Abstract
During the course of infection, Salmonella has to face several potentially lethal environmental conditions, one such being acidic pH. The ability to sense and respond to the acidic pH is crucial for the survival and replication of Salmonella. The physiological role of one gene (STM1485) involved in this response, which is upregulated inside the host cells (by 90- to 113-fold) is functionally characterized in Salmonella pathogenesis. In vitro, the ΔSTM1485 neither exhibited any growth defect at pH 4.5 nor any difference in the acid tolerance response. The ΔSTM1485 was compromised in its capacity to proliferate inside the host cells and complementation with STM1485 gene restored its virulence. We further demonstrate that the surface translocation of Salmonella pathogenicity island-2 (SPI-2) encoded translocon proteins, SseB and SseD were reduced in the ΔSTM1485. The increase in co-localization of this mutant with lysosomes was also observed. In addition, the ΔSTM1485 displayed significantly reduced competitive indices (CI) in spleen, liver and mesenteric lymph nodes in murine typhoid model when infected by intra-gastric route. Based on these results, we conclude that the acidic pH induced STM1485 gene is essential for intracellular replication of Salmonella.
Keywords: Salmonella containing vacuole, acid shock proteins, acid tolerance response, acidification, competitive indices, epithelial cells
Introduction
Salmonella enterica comprises a group of Gram-negative bacteria capable of causing clinical syndromes ranging from self-limiting diarrhea to severe fibrinopurulent necrotizing enteritis and life threatening systemic diseases.1,2 During the course of infection, Salmonella encounters several potentially lethal conditions such as the extremely low pH of the stomach, bile salts, reactive oxygen intermediates etc.3 One of the most frequently encountered hostile conditions by Salmonella is acid stress and hence the ability to sense and respond to the acid stress is crucial for its survival. Salmonella responds to the acidic pH changes through complex acid survival systems collectively called acid tolerance response (ATR).4 When S. Typhimurium cells are grown at sublethal pH (pH 4.5–5.5) in vitro for one generation, Salmonella gain the ability to survive at extreme acidic conditions (pH 3); this is known as ATR.5 ATR is characterized by synthesis of approximately 38–60 acid shock proteins (ASPs) upon exposure to moderate acidic conditions; in turn these ASPs appear to promote bacterial survival under extreme acidic stress conditions.6
Another crucial aspect of Salmonella pathogenesis is tolerance of episodes of low pH within the macrophage phagosome or phagolysosome.7 Following phagocytosis of Salmonella and its compartmentalization into the modified phagosome known as the Salmonella containing vacuole (SCV), a series of defense systems are initiated inside the host macrophages. In macrophages, phagosome acidification is considered as one of the major killing mechanism for clearing intracellular bacteria. Moreover, phagosomal acidification enhances the efficiency of other bactericidal mechanisms such as free radical formation (superoxide and nitric oxide radicals), increased fusion of phagosomes with lysosomes and activation of lysosomal acid hydrolases.8 Phagosomal pH is maintained between 4 to 5 by combined action of v-ATPases and chloride channels present in the vacuolar system.9 Salmonella are of particular interest as they reside in modified phagosomes called SCV and resist killing by modulating phagosomal maturation and also altering the vesicular trafficking of iNOS and NADPH oxidase vesicles.10-12 Furthermore, Salmonella uses acidic pH (~pH 4.5) for transcriptional regulation of a subset of PhoP regulated genes13 and assembly of the Salmonella pathogenicity island-2 (SPI-2) type three secretion system (T3SS),14-16 which is essential for survival inside the macrophages.
Several transcriptomic analyses of intracellular Salmonella showed that a set of acid inducible genes such as mgtC, cadB, cysB, adiY, pagC and marAB are upregulated inside the macrophage vacuole, which is consistent with the acidic nature of the SCV. However, none of the ATR regulators such as ompR, phoP or rpoS are significantly upregulated.17-19 STM1485 encoding putative acid shock protein of 82 amino acids was significantly upregulated inside the macrophages17,19 and to a very lesser extent in epithelial cells.18 The microarray data by Eriksson et al., depicted an interesting biphasic expression level of STM1485 in macrophages being 90-fold at 4 h, 26-fold at 8 h and 113-fold at 12 h after infection.17 Though the roles of mgtC, pagC,20 adiY21 and cadB18 acid inducible genes in Salmonella pathogenesis are known, the physiological role of one of the most highly induced genes STM1485 in Salmonella pathogenesis remains undetermined.
Therefore, the present study was undertaken to understand the possible role of STM1485 in the survival of Salmonella during acid stress, inside the host cells and in the mouse model. We report here that the STM1485 gene is fully dispensable for its in vitro growth. On the other hand, STM1485 plays a key role in intracellular survival of Salmonella by altering the phagolysosomal fusion of SCV through SPI-2 T3SS. This study is the first of its kind to show that STM1485, an acidic pH induced gene in Salmonella is required for its intracellular survival. It further fuels the possibility of an intricate relationship between STM1485 and SPI-2 and its importance in determining the course of Salmonella pathogenesis.
Results
STM1485 is not required for the log and stationary phase acid tolerance response in S. Typhimurium
It was reported that E. coli asr gene is highly expressed at low pH (pH 5.0)22 and is required for growth at moderate acidity (pH 4.5) and also for the log phase ATR of E. coli. Since Salmonella putative acid shock protein (STM1485) is having ~77% similarity at nucleotide level and ~68% identity at amino acid level compare with E. coli Asr protein, initially we have studied the role of STM1485 under acidic conditions.
