Abstract
Innate immune cells respond to microbial invaders using pattern recognition receptors that detect conserved microbial patterns. Among the cellular processes stimulated downstream of pattern recognition machinery is the initiation of autophagy, which plays protective roles against intracellular microbes. We have shown recently that Dictyostelium discoideum, which takes up bacteria for nutritive purposes, may employ pattern recognition machinery to respond to bacterial prey, as D. discoideum cells upregulate bactericidal activity upon stimulation by lipopolysaccharide (LPS). Here we extend these findings, showing that LPS treatment leads to induction of autophagosomal maturation in cells responding to the bacteria Staphylococcus aureus. Cells treated with the autophagy-inducing drug rapamycin clear internalized bacteria at an accelerated rate, while LPS-enhanced clearance of bacteria is reduced in cells deficient for the autophagy-related genes atg1 and atg9. These findings link microbial pattern recognition with autophagy in the social amoeba D. discoideum.
Keywords: Dictyostelium discoideum, autophagy, Staphylococcus aureus, innate immunity, LPS, pattern recognition, phagocytosis, lysosome
1. Introduction
Innate immune cells use evolutionarily-conserved pattern recognition molecules to recognize similarly conserved patterns on microbial invaders [1; 2]. The social amoeba Dictyostelium discoideum takes up bacteria for nutritional purposes [3], and recent evidence suggests that the amoebae detect and respond to bacterial prey using conserved pattern recognition machinery [4; 5]. Analysis of the D. discoideum genome reveals the existence of putative gene products bearing leucine-rich repeat regions (LRRs) and toll/interleukin-1 receptor (TIR) domains, which are conserved in pattern recognition receptors [4, 6–8]. Such gene products, including the TIR domain-containing protein TirA and the S-cell enriched with leucine-rich repeat-containing protein SlrA, are expressed at elevated levels in D. discoideum sentinel cells, which are highly phagocytic cells that differentiate during multicellular development and provide immune defense for the newly forming spores [4]. TirA, specifically, has been found to be critical for interactions between D. discoideum and bacteria, as cells deficient for TirA are unable to grow efficiently in the presence of gram-negative bacteria [4].
If D. discoideum cells use pattern recognition machinery to detect bacteria, then it would be anticipated that amoeboid cells would respond to known microbial patterns. We have reported previously that D. discoideum cells pre-treated with the microbial pattern LPS more efficiently clear certain bacteria, such as Staphylococcus aureus, following phagocytosis [5].
Among the cellular processes mediated in response to signaling downstream of conserved pattern recognition receptors is the induction of autophagy [9]. Macroautophagy (hereafter referred to as autophagy) is a conserved process by which cytoplasmic contents are engulfed in newly formed double-membraned vesicles that are subsequently delivered to lysosomes [10]. Autophagy can be used by cells for recycling of nutrients during starvation and for degradation of damaged organelles and misfolded protein aggregates [10; 11] Evidence is accumulating that autophagy also plays a role during innate immunity via the capture and elimination of intracellular bacteria and viruses [12; 13]. Various microbial stimuli, including LPS, induce autophagy in mammalian cells, thus linking autophagic processes with pattern recognition pathways [14–18].
Dictyostelium discoideum cells use conserved proteins to induce autophagy [19; 20] as a means to carry out various cellular processes [20–24], and recent results indicate that D. discoideum uses autophagy as a tool in its innate immune response to pathogens [25; 26]. The bacteria Salmonella typhimurium replicates more efficiently and causes increased cell death in D. discoideum cells deficient for the autophagy-related proteins Atg1, Atg6 and Atg7 [25]. In addition, D. discoideum infection with Legionella pneumophilia results in enhanced expression of the autophagy-related genes atg8, atg9 and atg16 [26]. Legionella multiplies more rapidly in D. discoideum cells deficient for Atg9, indicating a protective role for autophagy against Legionella infection [26].
Here we report that, like in mammalian cells, the microbial pattern LPS induces autophagy in D. discoideum cells. We further show that induction of autophagy promotes enhanced bactericidal activity by D. discoideum cells, and that expression of autophagy-related proteins is required for LPS-enhanced bacterial clearance. Taken together, these results link microbial pattern recognition with the induction of autophagy in D. discoideum.
2. Materials and Methods
2.1 Cell culture
D. discoideum DH1, Atg1− [24], and Atg9− cells [26], and HR87 cells expressing GFP-Atg8 [23] were received from the Dicty Stock Center and grown axenically at 22°C in HL-5 media supplemented with vitamins (400 µg/L biotin, 100 µg/L cyanocobalamin, 4 mg/L folic acid, 8 mg/L lipoic acid, 10 mg/L riboflavin and 1 mg/L thiamine) and 20 µg/mL uracil. Media used for HR87 cells was also supplemented with 5 µg/mL G418.
Klebsiella pneumoniae and Staphylococcus aureus were obtained from Carolina Biological Supply Co. (Burlington, NC). Salmonella enterica serovar Typhimurium (ATCC 29631) was obtained from the American Type Culture Collection (Manassas, VA).
