Abstract
Juvenile myelomonocytic leukemia is a lethal disease of children characterized by hypersensitivity of hematopoietic progenitors to granulocyte macrophage-colony stimulating factor. Mutations in PTPN11, the gene encoding the protein tyrosine phosphatase Shp2, are common in juvenile myelomonocytic leukemia and induce hyperactivation of the phosphoinositide-3-kinase pathway. We found that genetic disruption of Pik3r1, the gene encoding the Class IA phosphoinositide-3-kinase regulatory subunits p85α, p55α and p50α, significantly reduced hyperproliferation and hyperphosphorylation of Akt in gain-of-function Shp2 E76K-expressing cells. Elevated protein levels of the phosphoinositide-3-kinase catalytic subunit, p110δ, in the Shp2 E76K-expressing Pik3r1−/− cells suggest that p110δ may be a crucial mediator of mutant Shp2-induced phosphoinositide-3-kinase hyperactivation. Consistently, treatment with the p110δ-specific inhibitor, IC87114, or the clinical grade pan-phosphoinositide-3-kinase inhibitor, GDC-0941, reduced granulocyte macrophage-colony stimulating factor hypersensitivity. Treatment with the farnesyltransferase inhibitor, tipifarnib, showed that Shp2 E76K induces hyperactivation of phosphoinositide-3-kinase by both Ras-dependent and Ras-independent mechanisms. Collectively, these findings implicate Class IA phosphoinositide-3-kinase as a relevant molecular target in juvenile myelomonocytic leukemia.
Keywords: juvenile myelomonocytic leukemia, granulocyte macropage colony stimulating factor, phosphoinositide 3-kinase, PTPN11
Introduction
Juvenile myelomonocytic leukemia (JMML) is a rare myelodysplastic syndrome/myeloproliferative neoplasm (MDS/MPN) of young children, characterized by clonal expansion of monocyte/macrophage-lineage cells and by granulocyte macrophage-colony stimulating factor (GM-CSF) hypersensitivity.1 JMML responds poorly to standard chemotherapy, and allogeneic bone marrow transplant (the only curative therapy) has a high relapse rate approaching 40%.2 GM-CSF hypersensitivity is associated with hyperactivation of Ras signaling, and can result from loss-of-function mutations in NF1 (15% of JMML cases)3 or c-CBL (10–15%),4 or gain-of-function mutations in N-RAS or K-RAS (20%)5,6 or PTPN11 (35%).7
PTPN11 encodes the protein tyrosine phosphatase, Shp2, which is involved in many signaling processes and is known to promote activation of Ras-MAPK signaling through incompletely understood mechanisms.8 We previously demonstrated that the leukemia-associated mutations E76K and D61Y result in GM-CSF-stimulated hyperproliferation, Erk hyperphosphorylation, and Akt hyperphosphorylation.9,10 Although the role of Ras-MAPK signaling in gain-of-function Shp2-induced GM-CSF hypersensitivity is well-established, the contribution of phosphoinositide 3-kinase (PI3K)-Akt signaling remains to be clarified.
PI3Ks are a family of lipid kinases that generate lipid second messengers which promote proliferation, survival and migration. Class IA PI3Ks are heterodimers composed of one regulatory subunit, p85α (or its splice variants p55α or p50α), p85β or p55γ, and one catalytic subunit, p110α, p110β or p110δ. The regulatory subunits function to recruit the p110 catalytic subunits to phospho-tyrosine residues on membrane associated proteins via their SH2 domains as well as to stabilize the p110 catalytic subunits, preventing their rapid degradation.11 Previous mechanistic studies demonstrated that GM-CSF induces nucleation of a protein complex at GM-CSF receptor βc chain at tyrosine (Y) 595 and/or Y612 consisting of Shp2, Grb2 and/or Gab2, which are able to interact with p85 leading to activation of the PI3K-Akt pathway.12–14 This means of PI3K activation can function independently of Ras. On the other hand, GM-CSF also induces nucleation of an alternative protein complex consisting of Shp2, Grb2 and Sos, leading to Ras activation14 with subsequent PI3K activation by a direct interaction between Ras and the PI3K p110 catalytic subunit, making this means of PI3K activation Ras-dependent.15 Thus, gain-of-function Shp2 mutants may contribute to PI3K activation in both a Ras-independent and/or Ras-dependent manner.
