Abstract
Although new therapies have doubled the survival of multiple myeloma patients, this remains an incurable disease. It has been postulated that the so-called myeloma cancer stem cells would be responsible for tumor initiation and relapse but their unequivocal identification remains unclear. Here, we investigated in a panel of myeloma cell lines the presence of CD20+ cells harboring a stem-cell phenotype. Thus, only a small population of CD20dim+ cells (0.3%) in the RPMI-8226 cell line was found. CD20dim+ RPMI-8226 cells expressed the plasma cell markers CD38 and CD138 and were CD19−CD27−. Additionally, CD20dim+ RPMI-8226 cells did not exhibit stem-cell markers as shown by gene expression profiling and the aldehyde dehydrogenase assay. Furthermore, we demonstrated that CD20dim+ RPMI-8226 cells are not essential for CB17-SCID mice engraftment and show lower self-renewal potential than the CD20− RPMI-8226 cells. These results do not support CD20 expression for the identification of myeloma cancer stem cells.
Keywords: CD20, multiple myeloma, cell lines, stem cell, phenotype
Introduction
The introduction of high-dose therapy and the novel agents thalidomide, bortezomib and lenalidomide have improved complete response rates and doubled multiple myeloma (MM) patients’ survival,1 but unfortunately the majority of patients relapse. In MM, relapses have been attributed to MM Cancer Stem Cells (MM-CSC), although inconsistencies have emerged with respect to their frequency, clonogenicity and phenotype.2–5 It has been proposed that CD20 could be a hallmark of MM-CSC. In support of this hypothesis, Matsui et al. have shown the capacity of the anti-CD20 MoAb rituximab to inhibit the clonogenic growth of MM-CSC in vitro.3,6 However, clinical trials to test the effect of rituximab as maintenance therapy have failed to confirm a survival benefit.7,8 Furthermore, data from other groups do not support the hypothesis that B cells are the feeder cells in myeloma.9,10 In order to shed some light on this controversial area, we have searched for the presence and functionality of CD20+ putative MM-CSC in a panel of MM cell lines.
Design and Methods
The human MM cell lines used were: RPMI-8226 and U266 (from Dr W Dalton, Tampa, FL, USA); MM1S and MM1R (from Dr ST Rosen, Chicago, IL, USA); NCI-H929 (from Dr J Teixidó, Madrid, Spain); RPMI-LR5, U266-LR7 and U266-Dox4 (from Dr KC Anderson, Boston, MA, USA). The cells were cultured as previously described.11 Briefly, the cells were cultured in RPMI-1640 medium supplemented with 2 mM L-glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin and 10% fetal bovine serum at 37°C and 5% CO2/95% air.
MM cell lines were immunophenotyped using a 7-color immunofluorescence technique,12 with the following combination of monoclonal antibodies (Pacific Blue (PB)/anemonia majano cyan (AmCyan)/fluorescein isothiocyanate (FITC)/peridinin chlorophyll protein-cyanin 5.5 (PerCP-Cy5.5)/PE-cyanin 7 (PE-Cy7)/allophycocyanin (APC)/alexafluor 700 (AF700)): CD19/CD45/CD20/CD138/CD27/CD56/CD38. Data were stored for a minimum of 3×105 events. CD20dim+ and CD20− RPMI-8226 cells were sorted after incubation with CD20-APC/7AAD and acquisition on a FACSAria cytometer (Becton Dickison Biosciences). Sorting was performed only for viable cells (7AAD−) and debris were excluded by scatter properties. The CD20− and CD20dim+ RPMI-8226 sorted cells had a mean final purity of over 99% and 88%, respectively. The origin of the monoclonal antibodies was as follows: CD20-FITC (clone L27), CD20-APC (clone L27), CD138-PerCP-Cy5 (clone MI15), CD56-APC (clone NCAM16.2) and CD45-AmCyan (clone 2D1) were obtained from BD Biosciences (San Jose, CA, USA); CD19-PacificBlue (clone HIB19) and CD27-PE-Cy7 (clone O323) antibodies were purchased from eBioscience (San Diego CA, USA) and CD38-AlexaFluor700 antibody (clone HIT2) was obtained from Exbio (Vestec, Czech Republic).