2.2 Immunoblotting
HR87 cells expressing GFP-Atg8 were incubated at 22°C in shaking culture at 3×106 cells/mL with or without 1 µg/mL E. coli O55:B5 LPS (Sigma, St. Louis, MO) or Kdo-2 lipid A from E. coli (Enzo Life Sciences, Plymouth Meeting, MA) for 1 h and mixed with bacteria harvested at log phase. In experiments observing inhibition, cells were pre-treated with 5 mM 3-methyladenine (Acros Organics, Geel, Belgium) for 1 h. Cells were harvested 3 h after addition of bacteria and lysed with 1% NP-40 in TBS containing protease inhibitors (2.5 µg/ml each of chymostatin, leupeptin, antipain, and pepstatin A). Lysates were subjected to SDS-PAGE and immunoblotting using rabbit anti-GFP (Life Technologies, Grand Island, NY), followed by HRP-conjugated goat anti-rabbit Ig (Jackson ImmunoResearch, West Grove, PA) detected by chemiluminescence.
2.3 Confocal immunofluorescence microscopy
HR87 cells were pre-treated or not with 1 µg/mL LPS (E. coli O55:B5 LPS, Sigma) in shaking culture at 22°C and mixed with S. aureus at a ratio of 7.5:1 D. discoideum cell. After 1.5 h, non-internalized bacteria were washed from cells with 5 mM sodium azide in PBS [27] and cells were incubated on poly-lysine coated coverslips at 22°C. After 1 h, cells were fixed with 4% PFA in PBS, permeabilized with 0.1% Triton X-100 in PBS, and blocked with 0.2% BSA in PBS. To label bacteria, samples were incubated with biotin-labeled polyclonal rabbit antibodies specific for S. aureus (Thermo Scientific, Waltham, MA), followed by incubation with Alexa Fluor (AF) 594-conjugated streptavidin (Invitrogen). To amplify signals from GFP-Atg8, samples were incubated with mouse F(ab)’2 anti-GFP (MBL, Woburn, MA, F(ab)’2 fragments generated with Pierce F(ab)’2 micro preparation kit, Thermo Scientific) followed by incubation with AF488-conjugated F(ab)’2 anti-mouse IgG (Jackson ImmunoResearch). Samples were visualized using a Zeiss 510 Meta confocal laser scanning microscope (Jena, Germany) fitted with a 1.4 oil Plan-Apochromat. A 63x objective was used, and confocal sections acquired during channel mode, multi-track acquisition with Ex488/Em 505-530BP for AF488 and Ex543/Em 585LP for AF594. LSM software was used to calculate overlap coefficients (after Manders).
2.4 Bacterial intracellular survival assay
HR87 cells were incubated in 24-well plates at 3×105 cells/mL with or without 100 nM rapamycin (LC Laboratories, Woburn, MA) for 1 h at 22°C. Staphylococcus aureus, harvested from an overnight culture, was added at ratio of 10:1 D. discoideum cell. After 15 min each at 22°C and 4°C, cells were washed of non-internalized bacteria and treated with 30 µg/mL streptomycin to kill remaining external bacteria.
Survival of phagocytized S. aureus was assessed between 0–90 min by lysing cells with 0.1% Triton X-100 in PBS and plating on nutritive agar. Colonies were counted after 24 h at 37°C and percent remaining bacteria at each time point was determined relative to bacteria present at 0 min.
2.5 Bacterial clearance assay
DH1, Atg1− and Atg9− cells were incubated 1 h at 22°C in shaking culture at 2×106 cells/mL with or without E coli 055:B5 LPS (1 µg/mL). Staphylococcus aureus, taken from overnight cultures, were labeled with STYO-9® (Invitrogen, Carlsbad, CA), and added at a ratio of 15:1 D. discoideum cell. Cells were incubated 1 h at 22°C in the dark to allow for phagocytosis. Non-internalized bacteria were removed by centrifugation at 150g, and D. discoideum cells were resuspended at 2×106 cells/mL. Samples were harvested after an additional 0–45 min in shaking culture, washed and resuspended in PBS containing 1% paraformaldehyde. Samples were analyzed for bacterial clearance, using flow cytometry to measure the loss of fluorescence from cells at 45 min compared with the levels at 0 min.