To investigate the potential role of Class IA PI3K signaling and its potential cooperative interaction with Ras signaling in gain-of-function Shp2-induced GM-CSF hypersensitivity and JMML pathogenesis, we examined the consequence of genetic disruption of Pik3r1, which encodes the regulatory subunits p85α, p55α and p50α. We further tested the effect of PI3K catalytic subunit isoform-specific inhibitors alone and in combination with Ras inhibition using a farnesyltransferase inhibitor on GM-CSF-stimulated proliferation, PI3K-Akt signaling and Ras-MAPK signaling in gain-of-function Shp2 E76K-expressing cells.
Design and Methods
Information concerning the Design and Methods of this study are available in the Online Supplementary Appendix.
Results and Discussion
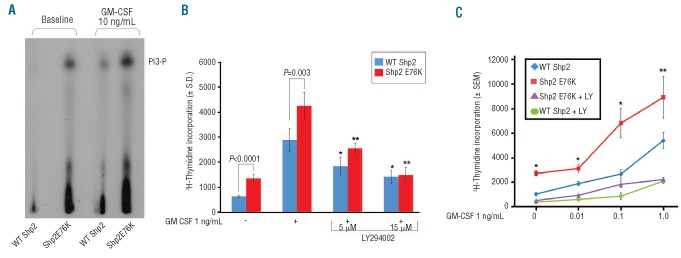
We previously demonstrated that gain-of-function Shp2-expressing macrophages exhibit enhanced GM-CSF-stimulated Akt activation.9 To confirm that this increased Akt activation results from enhanced PI3K activity, we performed PI3K assays using murine bone marrow-derived macrophages transduced with WT Shp2 or Shp2 E76K, and found increased [32P]-labeled PIP3 levels in Shp2 E76K-expressing cells compared to WT Shp2-expressing cells, both at baseline and following GM-CSF stimulation (Figure 1A). Functionally, GM-CSF-stimulated hyperproliferation of Shp2 E76K-expressing cells was reduced in a dose-dependent manner by the PI3K inhibitor, LY294002, to the level of WT Shp2 expressing cells as assessed by [3H]-thymidine-incorporation (Figure 1B). Furthermore, treatment with LY294002 significantly reduced hypersensitivity of Shp2 E76K-expressing cells to increasing concentrations of GM-CSF (Figure 1C, compare purple line to red line).
Figure 1.
Mutant Shp2 E76K-expressing cells demonstrate hyperactivation of PI3K and sensitivity to LY294002 treatment. (A) WT Shp2-and Shp2 E76K-transduced cells were differentiated into macrophage progenitors and assayed for PI3K activity at baseline and in response to 10 ng/mL GM-CSF for 60 min. Experiment repeated on 2 independent occasions. (B) Proliferation of WT Shp2- and Shp2 E76K-transduced bone marrow LDMNCs in response to GM-CSF 1 ng/mL in the presence of increasing concentrations of LY294002; representative of 2 independent experiments, n=4–5, *P=0.003 and *P<0.0001 comparing WT Shp2-expressing cells in the absence to the presence of LY294002 5 μM and 15 μM, respectively; **P<0.0001 comparing Shp2 E76K-expressing cells in the absence to the presence of LY294002 5 μM and 15 μM, statistics performed using unpaired, two-tailed Student’s t-test. (C) Proliferation of WT Shp2- and Shp2 E76K-transduced bone marrow LDMNCs in response to increasing GM-CSF concentrations in the presence and absence of 5 μM LY294002, n=3, *P<0.05 and **P=0.06 comparing Shp2 E76K-expressing cells in the presence (purple line) and absence of LY294002 (red line), statistics performed using unpaired, two-tailed student’s t test.