STM1485 gene was deleted from Salmonella enterica ser Typhimurium (WT) using lambda red recombination system23 and the mutant was confirmed by colony PCR with confirmatory primers against the sequence flanking the STM1485 gene and with a reverse primer specific for the CAT gene of pKD3 (Table S1 and Fig. S1A and B). We then compared the growth of ΔSTM1485 strain with that of the WT in rich medium (LB) defined medium (minimal media) at neutral pH 7.4, minimal medium F and ISM medium at pH 5.0. The growth of the WT and the ΔSTM1485 strains was comparable at pH 7.4 (LB and minimal media) and 5.0 (F- and ISM media) (Fig. S1C–F).
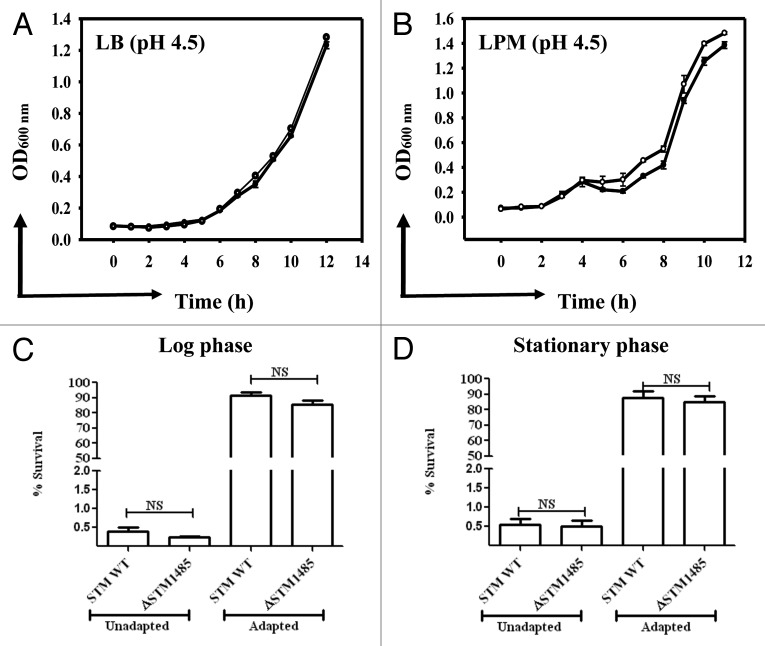
The WT and ΔSTM1485 were tested for their ability to grow in LB and LPM media buffered with sodium citrate buffer at pH 4.5, 4.0 and 3.5 for a period of 12 h as described in materials and methods. No difference was observed between growth rates of WT and ΔSTM1485 in either LB or LPM at pH 4.5 (Fig. 1A and B). However, at pH 4.0 and 3.5 both strains showed very poor growth (data not shown).
Figure 1. In vitro characterization of the ΔSTM1485. (A) and (B) Growth kinetics of the ΔSTM1485 and its parental strain at acidic pH. Growth kinetics was done in LB (A) and LPM (B) at pH 4.5 buffered with sodium citrate. Log (C) and stationary (D) phase acid tolerance response (ATR) of the ΔSTM1485 strain. Overnight grown cultures were sub cultured into LPM media at pH 7.0 to an OD600 nm of 0.3 (log phase) or 1.5 (stationary). For adaptation, bacteria were resuspended in LPM media at pH 4.5 for 2 h, followed by shifting of pH to 3.0 and growth for 2 h. Unadapted cells were directly shifted to pH 3.0. Number of viable bacteria was enumerated by plating on selective LB plates. Shown is the data of two independent experiments. Filled circles, WT; open circles, ΔSTM1485; NS, not significant.
Then, we have analyzed the role of STM1485 in the acid tolerance response of S. Typhimurium at pH 3.0 after an initial adaptation of both the WT and ΔSTM1485 at pH 4.5 in LPM medium. Direct acid challenge of the log phase cultures (without adaptation) of both the WT and ΔSTM1485 from pH 7.0 to 3.0 showed 0.4% and 0.25% survival respectively. In contrast, adapted WT and ΔSTM1485 exhibited 91% and 87% survival after two hours of incubation at pH 3.0 (Fig. 1C). Same is the case even with the stationary phase acid tolerance response (Fig. 1D). No significant difference was observed in acid tolerance response of the WT and ΔSTM1485 strains implying that STM1485 is not essential for log and stationary phase acid tolerance response in S. Typhimurium in vitro.
STM1485 expression is upregulated inside the host cells and is dependent on vacuolar acidification
Before investigating the physiological role of STM1485 gene, we studied the expression of STM1485 in vitro as well as in cell culture. Since STM1485 and its ortholog (STY1582) in S. Typhi are upregulated inside the host cells,17,19,24 semi-quantitative RT-PCR was done to check the expression level of STM1485 in vitro as well as in intracellular bacteria i.e., infected RAW264.7 cells with or without bafilomycin (BAF) pretreatment at 4 h post infection. We used BAF, a specific inhibitor of V-ATPases to block the vacuolar acidification.25 We first determined the minimal concentration required for inhibiting the acidification of phagosomes. Effect of BAF on acidification of phagosomes was tested by vital staining with acridine orange, a dye that shifts fluorescence emission upon protonation (data not shown).