3. Results
3.1 Autophagosomal maturation is induced by S. typhimurium and Klebsiella pneumoniae in D. discoideum
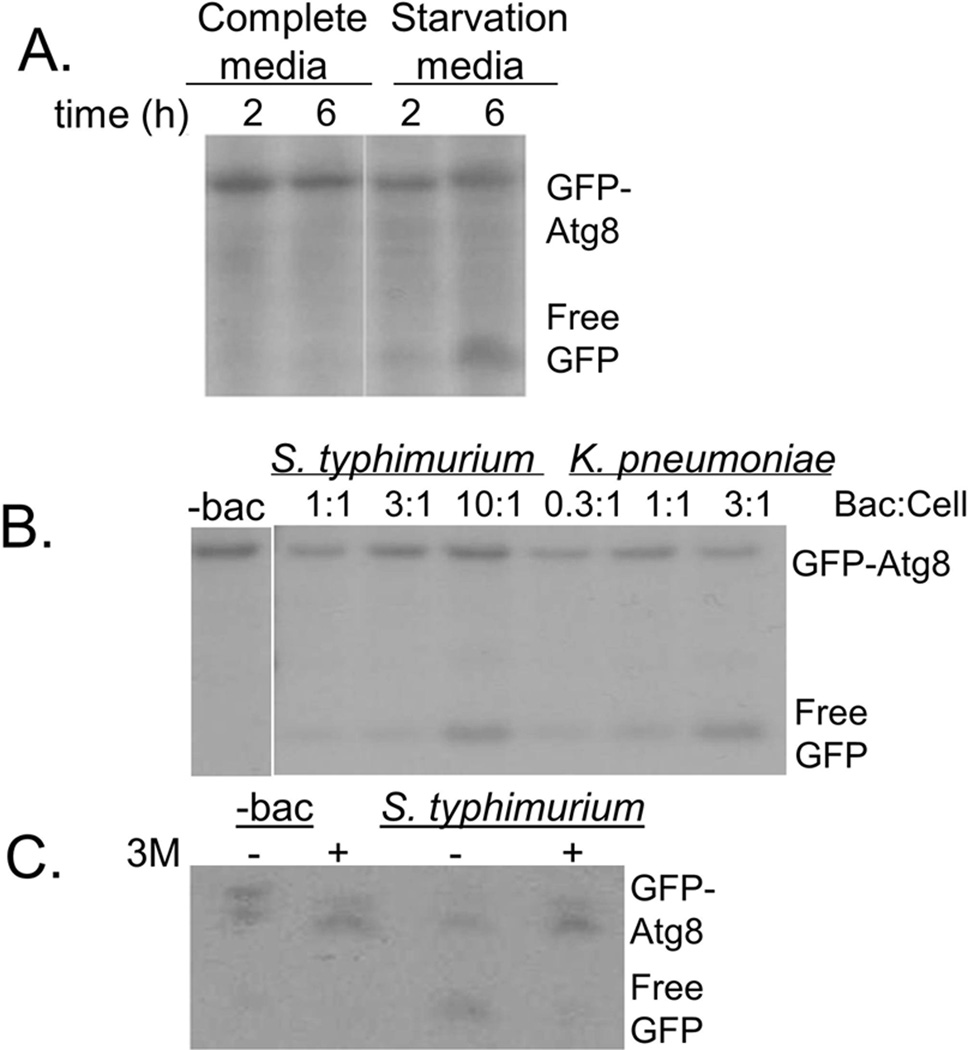
The autophagy-related protein Atg8 (as well as its mammalian homologue LC3) conjugated with GFP is a widely-used marker for autophagosomes [28]. Release of free GFP from GFP-Atg8 conjugates allows for monitoring of autophagosomal delivery to lysosomes, as Atg8 is rapidly degraded by lysosomal enzymes, while free GFP remains relatively stable [28; 29]. We tested whether the GFP-Atg8 cleavage assay could be used to monitor autophagosomal-lysosomal fusion in D. discoideum cells expressing GFP-Atg8. Free GFP was not detected in cells grown in nutritive media, while free GFP was seen after 2 h and further increased after 6 h of growth in amino acid-free starvation media (Fig 1A).
Fig. 1. Autophagosomal maturation is induced in D. discoideum exposed to S. typhimurium and K. pneumoniae.
A. HR87 cells were incubated in complete or amino-acid free media, harvested and lysed. Autophagosomal maturation was assayed using the GFP-Atg8 cleavage assay, immunoblotting with anti-GFP to detect GFP-Atg8 and free GFP, which is cleaved from GFP-Atg8 upon delivery of autophagosomes to lysosomes.
B. The effects of incubation with S. typhimurium and K. pneumoniae on autophagosomal maturation in HR87 cells were assessed using the GFP-Atg8 cleavage assay.
C. Cells incubated with or without S. typhimurium were treated with or without the autophagy inhibitor 3MA, and inhibition of autophagosomal maturation was assessed using the GFP-Atg8 cleavage assay. Shown are representative blots from at least three independent experiments.
Using the GFP-Atg8 cleavage assay, we tested whether phagocytosis of bacteria by D. discoideum could induce autophagosomal maturation. Consistent with previous reports tying autophagy to D. discoideum responses against bacteria [25; 26], we found that autophagosomal maturation was induced by incubation with S. typhimurium and K. pneumoniae in a dose-dependent manner (Fig1B). The GFP-Atg8 cleavage assay was specific for autophagosomal processes since no free GFP was detected in cells incubated with S. typhimurium but pre-treated with 3-methyladenine (3MA), a type III phosphatidylinositol 3-kinase inhibitor that has been shown to block autophagosome formation [30].