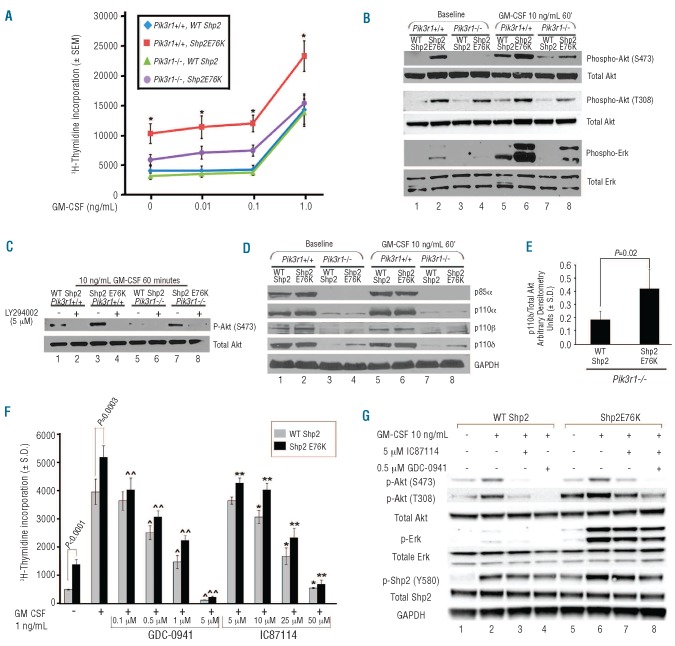
While LY294002 inhibition of proliferation provides useful insight toward the relevance of PI3K in gain-of-function Shp2-mediated GM-CSF hypersensitivity, interpretation of these data is limited as LY294002 is able to inhibit all classes of PI3K (IA, IB, II, and III) as well as to inhibit the function of PI3K-related protein kinases.16 To specifically evaluate the role of Class IA PI3K in activating Shp2-induced GM-CSF hypersensitivity, we utilized genetically modified mice generated by Terauchi et al. lacking expression of regulatory subunit p85α, yet permitting expression of the splice variants, p55α and p50α.17 In 3 independent experiments, we observed no correction in GM-CSF hypersensitivity of hematopoietic progenitors expressing Shp2 E76K in the absence of p85α alone (data not shown). Since p55α and p50α retain the SH2 domains which recruit the PI3K catalytic subunits (p110α, p110β and p110δ) to membrane-associated proteins, we hypothesized that ablation of p85α alone is inadequate to correct mutant Shp2-induced GM-CSF hypersensitivity. Using a knock-out model of Pik3r1 which results in ablation of the splice variants p55α and p50α, in addition to ablation of p85α,18 we observed significant but incomplete correction of GM-CSF hypersensitivity in Shp2 E76K-expressing cells in [3H]-thymidine-incorporation assays (Figure 2A, compare purple line to red and blue lines). Consistently, Akt phosphorylation at both Ser473 and Thr308 at baseline and following GM-CSF stimulation, was significantly reduced in Shp2 E76K-expressing Pik3r1−/− cells (Figure 2B, compare lanes 8 to 6 and lanes 4 to 2), but not completely eliminated. As the PI3K-Akt pathway has previously been shown to positively feed back to the Ras-MAPK pathway by direct phospholipid activation of Ras15,20,21 or by Rac1/2 and Pak1 activation leading to Mek phosphorylation,22,23 we also examined phospho-Erk levels in the Pik3r1−/− cells. As anticipated, genetic disruption of Pik3r1 resulted in a significant reduction in Shp2 E76K-induced Erk hyperphosphorylation (Figure 2B, compare lanes 8 to 6 and lanes 4 to 2), indicating that mutant Shp2-mediated PI3K signaling affects the MAPK pathway as well.
Figure 2.
Ablation of p85α, p55α, and p50α and inhibition with PI3K catalytic isoform-specific inhibitors normalizes gain-of-function Shp2-induced GM-CSF hypersensitivity. (A) Day 14.5 WT or Pik3r1−/− fetal liver cells were transduced with WT Shp2 or Shp2 E76K and subjected to [3H]-thymidine incorporation assays; 3 independent experiments combined with n=3 replicates per experiment, *P<0.01 for Pik3r1+/+, Shp2 E76K (red) versus Pik3r1−/−, Shp2 E76K (purple) at each concentration of GM-CSF; statistics performed using Prentice’s rank sum test for replicated block data.19 (B) Immunoblots demonstrating reduced phospho-Akt and phospho-Erk in Shp2 E76K-expressing Pik3r1−/− cells compared to Pik3r1+/+ cells, experiment repeated on 3 independent occasions. (C) Immunoblots demonstrating LY294002-mediated inhibition of Akt activation in Shp2 E76K-expressing Pik3r1−/− cells, experiment repeated on 2 independent occasions. (D) Immunoblots examining p110α, p110β, and p110δ levels in WT Shp2- and Shp2 E76K-expressing Pik3r1+/+ and Pik3r1−/− cells. (E) Quantitation using den-sitometry of immunoblot analyses comparing p110δ levels in WT Shp2- and Shp2 E76K-expressing Pik3r1−/− cells normalized to total Akt expression; n=5, P=0.02, statistics performed using unpaired, two-tailed students’ t test. (F) Proliferation of WT Shp2- and Shp2 E76K-transduced bone marrow LDMNCs in response to GM-CSF 1 ng/mL in the presence of increasing concentrations of GDC-0941 or of IC87114 (p110δ-specific); representative of 2 independent experiments, n=4–5, ^P=0.001, ^P<0.0001, and ^P<0.0001 comparing WT Shp2-expressing cells in the absence to the presence of GDC-0941 0.5 μM, 1 μM, and 5 μM, respectively; ^^P=0.002, ^^P<0.0001, ^^P<0.0001, and ^^P<0.0001 comparing Shp2 E76K-expressing cells in the absence to the presence of GDC-0941 0.1 μM, 0.5 μM, 1 μM, and 5 μM, respectively; *P=0.005, *P<0.0001, and *P<0.0001 comparing WT Shp2-expressing cells in the absence to the presence of IC87114 10 μM, 25 μM, and 50 μM, respectively; **P=0.002, **P=0.0005, **P<0.0001, and **P<0.0001 comparing Shp2 E76K-expressing cells in the absence to the presence of IC87114 5 μM, 10 μM, 25 μM, and 50 μM, respectively; statistics performed using unpaired, two-tailed students’ t-test. (G) Immunoblots demonstrating reduced phospho-Akt, phospho-Erk, and phospho-Shp2 in Shp2 E76K-expressing cells treated with 0.5 μM GDC-0941 or 5 μM IC87114, experiment repeated on 2 independent occasions.