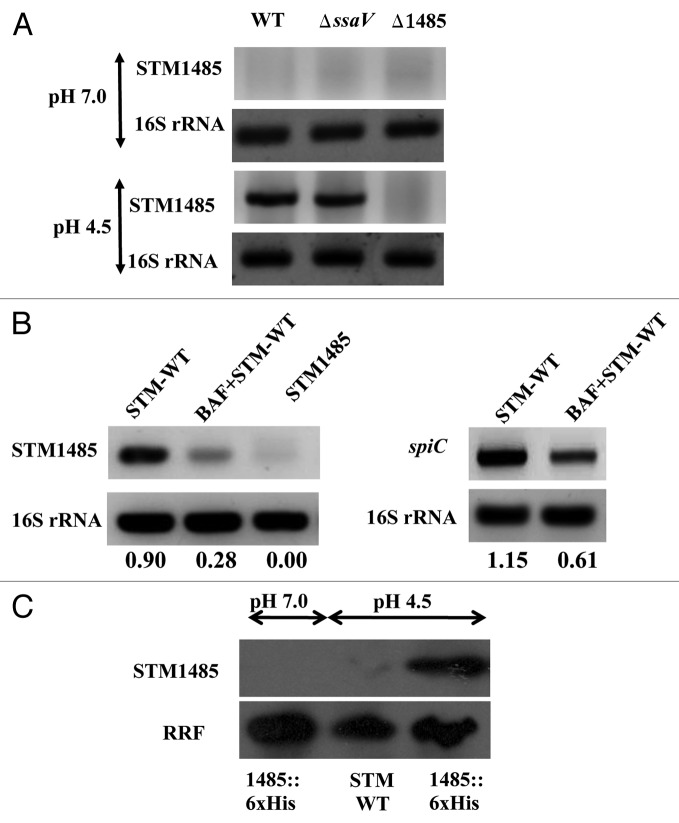
The results obtained by semi-quantitative RT-PCR showed that STM1485 expression was induced when the bacteria were grown in LB at pH 4.5 and not pH 7.0. More over the expression of STM1485 is not affected in SPI-2 null mutant STMΔssaV (Fig. 2A). Pretreatment of RAW264.7 cells with BAF reduced the expression of the gene STM1485 significantly in intracellular WT bacteria compared with the untreated one (Fig. 2B). The SPI-2 gene spiC is used as a positive control, whose expression was also reduced with BAF treatment, showing accordance with the previous report by Cirillo et al.26 Reduced expression of STM1485 in BAF pretreated cells could be attributed to the increase in the vacuolar pH. These results indicate that STM1485 is induced at low pH in vitro as well as inside the cells.
Figure 2. STM1485 gene expression in in vitro grown bacteria and intracellular bacteria. (A) and (B) cDNA was synthesized from the RNA isolated from in vitro grown bacteria (in LB pH 7.0 and 4.5 for 4 h) and from infected RAW264.7 cells (4 h post-infection) with and without BAF treatment. STM1485 and spiC (representative from SPI-2 island) were amplified by PCR. 16s rRNA served as internal control. (C) Expression of the His tagged STM1485 protein from STM1485::6xHis knock in strain grown in LB at pH (4.5 and 7.0) by immunoblot. Ribosome Recycling Factor (RRF) probing was done to equalize the amount of bacterial protein loaded for the SDS PAGE. Numbers below the gels indicate the values of densitometric image analysis using Multi Guage software. Images are representative of two independent experiments.
To check the STM1485 expression at protein level, we made a knock-in strain where six histidine amino acids were tagged to the C-terminal of the chromosomal STM1485. As shown in Figure 2C, no band was observed in the case of Salmonella grown in LB at pH 7.0. However, a 10 kDa band corresponding to His-tagged STM1485 protein was induced at pH 4.5. The WT strain has been used as control (Fig. 2C). The expression of STM1485 in different epithelial cells (INT-407, Caco-2 and HeLa) was assessed. STM1485 gene was upregulated in all the epithelial cell lines tested both at mRNA as well as protein level, when compared with its expression in bacteria grown in DMEM (Fig. 3A and B). These results suggest the importance of the STM1485 in epithelial cells, which Salmonella encounters during its course of infection.
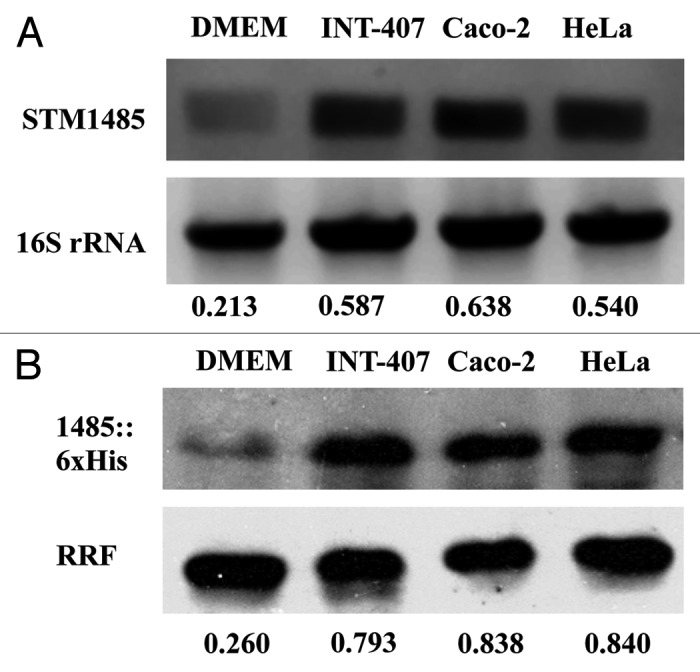
Figure 3. Evaluation of STM1485 expression in epithelial cells (INT-407, Caco-2 and HeLa). STM1485 expression was studied using (A) RT-PCR assay. (B) Immunoblot method using the STM1485::6xHis tagged strain. Bacteria were isolated from the infected epithelial cells at 6th h post infection. Bacteria grown in DMEM at 37°C under 5% CO2 were served as control to determine expression outside host cells. Numbers below the gels indicate the values of densitometric image analysis using Multi Guage software. Results are representative of two independent experiments.