CD20dim+ and CD20− RPMI-8226 cells were extensively characterized. For real time quantitative PCR (qRT-PCR), total RNA was extracted from CD20dim+ and CD20− RPMI-8226 cells using an RNeasy Mini Kit (Qiagen, Valencia, USA) following the manufacturer's protocol. RNA quality and quantity were assessed with the RNA Nano LabChip (Agilent Tech. Inc., Palo Alto, CA, USA). The retrotranscription reaction was performed with a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems Foster City, CA, USA) according to the manufacturer’s recommendations. Finally, real time quantitative PCR was performed using TaqMan gene expression assay kits (Applied Biosystems Foster City, CA, USA): Hs_00544819 for MS4A1 (CD20) and Hs99999905_m1 GAPDH as a control gene. Relative gene expression was calculated by the 2−ΔCt method, ΔCt=Ct (gene) − Ct (GAPDH). Morphological characterization was performed with May-Grünwald-Giemsa staining. May-Grünwald and Giemsa stains were obtained from Merck (Darmstadt, Germany). Characterization of VDJH and IGκ rearrangements was performed in genomic cDNA as described elsewhere.13 The expression of aldehyde dehydrogenase (ALDH) was assessed using the Aldefluor Kit (StemCell Technologies, Grenoble, France) following the manufacturer’s instructions with further staining with a CD20-APC antibody. For microarray studies, RNA from 3 independent CD20dim+ or CD20− RPMI-8226 samples was isolated, labeled and hybridized to Human Gene 1.0 ST array (Affymetrix) according to Affymetrix protocols.14 The arrays were analyzed using the DNA-Chip Analyzer software (DChip). Fold change of 2 or more was considered significant. All microarray data have been deposited with the Gene Expression Omnibus under accession number GSE33020.
For serial colony assays, 1000–1500 CD20dim+ or CD20− RPMI-8226 cells/mL were plated in Methocult® (StemCell Technologies, catalog n. H4230) and incubated at 37°C and 5% CO2. After 14 days, colonies (≥40 cells) were scored and subsequently collected, rinsed with PBS and plated again in fresh Methocult®. A sample was used to evaluate CD20 expression in the colonies. For engrafting assays, unfractionated or sorted CD20dim+ or CD20− RPMI-8226 cells were subcutaneously inoculated into 6–7 week old CB17-SCID mice (Charles River, Spain).11 In selected mice, tumors were isolated, mechanically minced and filtered through 40 μm cell-strainers to perform phenotypic studies and/or reinjection into secondary recipients. All animal experiments were performed according to the protocol previously approved by the ethical committee of the University of Salamanca, Spain.
Statistical analyses were performed using the SPSS-15.0 software (SPSS, Chicago, IL, USA); significant differences between groups were assessed by the Student’s t and Mann-Whitney U tests.
Results and Discussion
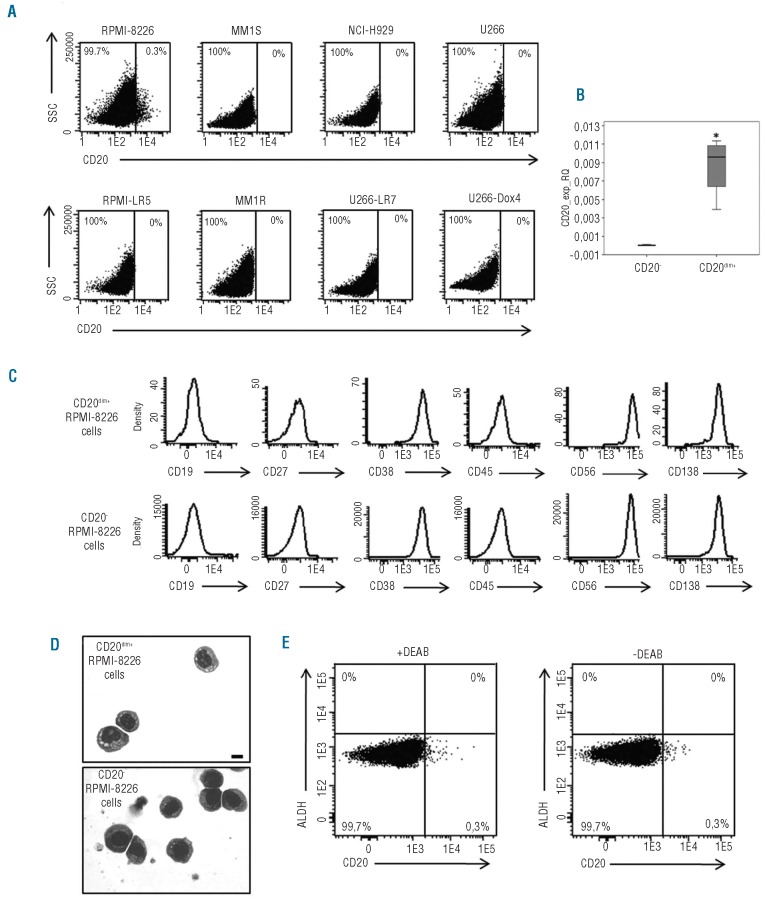
