | 22 | (71) | 42 | (62) | |
| Total | 99 | (100) | 31 | (100) | 68 | (100) | |
| MMRpredict (distal) | |||||||
| <0.5% | 2 | (2) | 0 | (0) | 2 | (3) | |
| 0.5–4% | 20 | (20) | 5 | (16) | 15 | (22) | |
| 5–19% | 20 | (20) | 6 | (19) | 14 | (21) | |
| 20–44% | 17 | (17) | 6 | (19) | 11 | (16) | |
| ≥45% | 40 | (41) | 14 | (45) | 26 | (38) | |
| Total | 99 | (100) | 31 | (100) | 68 | (100) | |
| Mean score (±SD) – proximal | 60 | (±35) | 64 | (±32) | 58 | (±36) | 0.41 |
| Mean score (±SD) – distal | 41 | (±36) | 44 | (±35) | 40 | (±37) | 0.65 |
| MMRpro | |||||||
| <5% | 9 | (7) | 5 | (10) | 4 | (5) | |
| 5–9% | 4 | (3) | 3 | (6) | 1 | (1) | |
| 10–19% | 9 | (7) | 6 | (12) | 3 | (4) | |
| 20–29% | 3 | (2) | 1 | (2) | 2 | (3) | |
| 30–39% | 5 | (4) | 1 | (2) | 4 | (5) | |
| ≥40% | 99 | (77) | 33 | (67) | 66 | (82) | |
| Total | 129 | (100) | 49 | (100) | 80 | (100) | |
| Mean score (±SD) | 74 | (±36) | 65 | (±40) | 79 | (±32) | 0.04 |
PREMM1,2,6 scores
Almost all (127/129) subjects garnered a PREMM1,2,6 score ≥ 5%. The mean PREMM1,2,6 score was 49 (SD = 29), with mutation carriers having a higher mean score compared with noncarriers (55 vs. 39, respectively, P = 0.001). Using a ≥5% cut-off score, the PREMM1,2,6 model had 99% sensitivity, 2% specificity, a PPV of 62, and an NPV of 50.
MMRpredict scores
Replacing a proximal CRC with EC, none of the clinic-based EC cases received an MMRpredict score <0.5%. The mean score was 60 (SD = 35). The mean score of mutation carriers was 58 and that of noncarriers was 64 (P = nonsignificant). MMRpredict also had 100% sensitivity, 0% specificity, and a PPV of 69% in the clinic-based cohort using a 0.5% threshold if the EC was treated as a proximal tumor. Using a 5% cut-off, MMRpredict had 91% sensitivity, 0% specificity, a PPV of 67%, and an NPV of 0%.
However, when EC was substituted for a distal CRC, an MMRpredict score cut-off of 0.5% would miss 2 (3%) mutation carriers, yielding 97% sensitivity and 0% specificity, whereas a 5% cut-off produced 75% sensitivity and 16% specificity.
MMRpro scores
In the clinic-based cohort, only 9 (7%) EC cases received MMRpro scores <5%. The mean MMRpro score was 74 (SD = 36), and the mean score was higher for the mutation carriers compared with the noncarriers in the clinic-based cohort (79 vs. 65, respectively, P = 0.04). Using a score cutoff of ≥5%, MMRpro yielded 95% sensitivity, 10% specificity, a PPV of 63%, and an NPV of 56%.
When molecular tumor testing results were incorporated into the MMRpro calculation, an MMRpro score >5% missed 6 (7%) mutation carriers and had a sensitivity of 93%, specificity of 22%, PPV of 66%, and NPV of 65%.
Supplementary Tables S1 and S2 online present the sensitivity, specificity, PPV, and NPV at the higher cut-off levels for all models in the population- and clinic-based cohorts, respectively.
DISCUSSION
We report on the performance of three prediction models in detecting LS among population- and clinic-based EC cases. Our analysis in these large cohorts reveals that these models were able to statistically distinguish mutation carriers from noncarriers, as reflected by their AUC, with the exception of MMRpredict, which had poor discrimination among clinic-based EC cases. The performance of PREMM, MMRpredict, and MMRpro was comparable in the population-based cohort, whereas PREMM and MMRpro had comparable discrimination in the clinic-based cohort. However, the discriminative ability of these models is much lower among probands with EC than among probands with CRC.