ΔSTM1485 strain is replication defective in the macrophage and epithelial cells
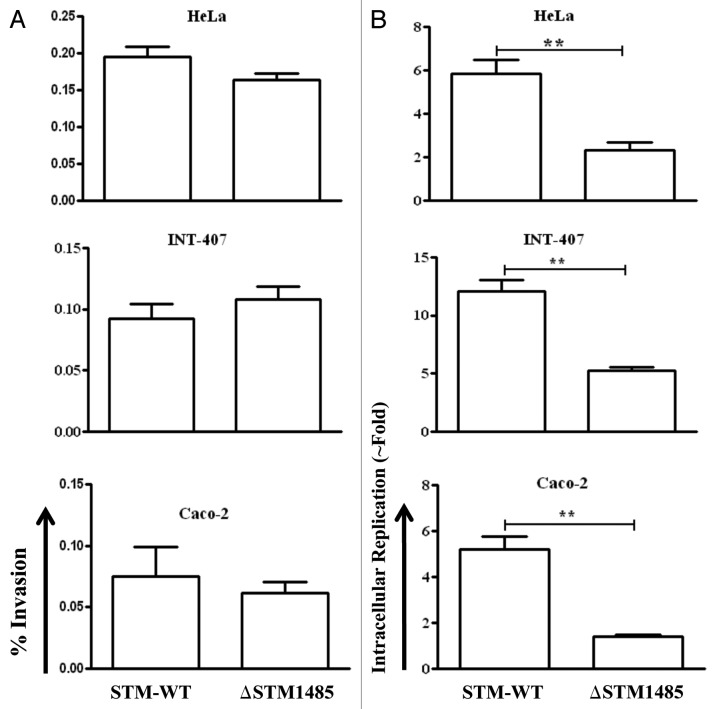
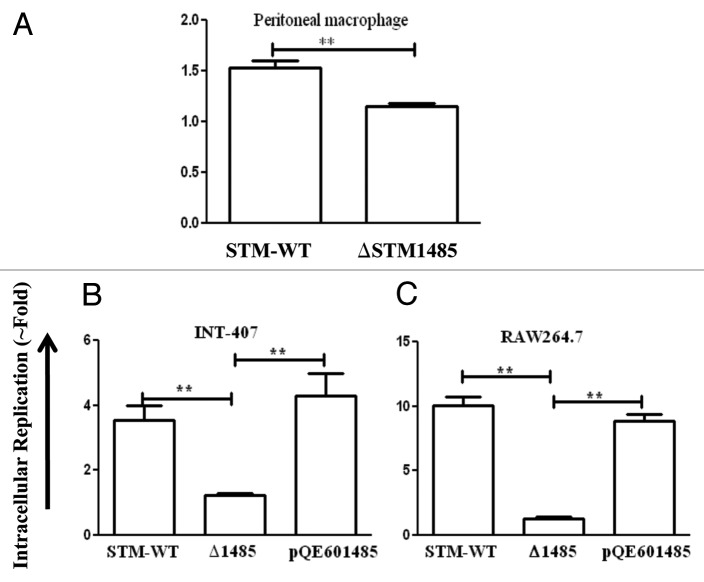
Salmonella encounters epithelial and macrophage cells during its course of pathogenesis.27 Therefore, the intracellular replication capacity of the ΔSTM1485 in human epithelial and murine macrophage cells was analyzed. The ΔSTM1485 is highly defective in intracellular replication compared with that of the WT in human epithelial (Fig. 4B) and murine macrophage cells (Fig. 5A) (p < 0.005). However, the number of ΔSTM1485 colonies obtained after 30 min of infection was comparable to that of the WT strain showing that the strain is not invasion defective (Fig. 4A). This decrease in the fold replication was not seen in the ΔSTM1485 complemented strain when tested in RAW264.7 and INT-407 cells (Fig. 5B and C). Complementation studies also suggested that the STM1485 gene deletion is indeed non polar.
Figure 4. Invasion and intracellular fold replication of the ΔSTM1485 strain inside epithelial cells (INT-407, Caco-2 and HeLa). (A) Invasion of ΔSTM1485 was determined by lysing the infected cells 30 min post-infection. (B) Fold replication was determined by lysing the cells infected with the WT and the ΔSTM1485 at 2 h and 16 h post infection. Graphs are representative of three independent experiments, each with triplicate samples. Standard error bars are shown. (**p < 0.005) (Student's t-test).
Figure 5. Intracellular survival assay of ΔSTM1485 in mouse macrophage cells (A) and complementation studies in INT-407 (B) and RAW264.7 (C). Cells were infected with WT or ΔSTM1485 or ΔSTM1485pQE601485 strains were lysed at 2 and 16 h post infection. Bacterial load was shown as fold increase in CFU from 2 h to 16 h. Graphs are representative of two independent experiments with similar results. Statistical significance was defined as follows (**p < 0.005) (Student's t-test).
Effect of vacuolar pH neutralization on survival and replication of the ΔSTM1485 strain
Salmonella containing vacuoles (SCVs) acidify soon after their formation and remain acidified for at least 5–6 h post infection. The vacuolar proton ATPases (V-ATPases) play critical role in vacuolar acidification and this pH drop is independent of phagosome-endosome/lysosomal fusion. The low vacuolar pH is required for the intracellular replication of Salmonella.7,28 Therefore we sought to determine the effect of neutralization of vacuolar pH on intracellular survival of the WT and the ΔSTM1485 in RAW264.7 macrophage cells.