One decade ago, several reports suggested the existence in MM of circulating clonotypic B cells that displayed drug resistance and could be responsible for spreading the disease, leading to their definition as putative MM-CSC.2,15 More recently, Matsui et al. identified CD138−CD20+CD27+ MM-CSC in two MM cell lines and in patient samples.3,6 Furthermore, their clonogenic growth was inhibited by the anti-CD20 MoAb rituximab.3,6 To confirm these results and to investigate MM-CSC more deeply, we examined the presence and characteristics of CD20+ cells in a panel of drug sensitive (RPMI-8226, MM1S, U266, NCI-H929) and resistant (RPMI-LR5, MM1R, U266-LR7, U266-Dox4) MM cell lines. In all except one (RPMI-8226), CD20+ cells were undetectable by flow cytometry with a sensitivity of 7×10−5 (Figure 1A).16 In line with our results, Rossi et al. reported that U266, NCI-H929 and MM1R cell lines are CD20−17 while Matsui et al. described a small population (2–5%) of CD138−CD20+ cells in the NCI-H929 and RPMI-8226 cell lines.3 In our hands, the RPMI-8226 cell line included a small CD20dim+ subset (0.3%) (Figure 1A). Data from qRT-PCR confirmed a significant higher expression of CD20 in the CD20dim+ RPMI-8226 cells with respect to the CD20−RPMI-8226 cells (Figure 1B). Furthermore, the CD20dim+ subset displayed a myelomatous plasma cell phenotype: CD38+CD138+CD19−CD27−CD45−(Figure 1C).16 Therefore, we did not confirm the B-cell phenotype of the putative CD20+ MM-CSC. Morphologically, CD20dim+ RPMI-8226 cells showed a higher number of vacuoles and a more relaxed chromatin (Figure 1D) suggesting that they represent a more immature compartment which would be concordant with data describing CD20 expression in MM associated with a plasmablast morphology.18 However, PCR-based clonal analysis showed that CD20dim+ and CD20− RPMI-8226 cells displayed equal IGH-IGL rearrangements, confirming their clonality. (The PCR-fragment size for CD20dim+ RPMI-8226 cells was: 341.91 bp for IGH-FR1, 286.21 bp for IGL-VJK and 269.58 bp for IGL-KDEL; the PCR-fragment size for CD20− RPMI-8226 cells was: 341.97 bp for IGH-FR1, 286.26 bp for IGL-VJK and 269.87 bp for IGL-KDEL).
Figure 1.
Study of CD20 expression in MM cell lines and characterization of the phenotype and the morphology of CD20dim+ and CD20− RPMI-8226 cells. (A) Bivariate dot plots showing the percentage of expression of CD20+ and CD20− cells in the RPMI-8226, MM1S, NCI-H929, U266, RPMI-LR5, MM1R, U266-LR7 and U266-Dox4 MM cell lines. (B) Expression of CD20 by real-time quantitative PCR in CD20− and CD20dim+ RPMI-8226 cells. Relative values were calculated by the 2−ΔCt method (ΔCt = Ct(Gene)− Ct(GAPDH)). The GAPDH gene was used as a control gene. Significance is expressed as *P<0.05 (Mann-Whitney U test). (C) Single parameter histograms illustrating the expression of CD19, CD27, CD38, CD45, CD56 and CD138 in CD20dim+ and CD20− RPMI-8226 cells. (D) May-Grünwald-Giemsa staining of CD20dim+ and CD20− RPMI-8226 cells. Cells were visualized with an Olympus BX51 microscope (Olympus, Japan). Images were captured with an Olympus DP70 camera (Olympus, Japan) using the software DP controller. Scale bar 10 μm. (E) Bivariate dot plots representing CD20 expression (x axis) versus ALDH expression (y axis) in the presence or absence of the ALDH-inhibitor, diethylaminobenzaldehyde (DEAB). The percentage of cells within each electronic gate is indicated.