Although mutant Shp2-induced GM-CSF hypersensitivity was significantly reduced in cells lacking p85α, p55α and p50α, the persistence of modest GM-CSF-stimulated hyperproliferation suggested residual PI3K activity in Pik3r1-deficient cells expressing oncogenic Shp2. In Pik3r1−/− cells expressing Shp2 E76K, Akt phosphorylation was almost completely abolished by LY294002 (Figure 2C, compare lane 8 to 7), suggesting that the residual hyper-proliferation of the Pik3r1−/− cells expressing Shp2 E76K was due to persistent PI3K activation.
One of the main functions of the PI3K regulatory subunits is to stabilize the p110 catalytic subunits and prevent their degradation.11 However, given the remaining Akt activation in the Pik3r1−/− cells which could be effectively eliminated by LY294002, we predicted residual expression of at least one of the catalytic subunits in the Shp2 E76K-expressing Pik3r1−/− cells. Upon immunoblot analysis, expression of p110β was ablated, while expression of both p110α and p110δ persisted, albeit at substantially reduced levels (Figure 2D, compare lanes 3 and 4 to 1 and 2, and 7 and 8 to 5 and 6). The residual expression of p110α and p110δ is potentially due to intact expression of the alternate Class IA regulatory subunit, p85β, although we did not observe a compensatory upregulation of p85β in the Pik3r1−/− cells (data not shown). Notably, however, p110δ expression was higher in the Shp2 E76K-expressing Pik3r1−/− cells compared to the WT Shp2-expressing Pik3r1−/− cells (Figure 2D, compare lanes 4 to 3 and 8 to 7). Compiling results from 5 independent experiments, we found p110δ expression to be significantly increased in the Shp2 E76K-expressing Pik3r1−/− cells (Figure 2E). These findings indicate preferential stabilization of p110δ in the Shp2 E76K-expressing cells and implicate p110δ as a potentially crucial mediator of gain-of-function Shp2-induced hyperactivation of PI3K signaling. Although we did not detect increased expression of p85β, it is possible that in the absence of p85α, p110δ preferentially interacts with p85β leading to its stabilization in Shp2 E76K expressing cells.
Given the increased expression of p110δ in mutant Shp2-expressing Pik3r1−/− cells, we examined the inhibitory activity of the PI3K catalytic isoform-specific p110δ inhibitor, IC8711424,25 on mutant Shp2-induced hypersensitivity to GM-CSF. IC87114 significantly reduced the proliferation of mutant Shp2-expressing hematopoietic cells in response to GM-CSF in a dose-dependent manner (Figure 2F), further implicating p110δ as a relevant mediator of mutant Shp2-induced hypersensitivity to GM-CSF. We next tested the effect of the PI3K inhibitor, GDC-0941, which has been developed into a clinical grade pharmaceutical product26,27 and has highest specificity for p110α and p110δ,28,29 and found inhibition of mutant Shp2-expressing cells in a dose-dependent manner between 0.1 and 5 μM (Figure 2F). Importantly, while both the WT Shp2- and Shp2 E76K-expressing cells demonstrate reduced proliferation in response to IC87114 and GDC-0941, higher concentrations of each drug were necessary to significantly reduce proliferation of the WT Shp2-expressing cells compared to the Shp2 E76K-expressing cells (0.5 μM vs. 0.1 μM for GDC-0941 and 10 μM vs. 5 μM for IC87114, respectively), suggesting that at lower concentrations, the mutant Shp2-expressing cells display increased sensitivity to the PI3K isoform-specific inhibitors. Consistent with reduced proliferation, both IC87114 and GDC-0941 reduced GM-CSF-stimulated hyper-phosphorylation of Akt, Erk, and Shp2 in mutant Shp2-expressing cells (Figure 2G, compare lanes 7 and 8 to lane 6).