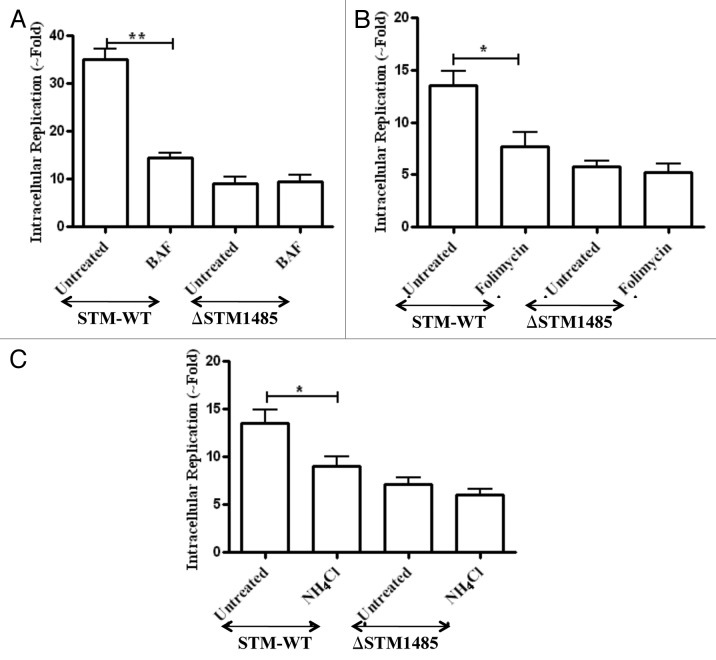
Pretreatment of RAW264.7 cells with BAF (50 nM), reduced the intracellular fold replication of the WT from 2 to 16 h. In contrast, the ΔSTM1485 did not show any further reduction in intracellular fold replication (Fig. 6A) (p < 0.005). In addition to BAF, we observed the same phenomenon with folimycin (Concanamycin A) and ammonium chloride pretreatment which inhibit the vacuolar pH through different mechanisms (Fig. 6B and C). These results suggest that Salmonella needs the acidification for induction of some of their genes protecting it from bactericidal effects different from the acidification. These results also hint that STM1485 might confer survival advantage to Salmonella inside the acidic phagosomes.
Figure 6. Effect of phagosomal pH neutralization on intracellular replication of ΔSTM1485 strain. RAW264.7 cells were pretreated with (A) bafilomycin A1 (50 nM), (B) Folimycin (100 nM) and (C) NH4Cl (100 mM) followed by infection as described in materials and methods. Infected cells were lysed at 2 h and 16 h post infection and bacterial load was shown as fold increase in CFU from 2 h to 16 h. Graphs are representative of three independent experiments with similar results. Statistical significance was defined as follows (*p < 0.05, **p < 0.005) (Student's t-test).
ΔSTMΔ1485 exhibited reduced surface translocation of SPI-2 encoded translocon proteins
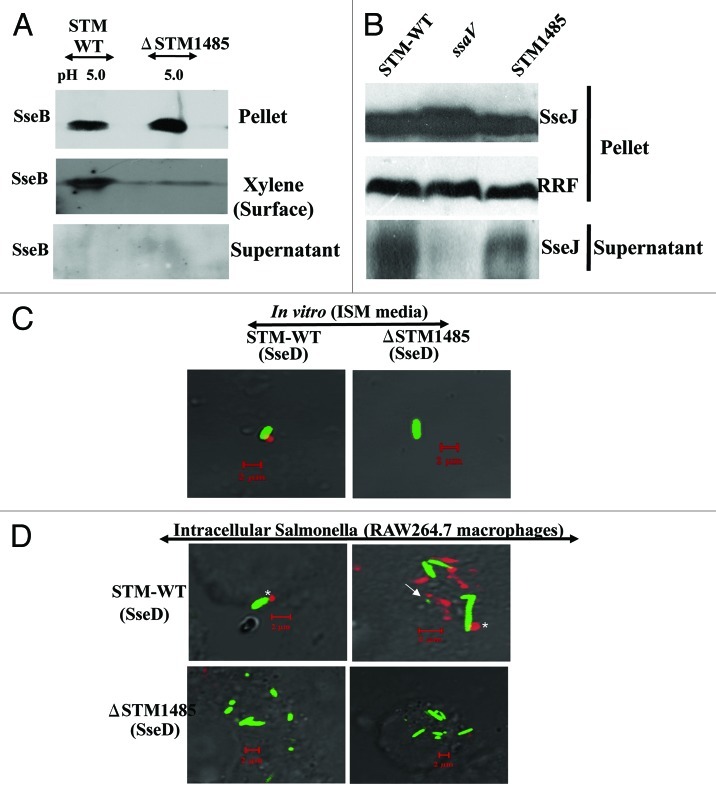
Low pH (between 4 and 5) is an environmental cue for the secretion of translocon proteins and formation of translocon encoded by SPI-2 type three secretion system of S. Typhimurium inside the macrophages.14-16 Furthermore, a recent study by Yoon et al. identified STM1485 as one of the genes which was co-regulated with SPI-2 genes.29 Therefore we hypothesized that STM1485 might be playing an important role in the formation of SPI-2 translocon at low pH. SseB is the first SPI-2 T3SS translocon protein to be identified whose surface secretion is controlled by pH of the medium.15,30 The surface translocation of SseB in vitro at low pH in ISM medium was studied. Proteins isolated from three fractions (pellet, surface and supernatant) were analyzed for the presence of SseB by immunoblotting. A band size corresponding to SseB (~23 kDa) was detected in the surface fraction and cell pellet fraction but not in the supernatant fraction. Compared with the WT, SseB translocation on to the cell surface was visibly reduced in the ΔSTM1485, when the bacteria were cultured at pH 5.0. In contrast, the level of SseB in the pellet fraction of the ΔSTM1485 strain was higher than the pellet fraction of the WT (Fig. 7A).