The expression of molecules related to drug resistance and self-renewal is a CSC-hallmark. Here we analyzed the expression of the MM-CSC marker ALDH6,19 in CD20dim+ and CD20− RPMI-8226 cells and we did not find overexpression of ALDH in the CD20dim+ versus the CD20− fraction (Figure 1E). To investigate other potential CSC-markers, we next studied the gene expression profile of CD20dim+ RPMI-8226 cells. This showed 48 genes up-regulated and one gene down-regulated versus CD20− RPMI-8226 cells (Online Supplementary Table S1). It should be mentioned that, among the 48 up-regulated genes, MS4A1, the CD20-encoding gene, displayed the highest fold change (37.27). When we focused on MM-CSC-associated genes such as Hedgehog,20 ABCG221 and SOX219 they were not expressed in CD20dim+ RPMI-8226 cells. Interestingly, the Notch-target gene, Hes1, was over-expressed in CD20dim+ RPMI-8226 cells (Online Supplementary Table S1). Although Hes genes have been generally associated with stem cell maintenance, this is apparently not the case for MM where Notch1 activation is related to growth inhibition and apoptosis.22
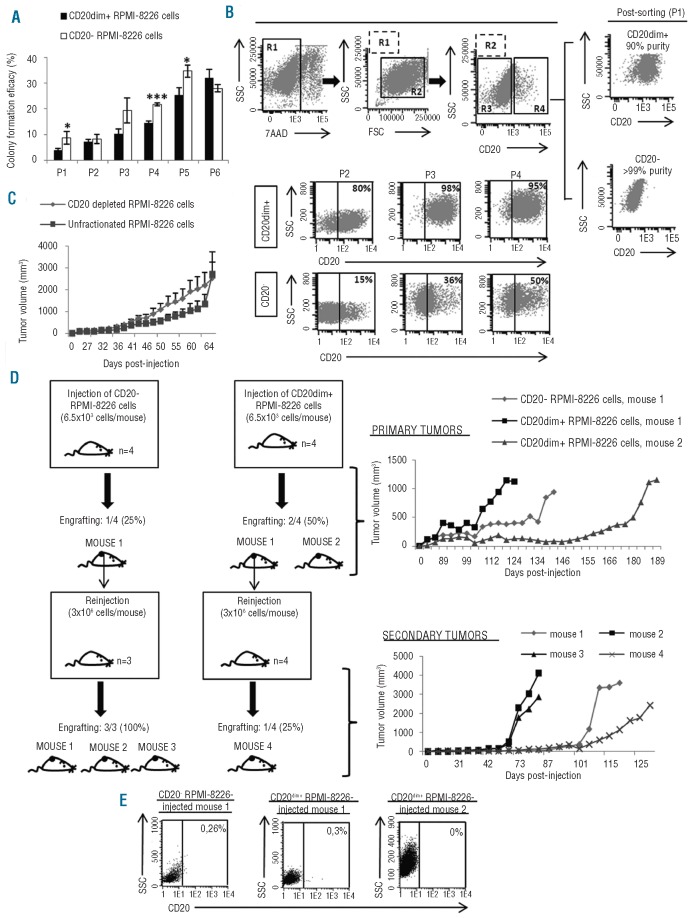
Finally, in vitro and in vivo functional characterization was tackled. Therefore both, CD20dim+ and CD20− RPMI-8226 cells, formed colonies after 6 rounds of replating (Figure 2A) suggesting their long-term proliferation capacity. Since differentiation potential is another characteristic of cancer stem cells, whether CD20dim+ cells give rise to CD20− cells or vice versa was studied during serial colony assays. We were, therefore, able to observe that the population of CD20− cells, initially sorted and plated (plating 1, P1) with a purity of over 99%, gave rise to CD20dim+ cells: 15% in plating 2 (P2), 36% in plating 3 (P3) and 50% in plating 4 (P4) (Figure 2B). Conversely, CD20dim+ RPMI-8226 cells, initially sorted and plated (P1) with a purity of 90%, did not seem to differentiate into CD20− cells since there was no great variation in the percentage of CD20dim+ cells (80%, 98% and 95% in P2, P3 and P4, respectively) (Figure 2B). Therefore, these data indicate a hierarchical order in which CD20dim+ cells derive from CD20− cells. It is well-known that RPMI-8226 cells promote plasmacytomas in SCID mice.23 However, whether this is due to a subpopulation of cells with the tumor-initiating capacity remains unknown. To clarify this we injected 1.5×106 unfractionated (n=4) or CD20 depleted (n=4) RPMI-8226 cells into 8 CB17-SCID mice. We found that 100% of the mice in each group developed plasmacytomas without any difference in the growth pattern (Figure 2C), suggesting that CD20dim+ cells are not essential for tumor formation. Nevertheless, since serial transplantation is the only in vivo assay to functionally measure stem cell self-renewal capacity,24 CB17-SCID mice were first injected with either CD20− or CD20dim+ RPMI-8226 cells (primary tumors) and, subsequently, secondary mice were injected with cells isolated from either a CD20− or a CD20dim+ derived tumor (secondary tumors) (Figure 2D, left). Despite differences in the primary engrafting