Abstract
Background
Allogeneic hematopoietic cell transplantation is the main curative therapy for patients with chronic myeloid leukemia who do not respond to tyrosine kinase inhibitors. It has been proposed that non-human leukocyte antigen gene polymorphisms influence outcome after hematopoietic cell transplantation and could be used alongside traditional patient-donor and transplant characteristics to create a recipient risk profile associated with allogeneic hematopoietic cell transplantation.
Design and Methods
A previous study from the European Group for Blood and Marrow Transplantation showed that the absence of recipient tumor necrosis factor receptor II, absence of donor interleukin 10 ATA/ACC and presence of donor interleukin 1 receptor antagonist allele 2 genotypes were associated with decreased survival and increased non-relapse mortality in adult patients with chronic myeloid leukemia undergoing myeloablative human leukocyte antigen-identical sibling transplantation. To explore these associations in unrelated donor transplantation, these polymorphisms were genotyped in 383 adult patients with chronic myeloid leukemia who underwent hematopoietic cell transplantation from unrelated donors matched for 10/10 human leukocyte antigens.
Results
The polymorphisms were not associated with overall survival, non-relapse mortality, relapse or acute graft-versus-host disease in the unrelated donor cohort. Comparison of the unrelated donor and human leukocyte antigen-identical sibling cohorts showed differences in survival and clinical characteristics.
Conclusions
We did not confirm that non-human leukocyte antigen polymorphisms were associated with outcomes in myeloablative unrelated donor hematopoietic cell transplantation for chronic myeloid leukemia, possibly because of the strong association between clinical variables and outcome which masked more subtle genetic effects.
Keywords: chronic myeloid leukemia, CML, polymorphisms, outcome, risk factors
Introduction
Although there has been a dramatic change in the role of allogeneic hematopoietic cell transplantation (HCT) for chronic myeloid leukemia (CML) since the introduction of tyrosine kinase inhibitors, HCT remains a curative option for patients in whom tyrosine kinase inhibitor therapy fails or who are intolerant of the drug.1 The main clinical factors influencing outcomes after HCT for CML have been identified through studies of large cohorts.2 The European Group for Blood and Marrow Transplantation (EBMT) derived a clinical risk score for CML, applicable to matched unrelated donor (URD) and human leukocyte antigen (HLA)-matched sibling patient/donor pairs.3 Five clinical variables (disease stage, recipient age, donor/recipient sex match, interval from time of diagnosis to HCT, donor type) are weighted and summed to calculate this risk score, which ranges from 0 to 7. The EBMT score has been further confirmed as valid for all patients undergoing HCT for a hematologic disorder4 and for patients undergoing reduced-intensity conditioning.5 Recognition of these important clinical factors associated with transplant success has led to different treatment recommendations, depending on the degree of risk associated with CML and HCT for each individual patient. The widespread availability of well-tolerated and effective tyrosine kinase inhibitors6–8 has made them first-line therapy for newly diagnosed CML patients today. For patients who do not respond, lose response, or cannot tolerate these drugs, HCT remains the therapy of choice.1,9 Unfortunately, many of these patients may have advanced disease prior to HCT. A better characterization of the risk profile associated with allogeneic HCT is, therefore, urgently warranted to help tailor treatment recommendations for patients with CML.
In addition to traditional patient-donor and transplant characteristics, several groups have demonstrated that non-HLA gene polymorphisms, including those of the innate immune system, pro- and anti-inflammatory cytokines, and steroid receptors, predict post-HCT outcomes.10 In general, these studies were based on small series and the results were not adjusted for the key risk factors for HCT outcomes. The role of non-HLA genetics was previously studied in a homogenous cohort of 228, primarily Caucasian, adult CML patients with HLA-matched sibling donors, transplanted from 1984–2003;11 the results were reported to the EBMT. Among nine targeted cytokines, cytokine receptors, or other genes of interest, an intronic variable number tandem repeat within the gene for interleukin 1 receptor antagonist (ILIRA), a single nucleotide polymorphism (SNP) within the tumor necrosis factor II receptor superfamily, member 1B (TNFRSF1B) gene, and the interleukin 10 (IL10) gene were associated with survival when analyzed alone and alongside the EBMT risk score. There was no evidence of association between these three genotypes and relapse or acute or chronic graft-versus-host disease (GVHD), but there was some evidence of association with non-relapse mortality.11
The present study was intended to assess the impact of these three high-risk genotypes on outcomes of adult URD HCT for CML, adjusting for the EBMT risk score using data provided by the Center for International Blood and Marrow Transplant Research (CIBMTR). The transplants were done in the era prior to tyrosine kinase inhibitor therapy.