Figure 7. STM1485 is required for the SPI-2 translocon formation. Surface translocation of SPI-2 encoded translocon proteins (SseB and SseD) in the ΔSTM1485 (A). WT and ΔSTM1485 were grown in ISM media at pH 5.0 and then cell pellet, surface and supernatant fractions were isolated as described in Materials and Methods. By immunoblotting, the presence of SseB protein in all three fractions was analyzed. Secretion of SPI-2 effector protein (B). STM-WT, ΔSTM1485 and ΔssaV mutant strains carrying the His-tagged SseJ construct were grown under SPI-2 inducing conditions (F-media, pH 5.0) for overnight and secreted proteins were precipitated from the culture supernatant. We evaluated the cell pellet and supernatant fractions for intracellular and secreted levels of the SseJ-His by immunoblotting. Detection of SseD in vitro and in intracellular bacteria (C and D). WT-GFP and ΔSTM1485-GFP were grown in ISM media at pH 5.0 for overnight. Bacteria were harvested and preceded for immunostaining (C). RAW264.7 cells were infected with WT-GFP and ΔSTM1485-GFP. Twelve hours post-infection cells were fixed and immunostained (D). Samples were analyzed using a confocal laser scanning microscope (LSM Meta, Zeiss). Note the appearance of SseD (red) on the surface of intracellular bacteria (green) (*) and also distant to the bacteria (arrow). Scale bar represent the 2 µm.
These results were confirmed by analyzing the localization of SseD on the Salmonella grown in in vitro and in vivo conditions using confocal microscopy. Antiserum raised against SseB was used for detecting surface appendages formed during the growth of the bacteria. A punctuate appearance of SSeD on one pole of the bacterial cell was observed in WT but not in ΔSTM1485 both under in vitro growth conditions (Fig. 7C) and in infected cells (Fig. 7D). These results indicate that SPI-2 translocon formation is affected in the absence of STM1485.
Further experiments were performed to establish that STM1485 is required for the SPI-2 translocon formation. Secretion of SPI-2 effector SseJ under in vitro growth conditions was studied by immunoblotting. STM-WT, ΔSTM1485 and ΔssaV mutant strains carrying the His-tagged SseJ construct were grown under SPI-2 inducing conditions (F media, pH 5.0) for overnight and secreted proteins were precipitated from the culture supernatant. We evaluated the cell pellet and supernatant fractions for intracellular and secreted levels of the SseJ-His by immunoblotting. A band size corresponding to SseJ (~50 kDa) was detected in the supernatant as well as in the cell pellet fraction. We found that SseJ secretion was noticeably reduced in the supernatant of ΔSTM1485 compared with WT. RRF was not observed in any of the secreted fractions suggesting that bacterial cell integrity was maintained throughout the experiment. No secretion of SseJ-His was observed in the supernatant of ΔssaV mutant (Fig. 7B). Taken together these observations suggest that STM1485 is important for SPI-2 translocon formation.
ΔSTM1485 containing phagosome progresses along the degradative pathway and fuses with lysosomes
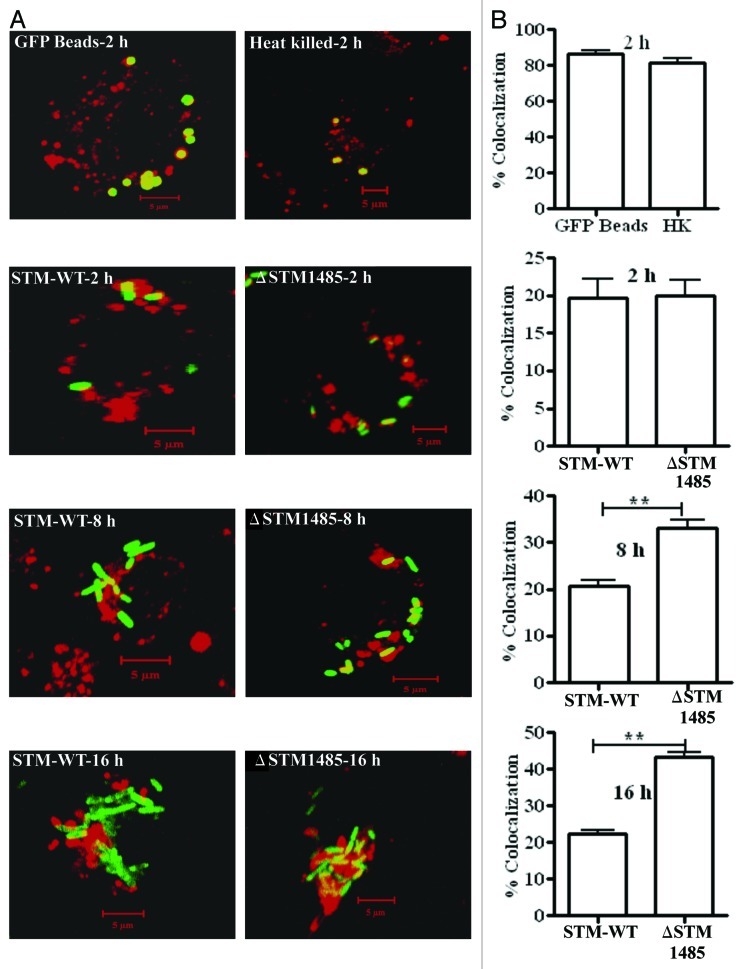
Salmonella is an intracellular pathogen that requires a set of virulence proteins encoded in the SPI-2 for its intracellular survival. The effector proteins encoded by SPI-2 allows Salmonella to establish SCV by preventing the fusion of SCV with lysosomes and interferes with the trafficking of NADPH oxidase and iNOS containing vesicles to the SCV inside the host cell.11,31-33 The reduction in the surface translocation of SPI-2 needle proteins in ΔSTM1485 might have an effect on the SPI-2 function and hence the intracellular fate of the bacteria. To test this, we investigated the co-localization of GFP bacteria with the endocytic probe texas red conjugated ovalbumin (TROv).34 TROv gets distributed throughout the endocytic network after pinocytosis and can be chased into lysosomes to serve as lysosomal marker. Approximately 20% of the WT bacteria were co-localized with TROv at each time point; in contrast, ΔSTM1485 showed ~19%, ~34% and ~44% of co-localization with TROv at 2, 8 and 16 h respectively. Co-localization of the ΔSTM1485 was significantly higher than the WT at 8 and 16 h post infection (p = < 0.05) (Fig. 8A and B). These results indicate that the lack of STM1485 results in the maturation of SCV along the degradative pathway.