capacity for CD20− and CD20dim+ cells (25% and 50%, respectively), a high variability in tumor growth curves was observed (Figure 2D, top right). However, whereas sorted CD20dim+ cells formed secondary tumors only in 25% of mice, sorted CD20− cells formed secondary tumors in 100% of the mice and, furthermore, the latter grew faster (Figure 2D, bottom right). These results suggest that CD20− RPMI-8226 cells have higher self-renewal capacity than CD20dim+ cells and do not support CD20 as a marker of MM-CSC.3,6 In fact, we found that more than 99% of cells isolated from primary tumors were negative for CD20 even when CD20dim+ cells had been injected (Figure 2E). Nevertheless, it would be interesting to carry out further experiments of serial dilution and subsequent transplantation of CD20dim+ and CD20− RPMI-8226 cells in order to evaluate the tumor-initiating ability of each fraction quantitatively.
Figure 2.
Functional characterization of CD20dim+ and CD20− RPMI-8226 cells. (A) Colony assay for sorted CD20dim+ and CD20− RPMI-8226 cells. Results are expressed as mean ± SEM of 3 independent experiments and represent the percentage of the number of colonies scored compared to the number of cells plated in each plating (P1, P2, P3, P4, P5 and P6). Statistically significant differences between CD20dim+ and CD20- RPMI-8226 cells are given as *P<0.05 and ***P<0.001. (B) (Top) viable cells (7AAD-) from the RPMI-8226 cell line were gated (R1) and subsequently debris were eliminated by scatter properties (R2). CD20− (R3) and CD20dim+ (R4) RPMI-8226 cells were gated and sorted and subsequently plated in a colony assay (P1, plating 1). (Bottom) The expression of CD20 in the CD20dim+ and CD20− derived colonies was analyzed by flow cytometry in plating 2, 3 and 4 (P2, P3 and P4). The percentage of CD20-positive cells is indicated in each dot plot. (C) Tumor growth curves for CB17-SCID mice subcutaneously inoculated with 1.5×106 unfractionated (n=4) or CD20 depleted (n=4) RPMI-8226 cells. Results are expressed as mean ± SEM values. Comparisons with the Mann Whitney U test showed no statistically significant differences between the distinct time points. (D) (Left) Diagram representing serial transplantation of CD20− and CD20dim+ RPMI-8226 cells. (Top right) Tumor growth curves for CB17-SCID mice inoculated with 6.5×103 CD20− or CD20dim+ RPMI-8226 cells which developed measurable primary tumors. (Bottom right) Tumor growth curves for CB17-SCID mice inoculated with 3×106 cells isolated from mouse 1 CD20− derived primary tumor (mouse 1, 2 and 3) or from mouse 1 CD20dim+ derived primary tumor (mouse 4) which developed measurable secondary tumors. (E) Bivariate dot plots illustrating the expression of CD20 in cells isolated from primary tumors in mice. The percentage of CD20-positive cells is indicated.
In conclusion, our results show that CD20 is undetectable in the majority of MM cell lines. Additionally, the expression of CD20 in the RPMI-8226 cell line is not associated with a cancer stem cell phenotype. Therefore, our results do not support CD20+ expression for the identification of MM-CSC.