Design and Methods
Data collection
Data used in this study were obtained from the Statistical Center of the CIBMTR. The CIBMTR is a research affiliation of the International Bone Marrow Transplant Registry (IBMTR) and the National Marrow Donor Program (NMDP), established in 2004, which comprises a voluntary working group of more than 450 transplantation centers worldwide which contribute detailed data on consecutive allogeneic and autologous hematopoietic HCT to the Statistical Center at the Medical College of Wisconsin in Milwaukee and the NMDP Coordinating Center in Minneapolis. Participating centers are required to report all transplants consecutively; compliance is monitored by on-site audits. Patients are followed longitudinally, with yearly follow-ups. Computerized checks for discrepancies, physicians’ review of submitted data and on-site audits of participating centers ensure data quality. Observational studies conducted by the CIBMTR are performed in compliance with the Privacy Rule (HIPAA) as a Public Health Authority, and in compliance with all applicable federal regulations pertaining to the protection of human research participants as determined by continuous review of the Institutional Review Boards of the NMDP and the Medical College of Wisconsin since 1985.
Inclusion criteria
The study population included 383 HCT recipients, primarily Caucasian, transplanted from 10/10 HLA-matched donors between 1989 and 2002. The study was restricted to adult patients (16 years or older), with a diagnosis of CML at any disease stage, with HCT performed using myeloablative conditioning. Informed consent was obtained in accordance with the Declaration of Helsinki. All surviving URD recipients included in this analysis were retrospectively contacted and provided informed consent to participation in the NMDP research program. Informed consent was waived by the NMDP Institutional Review Board for all deceased recipients. Approximately 10% of surviving patients did not consent to the use of the research data. To adjust for the potential bias introduced by exclusion of non-consenting surviving patients, a corrective action plan modeling process randomly excluded the same percentage of deceased patients using a biased coin randomization with exclusion probabilities based on characteristics associated with not providing consent for use of the data in survivors.12
Genotyping
All recipient-donor pairs were confirmed to be 10/10 HLA-matched at HLA-A, B, C, DRB1 and DQB1. DNA from the recipients and donors was analyzed at a single coordination center (Newcastle upon Tyne, UK) for SNP or microsatellites for IL1RA,13 IL10,14 and TNFRSF1B15 gene polymorphisms. Methods were carried out as previously described.11
Endpoints
The aim of the study was to validate the results from the CML HLA-matched sibling study using CML unrelated donors. The outcomes analyzed were overall survival, non-relapse mortality, relapse, and acute and chronic GVHD.
The acute GVHD endpoint was defined as the development of grades II-IV according to the Glucksberg criteria.16 Chronic GVHD was diagnosed following older definitions.17 Relapse was defined as leukemia recurrence, and non-relapse mortality was death resulting from any cause other than relapse. Competing risks were relapse and death.
Biostatistical analysis
The EBMT score was calculated based on five variables: disease stage (0 for early stage, 1 for intermediate stage and 2 for late stage), age of recipient (0 for < 20 years, 1 for 20–40 years, and 2 for > 40 years), donor-recipient sex matching (1 for female donor to male recipient and 0 for other gender combinations), interval from time of diagnosis to transplant (0 for ≤ 12 months, and 1 for > 12 months) and type of donor (1 for URD transplants and 0 for sibling transplants).3 For the multivariate analysis, Cox proportional hazards regression models were applied, adjusting for the EBMT risk score. The proportional hazards assumption was assessed for each covariate and found to be valid. SAS software version 9.1 (SAS Institute, Cary, NC, USA), NCSS and R were used in all the analyses. Since multiple SNP and multiple endpoints were tested, a P value less than 0.01 was considered statistically significant.
Patient-, disease-, and transplant-related variables were compared between the URD cohort and the previously published Chronic Leukemia Working Party HLA-identical sibling cohort11 using two-sample t-tests and tests of two proportions.