Figure 8.
Co-localization of GFP expressing bacteria, latex beads and heat killed bacteria with TROv in RAW264.7 cells (A). Prior to infection, RAW264.7 cells were pulse-chased with 50 μg/ml of Texas Red-Ovalbumin (TROv) for 30 min, washed and then incubated for another 30 min. Then cells were infected with GFP expressing WT and ΔSTM1485 and fixed at different time points. Cells were also incubated with latex GFP beads and heat killed bacteria. Heat killed bacteria were detected with antibody specific to Salmonella LPS followed by FITC-conjugated secondary antibody. Samples were analyzed using a confocal laser scanning microscope (LSM Meta, Zeiss). At least 50 cells were counted in each case in each experiment. (B) Results of three experiments in which SCVs from cells infected with each strain or phagosomes containing latex beads and heat killed bacteria were scored for TROv co-localization at 2, 8 and 16 h post infection. Shown are the representative images of a single XY section taken from Z-stack. The values are given as mean ± standard error in percentages. Statistical significance was defined as follows (**p < 0.005) (Student's t-test).
ΔSTM1485 strain failed to compete with the WT in mouse model
In order to evaluate the contribution of STM1485 gene in virulence of Salmonella in vivo, we tested the ability of the ΔSTM1485 strain to compete with the WT strain in mixed infections in murine typhoid model (Table 2). Competitive index is an alternative measure to LD50 to determine the degree of virulence attenuation. The WT and mutant strains were recovered 4 d after intra-gastric inoculation of 107 CFU/mouse at a ratio of 1:1. As shown in the Table 2, the ΔSTM1485 strain showed significantly reduced competitive indices (CI) in spleen, liver and mesenteric lymph nodes (MLN), suggesting that STM1485 gene is essential for the in vivo colonization of S. Typhimurium. The pQE601485 complemented strain was found to compete equally against the WT strain.
Table 2. Competitive Indices of mixed infections using the WT, ΔSTM1485 and the pQE601485 complemented strains.
| |
Strain-1 |
Strain-2 |
Competitive index of strain 2 with respect to strain 1 |
||
|---|---|---|---|---|---|
| Spleen | Liver | MLN¶ | |||
| 1 |
WT# |
WT§ |
1.121 ± 0.12 |
0.975 ± 0.17 |
1.124 ± 0.09 |
| 2 |
WT# |
ΔSTM1485 |
0.413 ± 0.05** |
0.341 ± 0.07** |
0.206 ± 0.13** |
| 3 | WT# | pQE601485 | 1.035 ± 0.14 | 1.367 ± 0.09 | 1.107 ± 0.12 |
# Resistant to nalidixic acid and the parent strain for knock out.
§ Resistant to Carbencillin. Both WT strains were isogenic.
¶ Mesentric lymph nodes.
p < 0.005 (Student's t-test); statistical difference between 1 and 2.
Discussion
Adaptations to environmental changes allow pathogens to survive and perpetuate during their encounter with nature’s extremes. One such natural stress situation confronted by Salmonella in host and non-host environments is acid stress. S. Typhimurium can survive lethal acidic conditions (pH between 2 and 3) if it is first adapted to mild acidic conditions and the phenomenon involves production of acid shock proteins.35 These ASPs belong to different groups including those of membrane bound transporters, periplasmic proteins, cytoplasmic enzymes and extracellular components.36 The physiological function of one such gene, STM1485, which is induced under acidic pH, in Salmonella pathogenesis is not known. Moreover, in spite of the acidic pH of the phagosome inside the host cells, Salmonella can multiply within the phagosomes of macrophages and use acidic pH as a cue for regulation of virulence genes or assembly of SPI-2 T3SS. We speculated if STM1485 induced at low pH has any role in counteracting the acidic pH inside the phagosomes.
STM1485 gene is induced when bacteria are grown in acidic pH (pH 4.5) in LB media but not at neutral pH. The expression of STM1485 in infected macrophages was reduced with BAF pretreatment indicating that the STM1485 gene might be required for survival at acidic pH inside the phagosomes of the host. The fact that the deletion of STM1485 did not affect the growth of S. Typhimurium at low pH in vitro and its acid tolerance response (log and stationary phase) suggests that STM1485 gene is dispensable for Salmonella survival at low pH or for acid tolerance response under in vitro conditions. The ΔSTM1485 was attenuated in all the cell lines tested and could not compete with the WT in mixed infections in murine typhoid model. Indispensability of STM1485 gene for survival and replication of S. Typhimurium is confirmed by complementation of STM1485 gene which fully restored the phenotype back to wild-type. These results clearly state that the STM1485 gene is essential for intracellular replication and Salmonella pathogenesis. Our data suggest that though STM1485 is induced at low pH conditions, it is not involved in counteracting acidic conditions in vitro but indirectly helps in counteracting the other anti-bactericidal mechanisms inside the host.