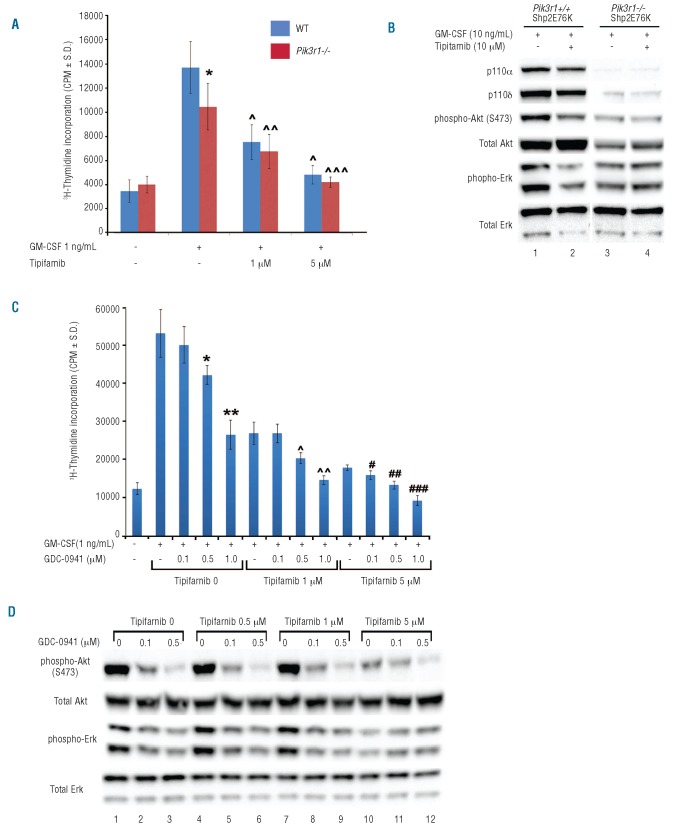
As one of the hallmarks of JMML is Ras hyperactivation, we next examined the contribution of Ras-dependent and Ras-independent PI3K activation to mutant Shp2-induced GM-CSF hypersensitivity and Erk and Akt hyperactivation. This distinction has clinical relevance as the finding of Ras-dependent only activation of PI3K would suggest that targeting the Ras-MAPK pathway alone would suffice to treat JMML, and that further inhibition with a PI3K inhibitor would be redundant. We first examined the effect of adding Ras inhibition to genetic disruption of Pik3r1. Consistent with previous studies, genetic disruption of Pik3r1 led to partial correction of GM-CSF-stimulated proliferation of Shp2 E76K-expressing cells (Figure 3A) as well as reduced phospho-Akt and phospho-Erk (Figure 3B, compare lane 1 to lane 3). In the presence of the Pik3r1-encoded regulatory subunits (WT cells), addition of the farnesyltransferase inhibitor, tipifarnib, resulted in reduced GM-CSF-stimulated hyperproliferation in a dose-dependent manner (Figure 3A, compare blue bars) as well as reduced phospho-Akt and phospho-Erk (Figure 3B, compare lane 1 to lane 2). However, in the absence of Pik3r1-encoded regulatory subunits, addition of tipifarnib did not result in a further reduction of Shp2 E76K-induced phospho-Erk or phospho-Akt beyond that achieved by loss of the PI3K regulatory subunits (Figure 3B, compare lane 3 to lane 4). These biochemical findings indicate that the Pik3r1−/− cells are less sensitive to Ras inhibition than the WT cells. Consistently, tipifarnib treatment reduced proliferation of Shp2 E76K-transduced Pik3r1−/− cells by only approximately 30%, while tipi-farnib treatment of the Shp2 E76K-transduced WT cells induced a reduction of 50% (Figure 3A), again indicating that the Pik3r1−/− cells are less sensitive to Ras inhibition than the WT cells. This reduced sensitivity to Ras inhibition suggests that the residual PI3K activity in the Pik3r1−/− cells functions at least partially in a Ras-independent manner. This finding is particularly significant given the observed increased expression of p110δ in Pik3r1−/− cells expressing Shp2 E76K (Figure 2D and E) as p110δ, in contrast to p110α, can induce oncogenic transformation in a Ras-independent manner.30
Figure 3.