Results
Patients’ characteristics
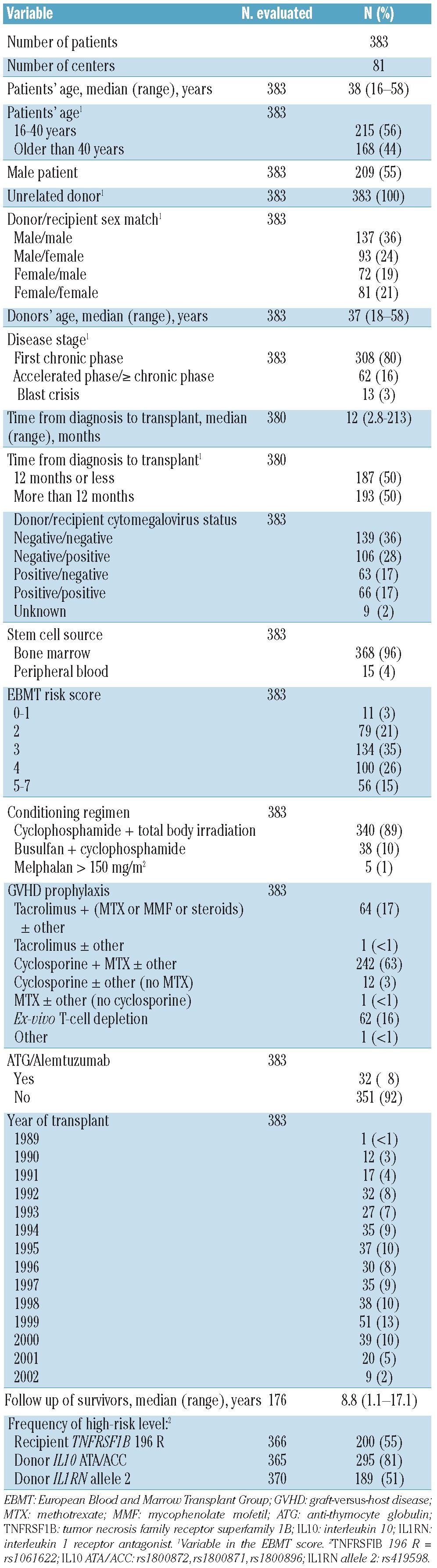
Table 1 shows the characteristics of the 383 URD recipients. The median age of the recipients was 38 years, while that of the donors was 37 years, 55% of the recipients were male and 80% were in first chronic phase at the time of HCT. Ninety-six percent received bone marrow; 89% were given cyclophosphamide and total body irradiation conditioning. The median follow-up of survivors was 8.8 years. The cumulative incidence of acute GVHD II-IV was 65% (95% CI 60%–70%) and the 2-year cumulative incidence of chronic GVHD was 62% (95% CI 56%–68%). The observed frequency of the high-risk level of the genotypes was similar to the pattern previously observed in the HLA-identical sibling cohort for recipient TNFRSF1B 196R (55% versus 55%, P=0.97). In contrast, URD donors showed different frequencies in the high-risk level of IL10 ATA/ACC (81% versus 88%, P=0.02) and IL1RN allele 2 (51% versus 20%, P<0.0005).
Table 1.
Characteristics of the patients and their transplants.
Analysis of the genetic variables
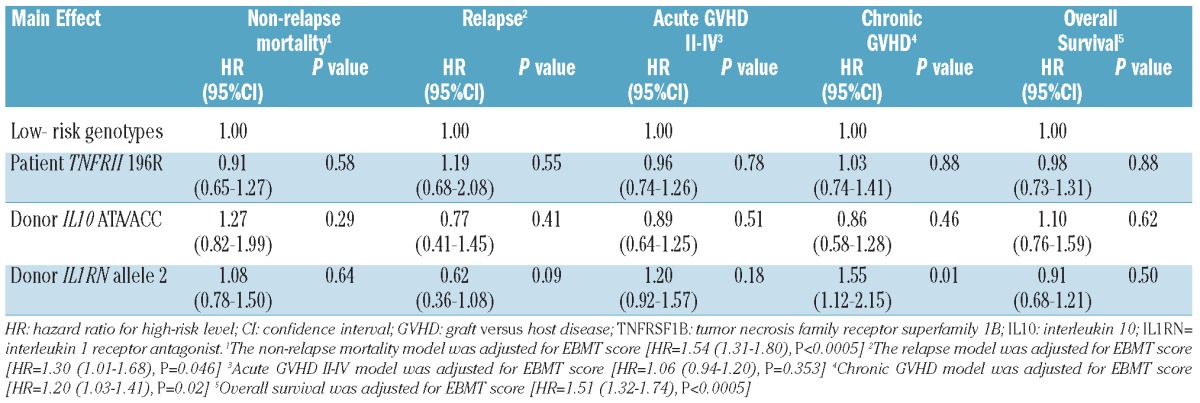
Table 2 shows that the EBMT clinical risk score was an important predictor but that there was no association between recipient TNFRSF1B 196R, donor IL10 ATA/ACC, or donor IL1RN allele 2 with most of the outcomes. Only donor IL1RN allele 2 was associated with an increased risk of chronic GVHD (HR 1.55, 95% CI 1.12–2.15; P=0.01). The three SNP also showed non-significance after repeating the analysis using only those components of the EBMT risk score which were predictive of each individual outcome.
Table 2.