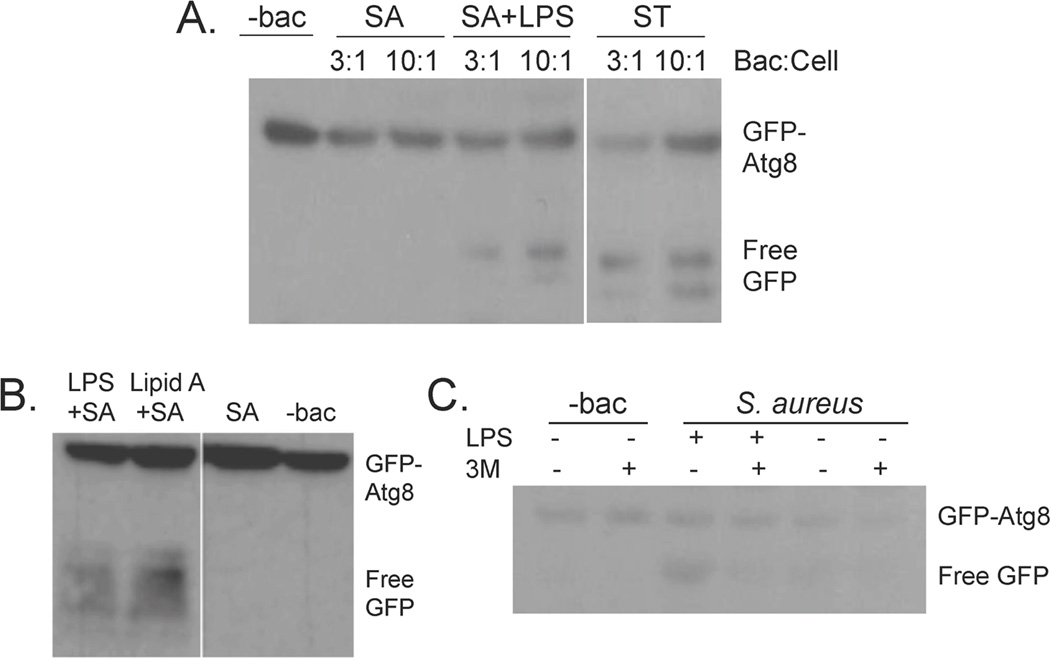
3.2 Staphylococcus aureus induces autophagosomal maturation only upon addition of LPS
Although incubation of D. discoideum with S. typhimurium and K. pneumoniae induced autophagosomal maturation, as measured by detection of free GFP in the GFP-Atg8 cleavage assay, incubation with S. aureus resulted in autophagosomal maturation only in cells pre-treated with LPS (Fig2A). To address the question of impurities often found in commercial preparations of LPS, we also tested the effects of the active lipid moiety of LPS, lipid A, on induction of autophagosomal maturation. Like LPS, treatment with lipid A resulted in induction of autophagosomal maturation in cells incubated with S. aureus (Fig 2B). LPS treatment did not result in release of free GFP in cells treated with the autophagosomal inhibitor 3MA, indicating that free GFP seen in the GFP-Atg8 cleavage assay was specific for autophagosomal maturation (Fig 2C).
Fig. 2. Autophagosomal maturation is induced in cells incubated with S. aureus only upon treatment with LPS.
A. HR87 cells were treated with or without 1 µg/mL LPS and incubated with varying ratios of S. aureus or S. typhimurium. The GFP-Atg8 cleavage assay was used to detect autophagosomal maturation.
B. HR87 cells were untreated, treated with LPS (1 µg/mL) or Lipid A (1 µg/mL), and incubated with S. aureus at a ratio of 3:1 HR87 cell. Autophagosomal induction was tested using the GFP-Atg8 cleavage assay.
C. HR87 cells incubated with or without LPS (1 µg/mL) and S. aureus at a ratio of 3:1 HR87 cell were pre-treated with or without the autophagosomal inhibitor 3MA (5 mM). The GFP-cleavage assay was used to detect autophagosomal maturation. Shown are representative blots from at least three independent experiments.
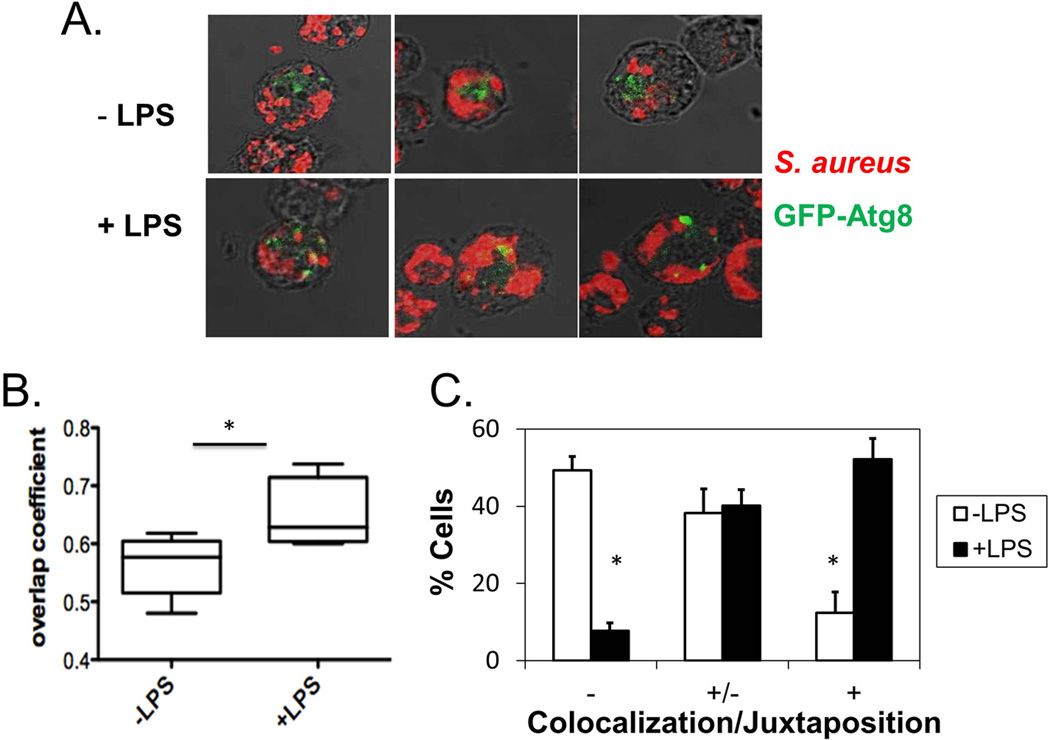
3.3 Autophagosomal localization of S. aureus in D. discoideum cells treated with LPS
To further characterize the effects of LPS on induction of autophagy, we used confocal fluorescence microscopy to observe the localization of internalized S. aureus in relation with autophagosomes in cells treated with or without LPS. GFP-Atg8 was recruited near S. aureus- containing vesicles in cells treated with LPS (Fig. 3A), while internalized S. aureus and GFP-Atg8 localized in different areas in untreated cells. The differences in colocalization were quantified by calculating the overlap coefficients (after Manders) between S. aureus and GFP-Atg8 in cells treated or not with LPS. Pre-treatment with LPS resulted in a statistically significant increase in average overlap coefficients from 0.56 without LPS to 0.65 with LPS (Fig. 3B). Furthermore, by quantification of the number of cells in which S. aureus and GFP-Atg8 were colocalized, it was determined that cells pre-treated with LPS were more likely than untreated cells to exhibit moderate or nearly complete juxtaposition or colocalization between S. aureus and GFP-Atg8 (Fig 3C).