Ras and PI3K inhibition cooperatively normalize gain-of-function Shp2-induced GM-CSF hypersensitivity. (A) Proliferation of WT Shp2- and Shp2 E76K-transduced Pik3r1+/+ or Pik3r1−/− fetal liver cells in response to GM-CSF 1 ng/mL in the presence of increasing concentrations of the farnesyltransferase inhibitor, tipifarnib; representative of 2 independent experiments, n=8, *P<0.02 comparing Pik3r1+/+, Shp2 E76K versus Pik3r1−/−, Shp2 E76K in response to 1 ng/mL GM-CSF in the absence of tipifarnib, ^P<0.001 comparing Pik3r1+/+, Shp2 E76K-expressing cells in the absence to the presence of tipifarnib at 1 μM and 5 μM; ^^P<0.005 comparing Pik3r1−/−, Shp2 E76K-expressing cells in the absence to the presence of 1 μM tipifarnib; ^^^P<0.001 comparing Pik3r1−/−, Shp2 E76K-expressing cells in the absence to the presence of 5 μM tipifarnib; statistics performed using unpaired, two-tailed students’ t-test. (B) Immunoblots demonstrating p110α, p110δ, phospho-Akt, and phospho-Erk levels in Shp2 E76K-expressing Pik3r1+/+ cells compared to Pik3r1−/−cells in the absence and presence of tipifarnib, experiment repeated on 3 independent occasions. (C) Proliferation of Shp2 E76K-transduced cells in response to GM-CSF 1 ng/mL in the presence of increasing concentrations of tipifarnib and GDC-0941; representative of 2 independent experiments, n=7, *P<0.01 and **P<0.001 comparing GDC-0941 0.5 μM and 1 μM to no GDC-0941 in the absence of tipifarnib; ^P<0.005 and ^^P<0.001 comparing GDC-0941 0.5 μM and 1 μM to no GDC-0941 in the presence of 1 μM tipifarnib; #P<0.01, ##P<0.001, and ###P<0.001 comparing GDC-0941 0.1 μM, 0.5 μM, and 1 μM to no GDC-0941 in the presence of 5 μM tipifarnib; statistics performed using unpaired, two-tailed students’ t-test. (D) Immunoblots demonstrating reduced phospho-Akt and phospho-Erk in Shp2 E76K-expressing cells treated with increasing concentrations of GDC-0941 in the absence and presence of increasing tipifarnib concentrations, experiment repeated on 2 independent occasions.
To further test the hypothesis that both Ras-dependent and Ras-independent activation of PI3K contributes to mutant Shp2-induced hyperproliferation and hyperactivation of Erk and Akt, we next treated Shp2 E76K-transduced cells with tipifarnib plus increasing concentrations of the PI3K inhibitor, GDC-0941 to determine if PI3K inhibition could further reduce the proliferation of tipifarnib-treated cells. We observed that treatment of Shp2 E76K-transduced cells with GDC-0941 in the presence of tipifarnib further reduced proliferation in a dose-dependent manner (Figure 3C). We found a corresponding reduction of Erk and Akt activation, also in a dose-dependent manner (Figure 3D). Collectively, these findings demonstrate that the PI3K-Akt and Ras-MAPK pathway work in concert to promote GM-CSF hypersensitivity of mutant Shp2-expressing cells.
Using genetic, pharmacological and biochemical approaches, these data demonstrate that hyperactivated PI3K signaling in the presence of leukemia-causing gain-of-function mutations in Shp2 contributes to GM-CSF hypersensitivity in both a Ras-dependent and a Ras-independent fashion. These findings also suggest that inhibition of PI3K using catalytic subunit-specific inhibitors is a feasible way to reduce GM-CSF-stimulated Akt and Erk hyperactivation as well as hyperproliferation in mutant Shp2-expressing hematopoietic cells. These results suggest that PI3K signaling may represent a novel therapeutic target for JMML, particularly using the newer classes of catalytic subunit-specific PI3K inhibitors that target p110δ.
Acknowledgments
The authors gratefully acknowledge the administrative assistance of Linda S. Henson. The authors have no conflicting financial interests.
Footnotes
Funding: this work was supported by the Riley Children’s Foundation and the U.S. National Institutes of Health (F30 HL104867, CBG; HL082981 and HL092524, RJC).
The online version of this article has a Supplementary Appendix.