Multivariate analysis for polymorphisms.
Comparison of data sets
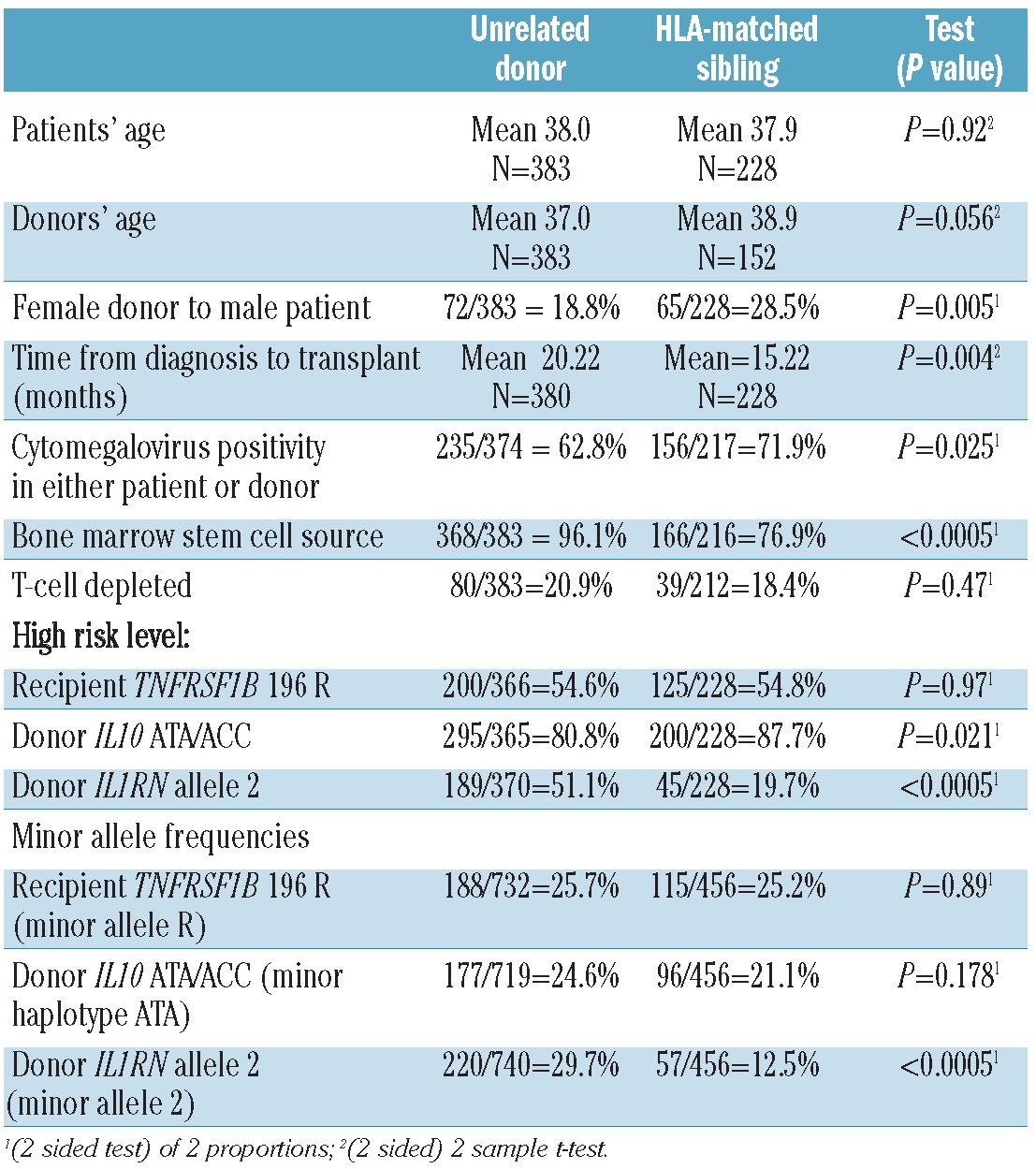
Given the different results seen in the previous sibling analysis and the current URD study, we tested for differences between the clinical characteristics and outcomes (Table 3). The URD cohort had some higher risk characteristics, e.g. longer time from diagnosis to transplant (mean 20.2 months versus 15.2 months, P=0.004), and some lower risk characteristics, e.g. fewer transplants from female donors to male patients (18.8% versus 28.5%, P=0.005), than the previously studied HLA-identical sibling group. The URD cohort was more likely to have received bone marrow (96.1% versus 76.9%, P<0.0005). The URD cohort was also less likely to have cytomegalovirus positivity in either the recipients or the donors (62.8% versus 71.9%, P=0.025). The patients’ age, donors’ age, and use of T-cell depletion were not statistically different. Estimated survival at 2 years was lower for the URD recipients (54%, 95% CI 49%–59%) than for the sibling recipients (68%, 95% CI 62%–74%) (P=0.002) and acute GVHD II-IV rates were higher (66% versus 42%, P<0.0005). The non-relapse mortality rate at 2 years was higher among the URD recipients (38%, 95% CI 33%–43%) than among the sibling HCT recipients (30%, 95% CI 24%–38%) (P=0.016). The 2-year relapse rate was lower for the URD recipients (13%, 95% CI 10%–17%) than for the sibling recipients (19%, 95% CI 14%–26%) (P=0.003). Individual EBMT risk factors were more predictive of outcomes in the URD cohort than in the HLA-identical sibling group (results not shown).