Fig. 3. Autophagosomes are recruited near S. aureus-containing vesicles upon treatment with LPS.
A. HR87 cells were pretreated with or without LPS (1 µg/mL) and incubated with S. aureus. Cells were fixed on poly-lysine slides and permeabilized. S. aureus was detected using biotin-labeled antibodies specific for S. aureus detected with AF594-conjugated streptavidin, while GFP-Atg8 was detected using mouse F(ab)’2 fragments specific for GFP detected with AF488-conjugated F(ab)’2 anti-mouse IgG. Images were acquired using a Zeiss LSM 510 laser scanning microscope and shown are the merged images of the optimal single planes at a magnification of 63x. Shown are representative images from five independent experiments.
B. The overlap coefficients (after Manders) of red and green pixels in cells treated with or without LPS were calculated using LSM software. Shown are means and SEM calculated from five independent experiments, analyzing at least 15 cells per condition for each experiment.
C. Depicted are the percentages of cells showing no colocalization (−), moderate juxtaposition (−/+), and almost complete juxtaposition or colocalization between S. aureus and GFP-Atg8 (+), as determined by a blinded observer using images acquired in 3B. * denotes significance (p<0.05) as measured by the paired t-test.
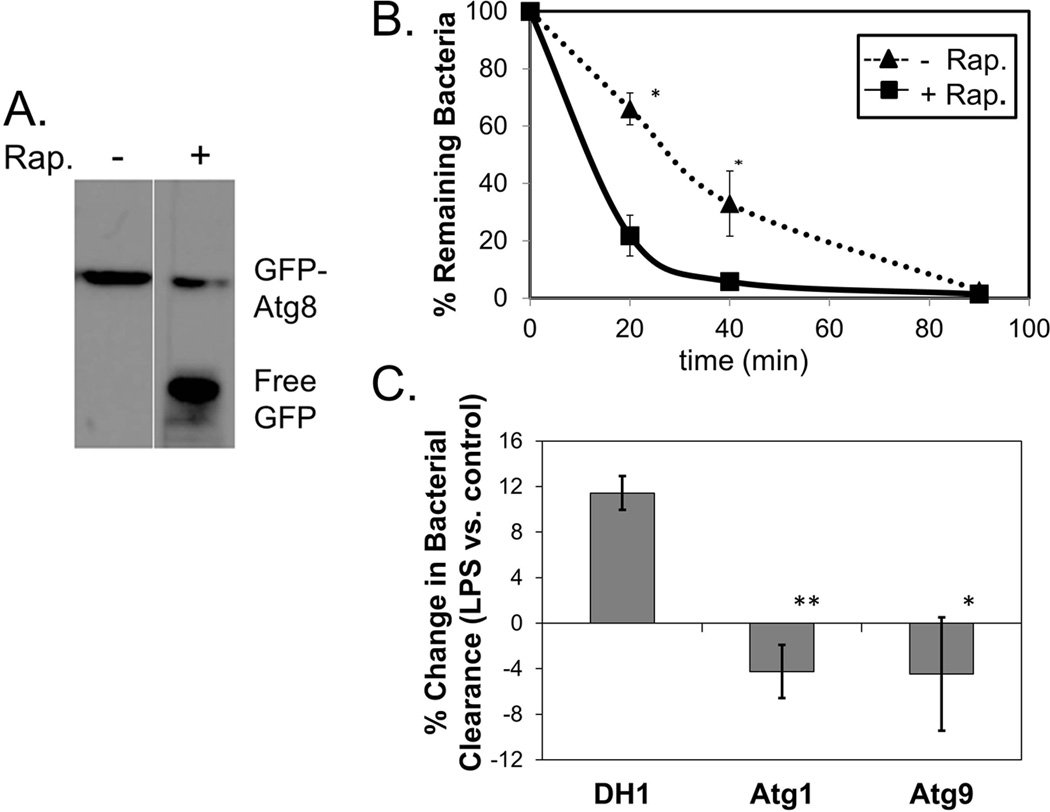
3.4 Induction of autophagy by rapamycin results in increased killing of phagocytized S. aureus
To determine if induction of autophagosomal maturation by LPS plays a role in the enhanced bactericidal activity observed in LPS-treated cells, we tested whether chemical induction of autophagy by treatment with the drug rapamycin affects bactericidal activity in D. discoideum. First we tested whether rapamycin, a drug that has been found to stimulate autophagy in a variety of cell types [31], had an effect on autophagosomal maturation in D. discoideum. Using the GFP-Atg8 cleavage assay, we found that treatment of D. discoideum cells with rapamycin leads to autophagosomal maturation (Fig. 4A).