Authorship and Disclosures
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
References
- 1.Emanuel PD, Bates LJ, Castleberry RP, Gualtieri RJ, Zuckerman KS. Selective hypersensitivity to granulocyte-macrophage colony-stimulating factor by juvenile chronic myeloid leukemia hematopoietic progenitors. Blood. 1991;77(5):925–9. [PubMed] [Google Scholar]
- 2.Locatelli F, Nollke P, Zecca M, Korthof E, Lanino E, Peters C, et al. Hematopoietic stem cell transplantation (HSCT) in children with juvenile myelomonocytic leukemia (JMML): results of the EWOG-MDS/EBMT trial. Blood. 2005;105(1):410–9. doi: 10.1182/blood-2004-05-1944. [DOI] [PubMed] [Google Scholar]
- 3.Shannon KM, O’Connell P, Martin GA, Paderanga D, Olson K, Dinndorf P, et al. Loss of the normal NF1 allele from the bone marrow of children with type 1 neurofibromatosis and malignant myeloid disorders. N Engl J Med. 1994;330(9):597–601. doi: 10.1056/NEJM199403033300903. [DOI] [PubMed] [Google Scholar]
- 4.Loh ML, Sakai DS, Flotho C, Kang M, Fliegauf M, Archambeault S, et al. Mutations in CBL occur frequently in juvenile myelomonocytic leukemia. Blood. 2009;114(9):1859–63. doi: 10.1182/blood-2009-01-198416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalra R, Paderanga DC, Olson K, Shannon KM. Genetic analysis is consistent with the hypothesis that NF1 limits myeloid cell growth through p21ras. Blood. 1994;84(10):3435–9. [PubMed] [Google Scholar]
- 6.Miyauchi J, Asada M, Sasaki M, Tsunematsu Y, Kojima S, Mizutani S. Mutations of the N-ras gene in juvenile chronic myelogenous leukemia. Blood. 1994;83(8):2248–54. [PubMed] [Google Scholar]
- 7.Tartaglia M, Niemeyer CM, Fragale A, Song X, Buechner J, Jung A, et al. Somatic mutations in PTPN11 in juvenile myelomonocytic leukemia, myelodysplastic syndromes and acute myeloid leukemia. Nat Genet. 2003;34(2):148–50. doi: 10.1038/ng1156. [DOI] [PubMed] [Google Scholar]
- 8.Neel BG, Gu H, Pao L. The ‘Shp’ing news: SH2 domain-containing tyrosine phosphatases in cell signaling. Trends Biochem Sci. 2003;28(6):284–93. doi: 10.1016/S0968-0004(03)00091-4. [DOI] [PubMed] [Google Scholar]
- 9.Yang Z, Li Y, Yin F, Chan RJ. Activating PTPN11 mutants promote hematopoietic progenitor cell-cycle progression and survival. Exp Hematol. 2008;36(10):1285–96. doi: 10.1016/j.exphem.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan RJ, Leedy MB, Munugalavadla V, Voorhorst CS, Li Y, Yu M, et al. Human somatic PTPN11 mutations induce hematopoietic-cell hypersensitivity to granulocyte-macrophage colony-stimulating factor. Blood. 2005;105(9):3737–42. doi: 10.1182/blood-2004-10-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu J, Zhang Y, McIlroy J, Rordorf-Nikolic T, Orr GA, Backer JM. Regulation of the p85/p110 phosphatidylinositol 3′-kinase: stabilization and inhibition of the p110alpha catalytic subunit by the p85 regulatory subunit. Mol Cell Biol. 1998;18(3):1379–87. doi: 10.1128/mcb.18.3.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dijkers PF, van Dijk TB, de Groot RP, Raaijmakers JA, Lammers JW, Koenderman L, et al. Regulation and function of protein kinase B and MAP kinase activation by the IL-5/GM-CSF/IL-3 receptor. Oncogene. 1999;18(22):3334–42. doi: 10.1038/sj.onc.1202678. [DOI] [PubMed] [Google Scholar]
- 13.Itoh T, Liu R, Yokota T, Arai KI, Watanabe S. Definition of the role of tyrosine residues of the common beta subunit regulating multiple signaling pathways of granulocyte-macrophage colony-stimulating factor receptor. Mol Cell Biol. 1998;18(2):742–52. doi: 10.1128/mcb.18.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X, Qu CK. Protein Tyrosine Phosphatase SHP-2 (PTPN11) in Hematopoiesis and Leukemogenesis. J Signal Transduct. 2011:195239. doi: 10.1155/2011/195239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodriguez-Viciana P, Warne PH, Dhand R, Vanhaesebroeck B, Gout I, Fry MJ, et al. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature. 1994;370(6490):527–32. doi: 10.1038/370527a0. [DOI] [PubMed] [Google Scholar]