Table 3.
Comparison of selected characteristics in the unrelated donor and HLA-matched sibling data sets.
Discussion
We studied 383 recipients of HLA-matched URD HCT with myeloablative conditioning for the treatment of CML, in order to validate previously described associations between non-HLA polymorphisms and outcomes. In our study, recipient TNFRSF1B 196 R, donor IL10 ATA/ACC, and donor IL1RN allele 2 were not associated with overall survival, non-relapse mortality, relapse, or acute GVHD. Only donor IL1RN allele 2 was marginally associated with an increased risk of chronic GVHD.
In the previous study of 228 HLA-identical sibling donor pairs, in which associations were identified between these polymorphisms and worse survival, the hazard ratios ranged from 1.82 (P=0.01) to 2.45 (P=0.001).11 An association of these polymorphisms with survival was biologically plausible since they are known to be involved in the inflammatory “cytokine storm” that characterizes acute GVHD. All have been reported to be associated with important transplant outcomes in previous studies. Absence of TNFRSF1B 196R in the recipient causes increased serum levels of soluble TNFRII and decreased circulating TNF. Absence of donor IL10 ATA/ACC is associated with higher levels of IL-10.18–20 Presence of donor IL1RA (allele 2) causes down-regulation of IL1 by IL1-receptor antagonist.13
In other studies, the presence of the TNFRSF1B 196R polymorphism was associated with autoimmune disease, increased incidence of acute GVHD and higher IL-6 levels.15 One URD study found that the absence of recipient TNFRSF1B 196R was associated with worse survival, consistent with Dickinson’s findings.21 The absence of recipient IL10-ATA in HLA-identical sibling transplants was associated with more severe acute GVHD.22 Xiao showed that even in heterogeneous related and unrelated donor cohorts, certain TGFB and IL10 polymorphisms (TGFB1-509T/T genotype, IL10 -1092, -819CC and -592 C/C genotypes) were associated with GVHD and survival.23 The absence of the IL10 genotype (ATA/ACC) in the donors, as shown in our sibling CML cohort,11 was associated with decreased survival and death in remission. Absence of the ATA haplotype in the donors was also associated with death in remission in an unrelated and sibling Chinese transplantation population.23
Ideally, populations used for validation have characteristics that are similar to the initial training dataset to minimize confounding effects. In the current study, we tested the hypothesis that genetic polymorphisms previously found to be of importance in sibling transplantation were also clinically significant in unrelated HCT. It is worth noting that some of the clinical characteristics of our unrelated cohort differed from those of the historical sibling population (Table 3), especially those incorporated into the EBMT risk score; however, the EBMT risk score was still found to be strongly associated with overall survival in both the unrelated cohort and historical sibling groups. The survival rate of the URD cohort was also worse and the acute GVHD II-IV rate higher than those of the HLA-identical sibling cohort. We do not have accurate data on other medical comorbidities24 or disease factors that could explain the differences and could, therefore, negate any influence of non-HLA genetic factors.
In other studies, donor HLA-DPB1 mismatching was associated with an increased risk of acute GVHD.25 An insufficient number of samples in the current analysis were genotyped for this locus and precluded consideration of DPB1 in the models. Whether HLA-DP mismatching might explain the differences in outcomes between the historic sibling cohort and the unrelated population remains to be tested. Regardless, HLA-DP mismatching is not likely to confound the analysis of the URD cohort, because the frequency of HLA-DP mismatching is independent of TNFRSF1B, IL10, and IL1RA genotypes. Finally, the frequencies of donor high-risk polymorphisms were very different for two of the SNP in the URD cohort compared to in the sibling cohort. Since the classical HLA loci are the only genetic determinants for which donors are selected, the degree of genetic variation elsewhere in the genome is predicted to be higher among a pool of unrelated individuals than among genotypically identical siblings.