Acknowledgments
We are grateful to Dr W Dalton for providing the RPMI-8226 and U266 cell lines, to Dr ST Rosen for providing the MM1S and MM1R cell lines, to Dr J Teixidó for providing the NCI-H929 cell line and to Dr KC Anderson for providing the RPMI-LR5, U266-LR7 and U266-Dox4 cell lines.
Footnotes
Funding: this work was supported by the Cooperative Research Thematic Network (RTICs; RD06/0020/0006), the “Junta de Castilla y León. Consejería de Sanidad” (GRS 391/B/09), the “Ministerio de Ciencia e Innovación” (PS09/01897) and the “Fundación Memoria D. Samuel Solórzano Barruso” (FS/2-2010).
The online version of this article has a Supplementary Appendix.
Authorship and Disclosures
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
References
- 1.San-Miguel JF, Mateos MV. Can multiple myeloma become a curable disease? Haematologica. 2011;96(9):1246–8. doi: 10.3324/haematol.2011.051169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pilarski LM, Giannakopoulos NV, Szczepek AJ, Masellis AM, Mant MJ, Belch AR. In multiple myeloma, circulating hyperdiploid B cells have clonotypic immunoglobulin heavy chain rearrangements and may mediate spread of disease. Clin Cancer Res. 2000;6(2):585–96. [PubMed] [Google Scholar]
- 3.Matsui W, Huff CA, Wang Q, Malehorn MT, Barber J, Tanhehco Y, et al. Characterization of clonogenic multiple myeloma cells. Blood. 2004;103(6):2332–6. doi: 10.1182/blood-2003-09-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robillard N, Pellat-Deceunynck C, Bataille R. Phenotypic characterization of the human myeloma cell growth fraction. Blood. 2005;105(12):4845–8. doi: 10.1182/blood-2004-12-4700. [DOI] [PubMed] [Google Scholar]
- 5.Yata K, Yaccoby S. The SCID-rab model: a novel in vivo system for primary human myeloma demonstrating growth of CD138-expressing malignant cells. Leukemia. 2004;18(11):1891–7. doi: 10.1038/sj.leu.2403513. [DOI] [PubMed] [Google Scholar]
- 6.Matsui W, Wang Q, Barber JP, Brennan S, Smith BD, Borrello I, et al. Clonogenic multiple myeloma progenitors, stem cell properties, and drug resistance. Cancer Res. 2008;68(1):190–7. doi: 10.1158/0008-5472.CAN-07-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim SH, Zhang Y, Wang Z, Varadarajan R, Periman P, Esler WV. Rituximab administration following autologous stem cell transplantation for multiple myeloma is associated with severe IgM deficiency. Blood. 2004;103(5):1971–2. doi: 10.1182/blood-2003-12-4331. [DOI] [PubMed] [Google Scholar]
- 8.Musto P, Carella AM, Jr, Greco MM, Falcone A, Sanpaolo G, Bodenizza C, et al. Short progression-free survival in myeloma patients receiving rituximab as maintenance therapy after autologous transplantation. Br J Haematol. 2003;123(4):746–7. doi: 10.1046/j.1365-2141.2003.04675.x. [DOI] [PubMed] [Google Scholar]
- 9.Pfeifer S, Perez-Andres M, Ludwig H, Sahota SS, Zojer N. Evaluating the clonal hierarchy in light-chain multiple myeloma: implications against the myeloma stem cell hypothesis. Leukemia. 2011;25(7):1213–6. doi: 10.1038/leu.2011.70. [DOI] [PubMed] [Google Scholar]
- 10.Rasmussen T, Haaber J, Dahl IM, Knudsen LM, Kerndrup GB, Lodahl M, et al. Identification of translocation products but not K-RAS mutations in memory B cells from patients with multiple myeloma. Haematologica. 2010;95(10):1730–7. doi: 10.3324/haematol.2010.024778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ocio EM, Maiso P, Chen X, Garayoa M, Alvarez-Fernandez S, San-Segundo L, et al. Zalypsis: a novel marine-derived compound with potent antimyeloma activity that reveals high sensitivity of malignant plasma cells to DNA double-strand breaks. Blood. 2009;113(16):3781–91. doi: 10.1182/blood-2008-09-177774. [DOI] [PubMed] [Google Scholar]