Fig. 4. LPS-enhanced bactericidal activity is linked with induction of autophagy.
A. HR87 cells incubated with S. aureus at a ratio of 3 bacteria: 1 cell were treated with 100 nM rapamycin and subjected to the GFP-Atg8 cleavage assay to test for induction of autophagosomal maturation.
B. HR87 cells were treated with or without rapamycin (100 nM) and incubated with S. aureus. At various times following phagocytosis, cells were lysed and released bacteria were plated on nutrient agar to test for viability. Shown are mean percent survival and SEMs from four independent experiments. The overall effects of rapamycin on clearance of bacteria were significant (p<0.0001) as measured by fixed-effects two-way ANOVA. * denotes significance (p<0.05) at specific time points as measured by Tukey’s HSD multiple comparison.
C. DH1, Atg1− and Atg9− cells were pre-treated with or without 1 µg/mL LPS and incubated with SYTO9©-labeled S. aureus. Cells were washed of non-phagocytized bacteria and clearance of internalized bacteria was assessed by measuring the loss of fluorescently-labeled bacteria in cells after 45 min. The percent differences in bacterial clearance in cells treated with vs. without LPS for each cell type were calculated. Shown are mean differences and SEMs from five independent experiments. * and ** denote significance (p<0.05 and p<0.01, respectively) compared with percent differences seen in DH1 cells as measured by the paired t-test.
To determine if rapamycin-induced autophagy leads to increased killing of phagocytized bacteria, we monitored the intracellular survival of phagocytized S. aureus in D. discoideum cells treated or not with rapamycin. We found a significant decrease in survival of bacteria in cells treated with rapamycin at 20–40 min following phagocytosis (Fig. 4B). While 66% and 33% of the bacteria were viable in untreated cells after 20 min and 40 min, respectively, only 22% and 6% remained in rapamycin-treated cells at these same times.
3.5 The autophagy-related proteins Atg1 and atg9 are required for LPS-enhanced bacterial clearance
The role of autophagosomal induction on LPS-enhanced bacterial clearance was analyzed by monitoring the clearance of fluorescently-labeled S. aureus after LPS stimulation in cells deficient for the autophagy-related proteins Atg1 and Atg9. As had been shown previously for WT AX2 cells (5), pre-treatment with LPS resulted in enhanced clearance of phagocytized S. aureus from WT DH1 cells, with an average 11.5% increase in bacterial clearance after 45 min in cells treated with LPS (Fig 4C). However, LPS pre-treatment did not enhance clearance of phagocytized S. aureus in cells deficient for either atg1 or atg9 (Fig 4C).
4. Discussion
Our previous results showed that D. discoideum cells respond to the microbial pattern LPS by increasing bactericidal activity against various bacterial species, including S. aureus [5]. Here we extend these findings, demonstrating that LPS induces autophagosomal maturation in D. discoideum cells that have taken up S. aureus. Induction of autophagy promotes enhanced bactericidal activity in D. discoideum, as cells treated with the autophagy-inducing drug rapamycin cleared phagocytized S. aureus more efficiently than control cells. Furthermore, induction of autophagy appears necessary for LPS-enhanced bactericidal activity, as LPS treatment failed to affect clearance of S. aureus in cells deficient for atg1 or atg9.
Our results linking LPS recognition with induction of autophagy in D. discoideum cells are consistent with findings that microbial pattern recognition upregulates autophagy in mammalian cells. Multiple groups have reported that LPS, as well as additional microbial ligands, stimulate autophagy in mammalian macrophages [14–18]. Although most studies linking microbial pattern recognition and autophagy have been carried out with mammalian cells, a relationship between pattern recognition and autophagy has been reported in the invertebrate Drosophila melongaster, as signaling via the Drosophila peptidoglycan-recognition protein (PGRP) induces the formation of autophagosomes in response to the intracellular bacteria Listeria monocytogenes [32]. Our studies here point to an even earlier emergence of a link between autophagy and microbial pattern recognition, with regulation of autophagy by microbial pattern recognition as a conserved process even in single-celled D. discoideum.
The induction of autophagy by microbial ligands provides an additional tool for innate immune defense. Genetic knockout or knockdown of autophagy-related proteins in a wide variety of organisms results in increased susceptibility to infections with various intracellular bacteria and viruses [13; 33]. In D. discoideum cells, deletion of the autophagy-related genes atg1, atg6 and atg7 results in increased survival of intracellular S. typhimurium [25], while deletion of atg9 leads to increased survival of intracellular Legionella [26]. A role for autophagy in the response of D. discoideum to intracellular bacteria is further supported by our observations here that autophagosomal maturation is induced in D. discoideum cells incubated along with S. typhimurium and K. pneumoniae.
Although evidence is accumulating that autophagy plays important protective roles against microbial invaders, perhaps not surprisingly, it has been found that bacterial species have evolved mechanisms to evade and even exploit autophagosomal processes to enhance their replication within host cells [34–36]. While some bacterial species escape from autophagosomes into the cytoplasm, others alter autophagosomal processes, inhibiting autophagosomal fusion with degradative lysosomes, and thus creating a replicative niche [37].