We further explored the clinical utility of these models by looking at the number of probands who would be referred for germline testing and the number of mutation carriers missed if a 5% cut-off was used. Using PREMM1,2,6 >5%, almost all (95–99%) EC cases would have been referred for germline testing and would have still missed 7% of mutation carriers among population-based EC cases and 1% of mutation carriers among cases from clinic-based families. By comparison, an MMRpredict score >5% would have selected only 14–38% (depending on whether the tumor was considered proximal vs. distal) of population-based EC cases for genetic testing, but would have missed 29–43% of mutation carriers. Among clinic-based cases, 78–94% would have been referred for genetic testing, yet 9–25% of mutation carriers would still have been missed. MMRpro would refer only 18% of population-based EC cases for genetic testing; however, it would miss 43% of mutation carriers. Conversely, MMRpro would have missed only 5% of mutation carriers among clinic-based EC cases, but almost all (93%) would have been referred for genetic testing. Incorporating the molecular tumor testing results into the MMRpro calculation yielded similar findings. These results indicate that although the sensitivity is high and comparable among the models, the specificity in clinic- and population-based cohorts was low. This limitation leads to a large pool of subjects eligible for genetic testing using a 5% cut-off and thereby hinders their clinical utility in effectively distinguishing gene mutation carriers from noncarriers.
To date, studies validating LS prediction models have only been conducted on CRC cohorts and have shown that prediction models perform remarkably well in discriminating mutation carriers from noncarriers, with AUCs ranging from 0.73 to 0.93 in clinic-based cohorts16–18 and 0.91 to 0.96 in population-based cohorts.14,15 High sensitivity and specificity were also demonstrated in these studies,14,15,18 indicating the clinical usefulness of these models as a potential screening tool for LS. The only other study looking at LS prediction models among EC cases examined the sensitivity of PREMM1,2, MMRpredict, and MMRpro in 13 population-based EC cases with LS and showed that these models performed reasonably well in EC cases, with sensitivities ranging from 64 to 100%.24 Our larger analysis supports the high yield in sensitivity but also reveals that the predictive models tend to assign greater weight to a diagnosis of EC compared with CRC (e.g., just the presence of an EC diagnosis in a proband results in a PREMM1,2,6 score above the 5% cut-off point).
We recognize that our study has several limitations. One limitation is the small number of mutation carriers (N = 14) in the population-based cohort. In addition, not all EC cases underwent germline mutation analysis. In the population-based cohort, the majority of EC cases that had normal tumor MMR results and no germline testing were classified as noncarriers. It is possible that a few of these cases could have had a mutation. Likewise, not all EC cases included in the clinic-based cohort underwent germline testing. For these cases, we had to extrapolate their mutation status by assigning the germline testing results of a first-degree relative. It is possible that the EC patients who did not have germline testing but were classified as mutation carriers may have had sporadic EC. Having information on EC features associated with LS or with sporadic cases, such as patient’s body mass index for the former and history of polycystic ovarian syndrome for the latter, could have further aided in the classification of whether these non–germline tested EC cases were mutation carriers. However, this information was not readily available. Another limitation of our study was that family cancer history for all population-based cancer cases was based on proband reports and largely unconfirmed. Nevertheless, several studies have shown that patient reports of family cancer history, particularly in first-degree relatives, are accurate.25,26
Despite these limitations, our study is the only one to date that presents a comprehensive assessment of the performance of LS prediction models among large cohorts of population-based and clinic-based EC cases. Our results show that when prediction models for LS are used in patients with EC, the discriminative ability for detecting MMR gene mutation carriers is lower than that seen in patients with CRC. Consequently, these models in their current form have limited clinical utility in determining which patients with EC should undergo clinical genetic testing for LS, irrespective of clinic- or population-based ascertainment.
The poorer performance of the LS prediction models among EC cases compared with CRC cases is not surprising considering that these models were developed and validated on CRC cohorts. As such, features associated with ECs in LS were not taken into account during model development. One of these features refers to the anatomic location of the endometrial tumor. Recent studies have shown that lower uterine segment anatomic location of the endometrial tumor may be associated with LS.27–29 Another feature of LS-associated EC not factored into the current models is the body mass index. It has been shown that LS EC patients had lower body mass index than their sporadic counterparts.5,8,30 Pathologic characteristics, including poorly differentiated tumors, higher stage disease, and deeper myometrial invasion, have also been associated with DNA MMR mutations in EC.1,30 The heterogeneity of LS ECs as evidenced by the predominance of MSH6 mutation carriers among population-based cases compared with mostly MLH1 and MSH2 carriers among high-risk clinic-based cases is another possible reason why the models at their current forms did not perform as well. These features must be considered to quantify the risk for LS among EC cases.