- 12.Paiva B, Perez-Andres M, Vidriales MB, Almeida J, de las Heras N, Mateos MV, et al. Competition between clonal plasma cells and normal cells for potentially overlapping bone marrow niches is associated with a progressively altered cellular distribution in MGUS vs myeloma. Leukemia. 2011;25(4):697–706. doi: 10.1038/leu.2010.320. [DOI] [PubMed] [Google Scholar]
- 13.van Dongen JJ, Langerak AW, Bruggemann M, Evans PA, Hummel M, Lavender FL, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17(12):2257–317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 14.Gutierrez NC, Lopez-Perez R, Hernandez JM, Isidro I, Gonzalez B, Delgado M, et al. Gene expression profile reveals deregulation of genes with relevant functions in the different subclasses of acute myeloid leukemia. Leukemia. 2005;19(3):402–9. doi: 10.1038/sj.leu.2403625. [DOI] [PubMed] [Google Scholar]
- 15.Rottenburger C, Kiel K, Bosing T, Cremer FW, Moldenhauer G, Ho AD, et al. Clonotypic CD20+ and CD19+ B cells in peripheral blood of patients with multiple myeloma post high-dose therapy and peripheral blood stem cell transplantation. Br J Haematol. 1999;106(2):545–52. doi: 10.1046/j.1365-2141.1999.01548.x. [DOI] [PubMed] [Google Scholar]
- 16.Rawstron AC, Orfao A, Beksac M, Bezdickova L, Brooimans RA, Bumbea H, et al. Report of the European Myeloma Network on multiparametric flow cytometry in multiple myeloma and related disorders. Haematologica. 2008;93(3):431–8. doi: 10.3324/haematol.11080. [DOI] [PubMed] [Google Scholar]
- 17.Rossi EA, Rossi DL, Stein R, Goldenberg DM, Chang CH. A bispecific antibody-IFNalpha2b immunocytokine targeting CD20 and HLA-DR is highly toxic to human lymphoma and multiple myeloma cells. Cancer Res. 2010;70(19):7600–9. doi: 10.1158/0008-5472.CAN-10-2126. [DOI] [PubMed] [Google Scholar]
- 18.San Miguel JF, Gutierrez NC, Mateo G, Orfao A. Conventional diagnostics in multiple myeloma. Eur J Cancer. 2006;42(11):1510–9. doi: 10.1016/j.ejca.2005.11.039. [DOI] [PubMed] [Google Scholar]
- 19.Brennan SK, Wang Q, Tressler R, Harley C, Go N, Bassett E, et al. Telomerase inhibition targets clonogenic multiple myeloma cells through telomere length-dependent and independent mechanisms. PLoS One. 2010;5(9) doi: 10.1371/journal.pone.0012487. pii: e12487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peacock CD, Wang Q, Gesell GS, Corcoran-Schwartz IM, Jones E, Kim J, et al. Hedgehog signaling maintains a tumor stem cell compartment in multiple myeloma. Proc Natl Acad Sci USA. 2007;104(10):4048–53. doi: 10.1073/pnas.0611682104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jakubikova J, Adamia S, Kost-Alimova M, Klippel S, Cervi D, Daley JF, et al. Lenalidomide targets clonogenic side population in multiple myeloma: pathophysiologic and clinical implications. Blood. 2011;117(17):4409–19. doi: 10.1182/blood-2010-02-267344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zweidler-McKay PA, He Y, Xu L, Rodriguez CG, Karnell FG, Carpenter AC, et al. Notch signaling is a potent inducer of growth arrest and apoptosis in a wide range of B-cell malignancies. Blood. 2005;106(12):3898–906. doi: 10.1182/blood-2005-01-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan Y, Gao Y, Chen L, Gao G, Dong H, Yang Y, et al. Targeting autophagy augments in vitro and in vivo antimyeloma activity of DNA-damaging chemotherapy. Clin Cancer Res. 2011;17(10):3248–58. doi: 10.1158/1078-0432.CCR-10-0890. [DOI] [PubMed] [Google Scholar]
- 24.Dick JE. Human stem cell assays in immune-deficient mice. Curr Opin Hematol. 1996;3(6):405–9. doi: 10.1097/00062752-199603060-00002. [DOI] [PubMed] [Google Scholar]