Whether the cumulative effects of such variation obscures potential effects of the genes of interest remains an interesting question for future studies. Finally, the sibling and unrelated donor cohorts differed with respect to the stem cell source – more patients in the URD cohort received bone marrow (Table 3). Whether the effects of the SNP of interest were attenuated in the setting of marrow compared to peripheral blood stem cells is an intriguing possibility that will require a larger transplant experience to clarify. It is interesting to note that after re-analysis using a sub-group of the historical sibling population, in which patients received bone marrow only, the relationship between overall survival and the three polymorphisms was still strong.
Our current study shows that the EBMT clinical risk score is still an important predictor of outcomes across variable cohorts, and should be used alongside SNP variables to enhance the understanding of factors that influence survival. Differences in the distribution of outcomes between the cohorts might have contributed to the lack of association between the polymorphisms and outcomes. We also found that the individual components of the EBMT risk score were more strongly associated with outcomes in the URD cohort than in the HLA-identical sibling cohort, especially for the survival outcome. We postulate that the strong association between clinical variables and outcome masked more subtle genetic effects for the URD cohort.
The challenge now is to identify those SNP that influence outcomes irrespective of the heterogeneity of the clinical cohorts. This can only be accomplished by the use of large cohorts of patients and genome-wide association studies. SNP analyses, as recently reviewed by Hansen et al., Mullighan et al. and Dickinson and Holler,26–28 have shown conclusively that SNP in the recipients and donors are associated with GVHD and survival. However, even current genome-wide association studies are inconclusive and difficult to replicate26 due to heterogeneity of cohorts of patients and clinical covariates.
For patients with CML, given the successful use of tyrosine kinase inhibitors and the consequent decreased use of transplantation, we would now need large collaborative studies to address future questions. Although we were not able to validate the hypothesized relationship between the evaluated SNP and transplant outcomes in unrelated donor transplantation, future studies should continue to refine HCT risk profiles for CML and other diseases so that treatment recommendations can be tailored accordingly. For example, certain subgroups of patients may benefit more from novel clinical trials, different standard treatment recommendations, or modifications of immunosuppressive therapy to improve their outcomes.
Footnotes
Funding: this work was supported by: EU grant ‘TRANSNET’ - contract number MRTN-CT-2004-512253; the grant ‘STEMDIAG-NOSTICS’ - contract number LSHB-CT-2007_037703; a Tyneside Leukaemia Research Association (TLRA) grant.
Funding
The CIBMTR is supported by Public Health Service Grant/Cooperative Agreement U24-CA76518 from the National Cancer Institute (NCI), the National Heart, Lung and Blood Institute (NHLBI) and the National Institute of Allergy and Infectious Diseases (NIAID); a Grant/Cooperative Agreement 5U01HL069294 from NHLBI and NCI; a contract HHSH234200637015C with Health Resources and Services Administration (HRSA/DHHS); two Grants N00014-06-1-0704 and N00014-08-1-0058 from the Office of Naval Research; and grants from Allos, Inc.; Amgen, Inc.; Angioblast; Anonymous donation to the Medical College of Wisconsin; Ariad; Be the Match Foundation; Blue Cross and Blue Shield Association; Buchanan Family Foundation; CaridianBCT; Celgene Corporation; CellGenix, GmbH; Children’s Leukemia Research Association; Fresenius-Biotech North America, Inc.; Gamida Cell Teva Joint Venture Ltd.; Genentech, Inc.; Genzyme Corporation; GlaxoSmithKline; Kiadis Pharma; The Leukemia & Lymphoma Society; The Medical College of Wisconsin; Millennium Pharmaceuticals, Inc.; Milliman USA, Inc.; Miltenyi Biotec, Inc.; National Marrow Donor Program; Optum Healthcare Solutions, Inc.; Otsuka America Pharmaceutical, Inc.; Seattle Genetics; Sigma-Tau Pharmaceuticals; Soligenix, Inc.; Swedish Orphan Biovitrum; THERAKOS, Inc.; and Wellpoint, Inc. The views expressed in this article do not reflect the official policy or position of the National Institute of Health, the Department of the Navy, the Department of Defense, or any other agency of the U.S. Government.