Our analysis showed that the sensitivity and specificity of MSI and IHC in identifying mutation carriers are considerably higher than that of any of the prediction models. This finding supports the use of molecular tumor testing in screening for LS among EC cases. A screening algorithm that has been proposed entails IHC testing in women younger than 50 years, in older women with tumors exhibiting features associated with MSI, and in cases where personal or family history is suggestive of hereditary nonpolyposis CRC. If loss of MSH2 or MSH6 is detected, the next step would be gene mutation analysis. Loss of MLH1 or PMS2 will warrant additional testing for DNA hypermethylation. If this is absent, the next step would be gene mutation analysis.8,28 Our data support this recommendation because there was a substantial proportion of EC patients with MSI and loss of MLH1 expression, where no MLH1 mutation was identified; these cases presumably represent somatic hypermethylation of MLH1. A recent study revealed that IHC triage of all ECs could identify the most mutation carriers and prevent the most CRCs but at considerable cost, whereas IHC triage of women with EC at any age having at least 1 first-degree relative with an LS-associated cancer is a cost-effective strategy for detecting LS.31
Identifying women who may have LS is important and remains a challenge. It is apparent that personal and family history, using either established clinical criteria6 or prediction models, is not robust in selecting those who should undergo predictive germline testing among EC cases. This finding implies that at the current time, universal IHC and MSI analysis is the only way available to screen for LS in patients with EC and should be implemented more widely in the pathologic evaluation of newly diagnosed EC. If the goal is to provide a quantitative risk estimate of having LS, new prediction models developed from EC populations will be needed.
ACKNOWLEDGMENTS
We gratefully acknowledge the work of Radhika Mopala, research volunteer at Dana-Farber Cancer Institute, who helped generate the pedigrees and MMRpro scores for all study subjects. We also gratefully acknowledge the research assistance provided by Victoria Schunemann and Lisa Schunemann from Ohio State University.
The study was supported by the National Cancer Institute through the following grants: R01CA132829 (S.S.), K24 CA113433 (S.S.), R01 CA67941 (A.d.l.C.), and P30 CA16058 (Ohio State University Comprehensive Cancer Center). This work was also supported by the National Cancer Institute, National Institutes of Health, under RFA CA-95-011, and through cooperative agreements with the members of the Colon Cancer Family Registry and principal investigators, including the Australasian Colorectal Cancer Family Registry (U01 CA097735); the Familial Colorectal Neoplasia Collaborative Group (U01 CA074799); and the Mayo Clinic Cooperative Family Registry for Colon Cancer Studies (U01 CA074800).
Footnotes
SUPPLEMENTARY MATERIAL
Supplementary material is linked to the online version of the paper at http://www.nature.com/gim
S.S. and R.C.M. had full access to all of the data in the study and take responsibility for the integrity of the data and accuracy of the data analysis. R.C.M. contributed to the conception and design, acquisition, analysis, and interpretation of data and prepared, revised, and approved the manuscript. H.H. contributed to the acquisition of data and drafted, revised, and approved the manuscript. F.K., E. Steyerberg, and J.B. contributed to the analysis and interpretation of data and prepared, revised, and approved the manuscript. E. Stoffel and S.S. contributed to the conception and design, analysis, and interpretation of data and prepared, revised, and approved the manuscript. D.E.C., F.J.B., J.L.H., M.A.J., N.M.L., G.C., R.H., S.M., and A.d.l.C. contributed to the acquisition of data and prepared, revised, and approved the manuscript.
The abstract of this study was presented as a poster at the 2010 ASCO Meeting, Chicago, Illinois, 4–8 June 2010, and as an oral presentation at the 4th Biennial Scientific Meeting of the International Society for Inherited Gastrointestinal Tumours, 30 March–2 April 2011.
The content of this presentation does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the Cancer Family Registries (CFR), nor does mention of trade names, commercial products, or organizations imply endorsement by the US government or the CFR.
DISCLOSURE
All authors have completed and submitted the International Committee of Medical Journal Editors form for the disclosure of potential conflicts of interest. S.S. reported having a compensated consultant or advisory relationship with Archimedes, Inc. H.H. reported having a compensated consultant or advisory relationship with Myriad Genetics Laboratories, Inc.