Recent studies in cultured mammalian cells reveal that S. aureus may alter autophagosomal machinery. S. aureus-containing vesicles in mammalian epithelial cells recruit autophagosomal markers; however, these vesicles never fuse with lysosomes [38; 39]. In our studies with D. discoideum, we also observe that phagocytosis of S. aureus does not result in autophagosomal/lysosomal fusion. However, we do not find evidence that S. aureus recruits autophagosomal proteins to create a protective niche in D. discoideum cells. High levels of GFP-Atg8 do not colocalize with intracellular S. aureus in D. discoideum unless the cells are pre-treated with LPS. In addition, rapamycin-mediated induction of autophagy in D. discoideum results in increased killing of S. aureus, pointing to a protective function for autophagy against S. aureus. It is worth noting that although Legionella has been found to exploit autophagosomal processes for enhanced replication in mammalian cells, results in D. discoideum cells deficient for the autophagosomal protein Atg9 reveal a protective effect of autophagy against Legionella [26]. Taken together these results may suggest differing evolution of microbial interactions with amoeboid vs. mammalian hosts. On the other hand, given that the mammalian studies with S. aureus were completed in non-professional phagocytes [38; 39], further studies with mammalian macrophages might reveal additional similarities among autophagosomal responses by professional phagocytes in these divergent hosts.
Whatever differences might be seen regarding induction of autophagosomal processes in mammalian and D. discoideum cells, our findings that LPS promotes autophagosomal maturation in D. discoideum cells and that the promotion of autophagosomal maturation is associated with enhanced bactericidal activity might prove interesting for therapeutic purposes. If stimulation with microbial ligands can also induce autophagosomal fusion with lysosomes in mammalian cells infected with S. aureus, one might envision development of treatments for infections associated with S. aureus, including methicillin resistant S. aureus (MRSA). It has been proposed previously that microbial ligands that stimulate autophagy might play therapeutic roles against intracellular pathogens, as incubation of mammalian macrophages with the microbial ligands ssRNA and imiquimod results in enhanced elimination of the M. tuberculosis var. bovis Bacille Calmette-Guérin (BCG) in an autophagy-dependent manner [14].
In conclusion, our results here indicate that LPS-enhanced bactericidal activity in D. discoideum cells is mediated, at least in part, by induction of autophagosomal processes. These results extend previous findings that autophagy plays a protective role against intracellular bacteria in D. discoideum, and for the first time link microbial pattern recognition with autophagosomal induction in D. discoideum, as previous reports have linked these processes in mammalian cells.
Autophagy is induced in D. discoideum exposed to S. typhimurium and K. pneumoniae.
LPS induces autophagosomal maturation in D. discoideum cells exposed to S. aureus.
Rapamycin-induced autophagy enhances bactericidal activity in D. discoideum.
LPS-enhanced bacterial clearance in D. discoideum is dependent on Atg1 and Atg9.
Acknowledgements
We would like to thank Drs. M. May and G. Snyder for discussions and careful reading of the manuscript, Drs. B. Margulies and B. Masters for helpful suggestions, and C. Kesler, O. Anochie and L. Winter for technical assistance. HR87, DH1, Atg1- and Atg9-deficient cells were obtained from the Dicty Stock Center. The project was supported by Award Number R15AI085503 from NIAID. K. Yovo was supported by an NIH Bridges to the Baccalaureate Award Number 2R25GM058264. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIAID or NIH. The project was also supported by grants to M. Snyder from the TU Faculty Development and Research Committee.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu.Rev.Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 2.Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. Int.Rev.Immunol. 2011;30:16–34. doi: 10.3109/08830185.2010.529976. [DOI] [PubMed] [Google Scholar]
- 3.Cosson P, Soldati T. Eat, kill or die: when amoeba meets bacteria. Curr.Opin.Microbiol. 2008;11:271–276. doi: 10.1016/j.mib.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 4.Chen G, Zhuchenko O, Kuspa A. Immune-like phagocyte activity in the social amoeba. Science. 2007;317:678–681. doi: 10.1126/science.1143991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walk A, Callahan J, Srisawangvong P, et al. Lipopolysaccharide enhances bactericidal activity in Dictyostelium discoideum cells. Dev.Comp Immunol. 2011;35:850–856. doi: 10.1016/j.dci.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benghezal M, Fauvarque MO, Tournebize R, et al. Specific host genes required for the killing of Klebsiella bacteria by phagocytes. Cell Microbiol. 2006;8:139–148. doi: 10.1111/j.1462-5822.2005.00607.x. [DOI] [PubMed] [Google Scholar]