Authorship and Disclosures
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
References
- 1.Baccarani M, Cortes J, Pane F, Niederwieser D, Saglio G, Apperley J, et al. Chronic myeloid leukemia: an update of concepts and management recommendations of European LeukemiaNet. J Clin Oncol. 2009;27(35):6041–51. doi: 10.1200/JCO.2009.25.0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Passweg JR, Walker I, Sobocinski KA, Klein JP, Horowitz MM, Giralt SA. Validation and extension of the EBMT Risk Score for patients with chronic myeloid leukaemia (CML) receiving allogeneic haematopoietic stem cell transplants. Br J Haematol. 2004;125(5):613–20. doi: 10.1111/j.1365-2141.2004.04955.x. [DOI] [PubMed] [Google Scholar]
- 3.Gratwohl A, Hermans J, Goldman JM, Arcese W, Carreras E, Devergie A, et al. Risk assessment for patients with chronic myeloid leukaemia before allogeneic blood or marrow transplantation. Lancet. 1998;352(9134):1087–92. doi: 10.1016/s0140-6736(98)03030-x. [DOI] [PubMed] [Google Scholar]
- 4.Gratwohl A, Stern M, Brand R, Apperley J, Baldomero H, de Witte T, et al. Risk score for outcome after allogeneic hematopoietic stem cell transplantation: a retrospective analysis. Cancer. 2009;115(20):4715–26. doi: 10.1002/cncr.24531. [DOI] [PubMed] [Google Scholar]
- 5.Crawley C, Szydlo R, Lalancette M, Bacigalupo A, Lange A, Brune M, et al. Outcomes of reduced-intensity transplantation for chronic myeloid leukemia: an analysis of prognostic factors from the Chronic Leukemia Working Party of the EBMT. Blood. 2005;106(9):2969–76. doi: 10.1182/blood-2004-09-3544. [DOI] [PubMed] [Google Scholar]
- 6.Cortes JE, Baccarani M, Guilhot F, Druker BJ, Branford S, Kim DW, et al. Phase III, randomized, open-label study of daily imatinib mesylate 400 mg versus 800 mg in patients with newly diagnosed, previously untreated chronic myeloid leukemia in chronic phase using molecular end points: tyrosine kinase inhibitor optimization and selectivity study. J Clin Oncol. 2010;28(3):424–30. doi: 10.1200/JCO.2009.25.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kantarjian H, Shah NP, Hochhaus A, Cortes J, Shah S, Ayala M, et al. Dasatinib versus imatinib in newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med. 2010;362(24):2260–70. doi: 10.1056/NEJMoa1002315. [DOI] [PubMed] [Google Scholar]
- 8.Saglio G, Kim DW, Issaragrisil S, le Coutre P, Etienne G, Lobo C, et al. Nilotinib versus imatinib for newly diagnosed chronic myeloid leukemia. N Engl J Med. 2010;362(24):2251–9. doi: 10.1056/NEJMoa0912614. [DOI] [PubMed] [Google Scholar]
- 9.Hehlmann R, Berger U, Pfirrmann M, Heimpel H, Hochhaus A, Hasford J, et al. Drug treatment is superior to allografting as first-line therapy in chronic myeloid leukemia. Blood. 2007;109(11):4686–92. doi: 10.1182/blood-2006-11-055186. [DOI] [PubMed] [Google Scholar]
- 10.Dickinson AM, Charron D. Non-HLA immunogenetics in hematopoietic stem cell transplantation. Curr Opin Immunol. 2005;17(5):517–25. doi: 10.1016/j.coi.2005.07.017. [DOI] [PubMed] [Google Scholar]
- 11.Dickinson AM, Pearce KF, Norden J, O’Brien SG, Holler E, Bickeboller H, et al. Impact of genomic risk factors on outcome after hematopoietic stem cell transplantation for patients with chronic myeloid leukemia. Haematologica. 2010;95(6):922–7. doi: 10.3324/haematol.2009.016220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee SJ, Klein J, Haagenson M, Baxter-Lowe LA, Confer DL, Eapen M, et al. High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood. 2007;110(13):4576–4583. doi: 10.1182/blood-2007-06-097386. [DOI] [PubMed] [Google Scholar]
- 13.Cullup H, Dickinson AM, Jackson GH, Taylor PR, Cavet J, Middleton PG. Donor interleukin-1 receptor antagonist genotype associated with acute graft-versus-host disease in human leukocyte antigen-matched sibling allogeneic transplants. Br J Haematol. 2001;113(3):807–13. doi: 10.1046/j.1365-2141.2001.02811.x. [DOI] [PubMed] [Google Scholar]
- 14.Morse HR, Olomolaiye OO, Wood NA, Keen LJ, Bidwell JL. Induced heteroduplex genotyping of TNF-alpha, IL-1beta, IL-6 and IL-10 polymorphisms associated with transcriptional regulation. Cytokine. 1999;11(10):789–95. doi: 10.1006/cyto.1999.0491. [DOI] [PubMed] [Google Scholar]