REFERENCES
- 1.Walsh MD, Cummings MC, Buchanan DD, et al. Molecular, pathologic, and clinical features of early-onset endometrial cancer: identifying presumptive Lynch syndrome patients. Clin Cancer Res. 2008;14:1692–1700. doi: 10.1158/1078-0432.CCR-07-1849. [DOI] [PubMed] [Google Scholar]
- 2.Lynch HT, Boland CR, Gong G, et al. Phenotypic and genotypic heterogeneity in the Lynch syndrome: diagnostic, surveillance and management implications. Eur J Hum Genet. 2006;14:390–402. doi: 10.1038/sj.ejhg.5201584. [DOI] [PubMed] [Google Scholar]
- 3.Resnick K, Straughn JM, Jr, Backes F, Hampel H, Matthews KS, Cohn DE. Lynch syndrome screening strategies among newly diagnosed endometrial cancer patients. Obstet Gynecol. 2009;114:530–536. doi: 10.1097/AOG.0b013e3181b11ecc. [DOI] [PubMed] [Google Scholar]
- 4.Vasen HF, Wijnen JT, Menko FH, et al. Cancer risk in families with hereditary nonpolyposis colorectal cancer diagnosed by mutation analysis. Gastroenterology. 1996;110:1020–1027. doi: 10.1053/gast.1996.v110.pm8612988. [DOI] [PubMed] [Google Scholar]
- 5.Lu KH, Schorge JO, Rodabaugh KJ, et al. Prospective determination of prevalence of lynch syndrome in young women with endometrial cancer. J Clin Oncol. 2007;25:5158–5164. doi: 10.1200/JCO.2007.10.8597. [DOI] [PubMed] [Google Scholar]
- 6.Hampel H, Frankel W, Panescu J, et al. Screening for Lynch syndrome (hereditary nonpolyposis colorectal cancer) among endometrial cancer patients. Cancer Res. 2006;66:7810–7817. doi: 10.1158/0008-5472.CAN-06-1114. [DOI] [PubMed] [Google Scholar]
- 7.Meyer LA, Broaddus RR, Lu KH. Endometrial cancer and Lynch syndrome: clinical and pathologic considerations. Cancer Control. 2009;16:14–22. doi: 10.1177/107327480901600103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Resnick KE, Hampel H, Fishel R, Cohn DE. Current and emerging trends in Lynch syndrome identification in women with endometrial cancer. Gynecol Oncol. 2009;114:128–134. doi: 10.1016/j.ygyno.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balmaña J, Stockwell DH, Steyerberg EW, et al. Prediction of MLH1 and MSH2 mutations in Lynch syndrome. JAMA. 2006;296:1469–1478. doi: 10.1001/jama.296.12.1469. [DOI] [PubMed] [Google Scholar]
- 10.Kastrinos F, Steyerberg EW, Mercado R, et al. The PREMM(1,2,6) model predicts risk of MLH1, MSH2, and MSH6 germline mutations based on cancer history. Gastroenterology. 2011;140:73–81. doi: 10.1053/j.gastro.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barnetson RA, Tenesa A, Farrington SM, et al. Identification and survival of carriers of mutations in DNA mismatch-repair genes in colon cancer. N Engl J Med. 2006;354:2751–2763. doi: 10.1056/NEJMoa053493. [DOI] [PubMed] [Google Scholar]
- 12.Chen S, Wang W, Lee S, Nafa K, et al. Prediction of germline mutations and cancer risk in the Lynch syndrome. JAMA. 2006;296:1479–1487. doi: 10.1001/jama.296.12.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balaguer F, Balmaña J, Castellví-Bel S, et al. Validation and extension of the PREMM1,2 model in a population-based cohort of colorectal cancer patients. Gastroenterology. 2008;134:39–46. doi: 10.1053/j.gastro.2007.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balmaña J, Balaguer F, Castellví-Bel S, et al. Comparison of predictive models, clinical criteria and molecular tumour screening for the identification of patients with Lynch syndrome in a population-based cohort of colorectal cancer patients. J Med Genet. 2008;45:557–563. doi: 10.1136/jmg.2008.059311. [DOI] [PubMed] [Google Scholar]
- 15.Green RC, Parfrey PS, Woods MO, Younghusband HB. Prediction of Lynch syndrome in consecutive patients with colorectal cancer. J Natl Cancer Inst. 2009;101:331–340. doi: 10.1093/jnci/djn499. [DOI] [PubMed] [Google Scholar]