- 16.Vanhaesebroeck B, Waterfield MD. Signaling by distinct classes of phosphoinositide 3-kinases. Exp Cell Res. 1999;253(1):239–54. doi: 10.1006/excr.1999.4701. [DOI] [PubMed] [Google Scholar]
- 17.Terauchi Y, Tsuji Y, Satoh S, Minoura H, Murakami K, Okuno A, et al. Increased insulin sensitivity and hypoglycaemia in mice lacking the p85 alpha subunit of phosphoinositide 3-kinase. Nat Genet. 2002;21(2):230–5. doi: 10.1038/6023. [DOI] [PubMed] [Google Scholar]
- 18.Fruman DA, Mauvais-Jarvis F, Pollard DA, Yballe CM, Brazil D, Bronson RT, et al. Hypoglycaemia, liver necrosis and perinatal death in mice lacking all isoforms of phosphoinositide 3-kinase p85 alpha. Nat Genet. 2000;26(3):379–82. doi: 10.1038/81715. [DOI] [PubMed] [Google Scholar]
- 19.Prentice MJ. On the Problem of m Incomplete Rankings. Biometrika. 1979;66:167–70. [Google Scholar]
- 20.Chan TO, Rodeck U, Chan AM, Kimmelman AC, Rittenhouse SE, Panayotou G, et al. Small GTPases and tyrosine kinases coregulate a molecular switch in the phosphoinositide 3-kinase regulatory subunit. Cancer Cell. 2002;1(2):181–91. doi: 10.1016/s1535-6108(02)00033-8. [DOI] [PubMed] [Google Scholar]
- 21.Rodriguez-Viciana P, Warne PH, Vanhaesebroeck B, Waterfield MD, Downward J. Activation of phosphoinositide 3-kinase by interaction with Ras and by point mutation. Embo J. 1996;15(10):2442–51. [PMC free article] [PubMed] [Google Scholar]
- 22.Ingram DA, Hiatt K, King AJ, Fisher L, Shivakumar R, Derstine C, et al. Hyperactivation of p21(ras) and the hematopoietic-specific Rho GTPase, Rac2, cooperate to alter the proliferation of neurofibromin-deficient mast cells in vivo and in vitro. J Exp Med. 2001;194(1):57–69. doi: 10.1084/jem.194.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDaniel AS, Allen JD, Park SJ, Jaffer ZM, Michels EG, Burgin SJ, et al. Pak1 regulates multiple c-Kit mediated Ras-MAPK gain-in-function phenotypes in Nf1+/− mast cells. Blood. 2008;112(12):4646–54. doi: 10.1182/blood-2008-04-155085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sadhu C, Masinovsky B, Dick K, Sowell CG, Staunton DE. Essential role of phosphoinositide 3-kinase delta in neutrophil directional movement. J Immunol. 2003;170(5):2647–54. doi: 10.4049/jimmunol.170.5.2647. [DOI] [PubMed] [Google Scholar]
- 25.Ikeda H, Hideshima T, Fulciniti M, Perrone G, Miura N, Yasui H, et al. PI3K/p110{delta} is a novel therapeutic target in multiple myeloma. Blood. 2010;116(9):1460–8. doi: 10.1182/blood-2009-06-222943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarker D, Kristeleit K, Mazina KE, Ware JA, Yan Y, Dresser M, et al. A phase I study evaluating the pharmacokinetics (PK) and pharmacodynamics (PD) of the oral panphosphoinositide-3 kinase (PI3K) inhibitor GDC-0941. J Clin Onc. 2009;27:15s. [Google Scholar]
- 27.Wagner AJ, Von Hoff DH, LoRusso M, Tibes R, Mazina KE, Ware JA, et al. A first-in-human phase I study to evaluate the pan-PI3K inhibitor GDC-0941 administered QD or BID in patients with advanced solid tumors. J Clin Onc. 2009;27:15s. [Google Scholar]
- 28.Folkes AJ, Ahmadi K, Alderton WK, Alix S, Baker SJ, Box G, et al. The identification of 2-(1H-indazol-4-yl)-6-(4-methanesulfonyl-piperazin-1-ylmethyl)-4-morpholin-4-yl-thieno[3,2-d]pyrimidine (GDC-0941) as a potent, selective, orally bioavailable inhibitor of class I PI3 kinase for the treatment of cancer. J Med Chem. 2008;51(18):5522–32. doi: 10.1021/jm800295d. [DOI] [PubMed] [Google Scholar]
- 29.Raynaud FI, Eccles SA, Patel S, Alix S, Box G, Chuckowree I, et al. Biological properties of potent inhibitors of class I phosphatidylinositide 3-kinases: from PI-103 through PI-540, PI-620 to the oral agent GDC-0941. Mol Cancer Ther. 2009;8(7):1725–38. doi: 10.1158/1535-7163.MCT-08-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao L, Vogt PK. Class I PI3K in oncogenic cellular transformation. Oncogene. 2008;27(41):5486–96. doi: 10.1038/onc.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]