- 7.Eichinger L, Pachebat JA, Glockner G, et al. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sillo A, Bloomfield G, Balest A, et al. Genome-wide transcriptional changes induced by phagocytosis or growth on bacteria in Dictyostelium. BMC.Genomics. 2008;9:291. doi: 10.1186/1471-2164-9-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delgado M, Singh S, De HS, et al. Autophagy and pattern recognition receptors in innate immunity. Immunol.Rev. 2009;227:189–202. doi: 10.1111/j.1600-065X.2008.00725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat.Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deretic V. Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol.Rev. 2011;240:92–104. doi: 10.1111/j.1600-065X.2010.00995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delgado MA, Elmaoued RA, Davis AS, et al. Toll-like receptors control autophagy. EMBO J. 2008;27:1110–1121. doi: 10.1038/emboj.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanjuan MA, Dillon CP, Tait SW, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 16.Shi CS, Kehrl JH. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J. Biol.Chem. 2008;283:33175–33182. doi: 10.1074/jbc.M804478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shin DM, Yuk JM, Lee HM, et al. Mycobacterial lipoprotein activates autophagy via TLR2/1/CD14 and a functional vitamin D receptor signalling. Cell Microbiol. 2010;12:1648–1665. doi: 10.1111/j.1462-5822.2010.01497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Y, Jagannath C, Liu XD, et al. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007;27:135–144. doi: 10.1016/j.immuni.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.King JS, Veltman DM, Insall RH. The induction of autophagy by mechanical stress. Autophagy. 2011;7 doi: 10.4161/auto.7.12.17924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calvo-Garrido J, Carilla-Latorre S, Kubohara Y, et al. Autophagy in Dictyostelium: genes and pathways, cell death and infection. Autophagy. 2010;6:686–701. doi: 10.4161/auto.6.6.12513. [DOI] [PubMed] [Google Scholar]
- 21.Duran JM, Anjard C, Stefan C, et al. Unconventional secretion of Acb1 is mediated by autophagosomes. J. Cell Biol. 2010;188:527–536. doi: 10.1083/jcb.200911154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giusti C, Tresse E, Luciani MF, Golstein P. Autophagic cell death: analysis in Dictyostelium. Biochim.Biophys.Acta. 2009;1793:1422–1431. doi: 10.1016/j.bbamcr.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 23.Otto GP, Wu MY, Kazgan N, et al. Macroautophagy is required for multicellular development of the social amoeba Dictyostelium discoideum. J.Biol.Chem. 2003;278:17636–17645. doi: 10.1074/jbc.M212467200. [DOI] [PubMed] [Google Scholar]
- 24.Otto GP, Wu MY, Kazgan N, et al. Dictyostelium macroautophagy mutants vary in the severity of their developmental defects. J. Biol.Chem. 2004;279:15621–15629. doi: 10.1074/jbc.M311139200. [DOI] [PubMed] [Google Scholar]
- 25.Jia K, Thomas C, Akbar M, et al. Autophagy genes protect against Salmonella typhimurium infection and mediate insulin signaling-regulated pathogen resistance. Proc.Natl.Acad.Sci.U.S.A. 2009;106:14564–14569. doi: 10.1073/pnas.0813319106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tung SM, Unal C, Ley A, et al. Loss of Dictyostelium ATG9 results in a pleiotropic phenotype affecting growth, development, phagocytosis and clearance and replication of Legionella pneumophila. Cell Microbiol. 2010;12:765–780. doi: 10.1111/j.1462-5822.2010.01432.x. [DOI] [PubMed] [Google Scholar]
- 27.Maselli A, Laevsky G, Knecht DA. Kinetics of binding, uptake and degradation of live fluorescent (DsRed) bacteria by Dictyostelium discoideum. Microbiology. 2002;148:413–420. doi: 10.1099/00221287-148-2-413. [DOI] [PubMed] [Google Scholar]
- 28.Klionsky DJ, Cuervo AM, Seglen PO. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;3:181–206. doi: 10.4161/auto.3678. [DOI] [PubMed] [Google Scholar]
- 29.Calvo-Garrido J, Carilla-Latorre S, Mesquita A, Escalante R. A proteolytic cleavage assay to monitor autophagy in Dictyostelium discoideum. Autophagy. 2011;7:1063–1068. doi: 10.4161/auto.7.9.16629. [DOI] [PubMed] [Google Scholar]
- 30.Seglen PO, Gordon PB. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc.Natl.Acad.Sci.U.S.A. 1982;79:1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cutler NS, Heitman J, Cardenas ME. TOR kinase homologs function in a signal transduction pathway that is conserved from yeast to mammals. Mol.Cell Endocrinol. 1999;155:135–142. doi: 10.1016/s0303-7207(99)00121-5. [DOI] [PubMed] [Google Scholar]
- 32.Yano T, Mita S, Ohmori H, et al. Autophagic control of listeria through intracellular innate immune recognition in Drosophila. Nat.Immunol. 2008;9:908–916. doi: 10.1038/ni.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host.Microbe. 2009;5:527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knodler LA, Celli J. Eating the strangers within: host control of intracellular bacteria via xenophagy. Cell Microbiol. 2011;13:1319–1327. doi: 10.1111/j.1462-5822.2011.01632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lerena MC, Vazquez CL, Colombo MI. Bacterial pathogens and the autophagic response. Cell Microbiol. 2010;12:10–18. doi: 10.1111/j.1462-5822.2009.01403.x. [DOI] [PubMed] [Google Scholar]
- 36.Ogawa M, Mimuro H, Yoshikawa Y, et al. Manipulation of autophagy by bacteria for their own benefit. Microbiol.Immunol. 2011;55:459–471. doi: 10.1111/j.1348-0421.2011.00343.x. [DOI] [PubMed] [Google Scholar]
- 37.Joshi AD, Swanson MS. Secrets of a successful pathogen: Legionella resistance to progression along the autophagic pathway. Front Microbiol. 2011;2:138. doi: 10.3389/fmicb.2011.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schnaith A, Kashkar H, Leggio SA, et al. Staphylococcus aureus subvert autophagy for induction of caspase-independent host cell death. J. Biol.Chem. 2007;282:2695–2706. doi: 10.1074/jbc.M609784200. [DOI] [PubMed] [Google Scholar]
- 39.Mestre MB, Fader CM, Sola C, Colombo MI. Alpha-hemolysin is required for the activation of the autophagic pathway in Staphylococcus aureus-infected cells. Autophagy. 2010;6:110–125. doi: 10.4161/auto.6.1.10698. [DOI] [PubMed] [Google Scholar]