- 16.Monzon JG, Cremin C, Armstrong L, et al. Validation of predictive models for germline mutations in DNA mismatch repair genes in colorectal cancer. Int J Cancer. 2010;126:930–939. doi: 10.1002/ijc.24808. [DOI] [PubMed] [Google Scholar]
- 17.Pouchet CJ, Wong N, Chong G, et al. A comparison of models used to predict MLH1, MSH2 and MSH6 mutation carriers. Ann Oncol. 2009;20:681–688. doi: 10.1093/annonc/mdn686. [DOI] [PubMed] [Google Scholar]
- 18.Ramsoekh D, van Leerdam ME, Wagner A, Kuipers EJ, Steyerberg EW. Mutation prediction models in Lynch syndrome: evaluation in a clinical genetic setting. J Med Genet. 2009;46:745–751. doi: 10.1136/jmg.2009.066589. [DOI] [PubMed] [Google Scholar]
- 19.Newcomb PA, Baron J, Cotterchio M, et al. Colon Cancer Family Registry: an international resource for studies of the genetic epidemiology of colon cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:2331–2343. doi: 10.1158/1055-9965.EPI-07-0648. [DOI] [PubMed] [Google Scholar]
- 20.Lindor NM, Burgart LJ, Leontovich O, et al. Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J Clin Oncol. 2002;20:1043–1048. doi: 10.1200/JCO.2002.20.4.1043. [DOI] [PubMed] [Google Scholar]
- 21.Poynter JN, Siegmund KD, Weisenberger DJ, et al. Molecular characterization of MSI-H colorectal cancer by MLHI promoter methylation, immunohistochemistry, and mismatch repair germline mutation screening. Cancer Epidemiol Biomarkers Prev. 2008;17:3208–3215. doi: 10.1158/1055-9965.EPI-08-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen S, Wang W, Broman KW, Katki HA, Parmigiani G. BayesMendel: an R environment for Mendelian risk prediction. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1063. Article21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.BayesMendel Lab. The BayesMendel R package. [Accessed 26 September 2011]; http://www.cancerbiostats.onc.jhmi.edu/BayesMendel/.
- 24.Backes FJ, Hampel H, Backes KA, et al. Are prediction models for Lynch syndrome valid for probands with endometrial cancer? Fam Cancer. 2009;8:483–487. doi: 10.1007/s10689-009-9273-5. [DOI] [PubMed] [Google Scholar]
- 25.Murff HJ, Spigel DR, Syngal S. Does this patient have a family history of cancer? An evidence-based analysis of the accuracy of family cancer history. JAMA. 2004;292:1480–1489. doi: 10.1001/jama.292.12.1480. [DOI] [PubMed] [Google Scholar]
- 26.Douglas FS, O’Dair LC, Robinson M, Evans DG, Lynch SA. The accuracy of diagnoses as reported in families with cancer: a retrospective study. J Med Genet. 1999;36:309–312. [PMC free article] [PubMed] [Google Scholar]
- 27.Masuda K, Banno K, Yanokura M, et al. Carcinoma of the lower uterine segment (LUS): clinicopathological characteristics and association with Lynch syndrome. Curr Genomics. 2011;12:25–29. doi: 10.2174/138920211794520169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garg K, Soslow RA. Lynch syndrome (hereditary non-polyposis colorectal cancer) and endometrial carcinoma. J Clin Pathol. 2009;62:679–684. doi: 10.1136/jcp.2009.064949. [DOI] [PubMed] [Google Scholar]
- 29.Westin SN, Lacour RA, Urbauer DL, et al. Carcinoma of the lower uterine segment: a newly described association with Lynch syndrome. J Clin Oncol. 2008;26:5965–5971. doi: 10.1200/JCO.2008.18.6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shih KK, Garg K, Levine DA, et al. Clinicopathologic significance of DNA mismatch repair protein defects and endometrial cancer in women 40years of age and younger. Gynecol Oncol. 2011;123:88–94. doi: 10.1016/j.ygyno.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 31.Kwon JS, Scott JL, Gilks CB, Daniels MS, Sun CC, Lu KH. Testing women with endometrial cancer to detect Lynch syndrome. J Clin Oncol. 2011;29:2247–2252. doi: 10.1200/JCO.2010.32.9979. [DOI] [PMC free article] [PubMed] [Google Scholar]