- 15.Stark GL, Dickinson AM, Jackson GH, Taylor PR, Proctor SJ, Middleton PG, et al. Tumour necrosis factor receptor type II 196M/R genotype correlates with circulating soluble receptor levels in normal subjects and with graft-versus-host disease after sibling allogeneic bone marrow transplantation. Transplantation. 2003;76(12):1742–9. doi: 10.1097/01.TP.0000092496.05951.D5. [DOI] [PubMed] [Google Scholar]
- 16.Glucksberg H, Storb R, Fefer A, Buckner CD, Neiman PE, Clift RA, et al. Clinical manifestations of graft-versus-host disease in human recipients of marrow from HL-A-matched sibling donors. Transplantation. 1974;18(4):295–304. doi: 10.1097/00007890-197410000-00001. [DOI] [PubMed] [Google Scholar]
- 17.Shulman HM, Sullivan KM, Weiden PL, McDonald GB, Striker GE, Sale GE, et al. Chronic graft-versus-host syndrome in man. A long-term clinicopathologic study of 20 Seattle patients. Am J Med. 1980;69(2):204–17. doi: 10.1016/0002-9343(80)90380-0. [DOI] [PubMed] [Google Scholar]
- 18.Edwards-Smith CJ, Jonsson JR, Purdie DM, Bansal A, Shorthouse C, Powell EE. Interleukin-10 promoter polymorphism predicts initial response of chronic hepatitis C to interferon alfa. Hepatology. 1999;30(2):526–30. doi: 10.1002/hep.510300207. [DOI] [PubMed] [Google Scholar]
- 19.Reuss E, Fimmers R, Kruger A, Becker C, Rittner C, Hohler T. Differential regulation of interleukin-10 production by genetic and environmental factors--a twin study. Genes Immun. 2002;3(7):407–13. doi: 10.1038/sj.gene.6363920. [DOI] [PubMed] [Google Scholar]
- 20.Turner DM, Williams DM, Sankaran D, Lazarus M, Sinnott PJ, Hutchinson IV. An investigation of polymorphism in the interleukin-10 gene promoter. Eur J Immunogenet. 1997;24(1):1–8. doi: 10.1111/j.1365-2370.1997.tb00001.x. [DOI] [PubMed] [Google Scholar]
- 21.Keen LJ, DeFor TE, Bidwell JL, Davies SM, Bradley BA, Hows JM. Interleukin-10 and tumor necrosis factor alpha region haplotypes predict transplant-related mortality after unrelated donor stem cell transplantation. Blood. 2004;103(9):3599–602. doi: 10.1182/blood-2002-11-3568. [DOI] [PubMed] [Google Scholar]
- 22.Lin MT, Storer B, Martin PJ, Tseng LH, Gooley T, Chen PJ, et al. Relation of an interleukin-10 promoter polymorphism to graft-versus-host disease and survival after hematopoietic-cell transplantation. N Engl J Med. 2003;349(23):2201–10. doi: 10.1056/NEJMoa022060. [DOI] [PubMed] [Google Scholar]
- 23.Xiao H, Cao W, Lai X, Luo Y, Shi J, Tan Y, et al. Immunosuppressive cytokine gene polymorphisms and outcome after related and unrelated hematopoietic cell transplantation in a Chinese population. Biol Blood Marrow Transplant. 2011;17(4):542–9. doi: 10.1016/j.bbmt.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 24.Sorror ML, Storer B, Storb RF. Validation of the hematopoietic cell transplantation-specific comorbidity index (HCT-CI) in single and multiple institutions: limitations and inferences. Biol Blood Marrow Transplant. 2009;15(6):757–8. doi: 10.1016/j.bbmt.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shaw BE, Gooley TA, Malkki M, Madrigal JA, Begovich AB, Horowitz MM, et al. The importance of HLA-DPB1 in unrelated donor hematopoietic cell transplantation. Blood. 2007;110(13):4560–6. doi: 10.1182/blood-2007-06-095265. [DOI] [PubMed] [Google Scholar]
- 26.Hansen JA, Chien JW, Warren EH, Zhao LP, Martin PJ. Defining genetic risk for graft-versus-host disease and mortality following allogeneic hematopoietic stem cell transplantation. Curr Opin Hematol. 2010;17(6):483–92. doi: 10.1097/MOH.0b013e32833eb770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mullighan CG, Bardy PG. New directions in the genomics of allogeneic hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2007;13(2):127–44. doi: 10.1016/j.bbmt.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 28.Dickinson AM, Holler E. Polymorphisms of cytokine and innate immunity genes and GVHD. Best Pract Res Clin Haematol. 2008;21(2):149–64. doi: 10.1016/j.beha.2008.03.004. [DOI] [PubMed] [Google Scholar]