Intracellular replication of WT and not ΔSTM1485 was compromised by vacuolar pH neutralizing agents (BAF, folimycin and ammonium chloride). Loss of WT intracellular replication with phagosomal pH neutralization may be due to the absence of expression of virulence genes or lack of SPI-2 encoded needle which are required for survival and growth within the macrophages.14,15 Our RT-PCR studies of intracellular bacteria with and without BAF treatment shows that STM1485 gene expression is reduced upon BAF treatment compared with the untreated ones. So we reason that reduction in the STM1485 gene expression in WT with BAF has an equivalent effect as that of ΔSTM1485.
Infection of macrophages by Salmonella leads to expression of SPI-2, which allows Salmonella to establish replicative vacuoles, called SCV. The type three secretion system encoded by SPI-2 is used to translocate cohorts of bacterial effector proteins directly into the host cell. There have been several studies to identify environmental cues that regulate either expression or assembly of SPI-2 T3SS. Acidic pH (pH 4.5–5.0) of SCV is an important cue, in part required for the assembly of T3SS.14-16 SPI-2 T3SS secretes translocon proteins (SseB, SseC and SseD) that form translocon or needle through which SPI-2 T3SS effector proteins (SpiC, SseF, SseG, SifA, SseI and SseJ) are delivered into the host cytosol. Some of the SPI-2 proteins forming needle components are present in the periplasmic region of bacteria, e.g., SsaV, SsaJ and SsaC.30
When the Salmonella are exposed to the harsh environmental conditions like low pH inside the phagosome, proteins present in the periplasm are more vulnerable to such conditions due to the porous nature of the periplasm, thus requiring greater protection than the cytoplasmic proteins. Periplasmic proteins or chaperons play a crucial role in the biogenesis of outer membrane proteins or formation of functional type three secretion systems, when Salmonella encounters harsh environmental conditions inside the host cells.37,38 We speculate that STM1485 encoded protein might be located in the periplasm based on the conservation of signal sequence of STM1485 with that of E. coli asr signal sequence.22 STM1485 encoded protein may play an important role in outer membrane protein biogenesis and formation of functional T3SS, which are important for the survival of Salmonella inside the host cells. Interestingly, in our study we have observed that the surface secretion of SseB and SseD and secretion of SseJ effector protein are affected in ΔSTM1485 compared with WT, which implies that STM1485 encoded protein is playing a crucial role in the secretion of translocon proteins. The possible reasons for this decreased surface secretion are either defect in needle formation or loss of membrane integrity in the ΔSTM1485.
Co-localization studies of GFP bacteria with the endocytic fluid phase marker texas red conjugated ovalbumin (TROv) showed increased co-localization of ΔSTM1485 with the TROv (lysosomes) compared with the WT. This can be explained by the reduced SPI-2 translocon formation as reported earlier that the reduction in the SPI-2 expression leads to efficient interaction of SCV with the components of the late endosomal or lysosomal system.31,32 Therefore, the inability of ΔSTM1485 in assisting the formation of functional SPI-2 T3SS translocon explains its high level of attenuation inside the cell line and its incapability to compete with the WT in murine model.
Based on our results, we conclude that perhaps Salmonella senses low pH using unknown sensors or two component regulatory systems like PhoPQ and synthesize the STM1485 encoded protein. STM1485 encoded protein in turn regulates the assembly of SPI-2 encoded T3SS at low pH inside the host cell. Absence of STM1485 results in the decreased SPI-2 T3SS needle formation thereby increasing the fusion of SCV with the degradative compartments. Hence, STM1485 is one of the key players in regulating the SPI-2 translocon thereby contributing to Salmonella pathogenesis.
Materials and Methods
Bacterial strains and growth conditions
The bacterial strains and plasmids used in this study are listed in the Table 1. Antibiotics namely kanamycin, carbenicillin, nalidixic acid (each at 50 μg/ml) and chloramphenicol (20 μg/ml) were added when needed for the selection of strains. Except where indicated, bacteria were grown at 37°C with aeration. Luria broth (LB) at different pH values (4.5, 4.0 and 3.5) buffered by sodium citrate (0.05 M); F-media with 50 mM KCl, 75 mM (NH4)2SO4, 5 mM K2SO4, 10 mM K2HPO4, 1 mM Bis-tris, 30 μM MgSO4, 38 mM glycerol and 0.1% casamino acids;15 Low phosphate salt glucose media (LPM) with 50 mM Tris-base, 20 mM KCl, 75 mM (NH4)2SO4, 240 μM MgCl2, 18.5 μM CaCl2, 6.5 mΜ FeCl3, and 0.2% Glucose Bactopeptone-0.6%);22 ISM media (170 mM K2PO4/KH2PO4 0.5 mM MgSO4, 1 mM CaCl2, 6 mM K2SO4, 5 mM NH4Cl, 5 mM NaCl, 0.4% glucose, 2 mg/mL nicotinic acid at pH 5 or 7.5).16 Heat killed bacteria were prepared by heating the 0.3 OD adjusted cultures at 65°C for 20 min.
Table 1. Strains and plasmids used in this study.
| Strain | Description | Reference |
|---|---|---|
| STM-WT 12023 |
Wild-type 12023, parental strain for all mutants, resistant to nalidixic acid |
15 |
| ΔSTM1485 |
Δ1485::CAT S. Typhimurium |
This study |
| WT-GFP |
WT 12023 with pFPV25.1, resistant to carbenicillin (used in confocal and competition experiments) |
This study |
| ΔSTM1485 GFP |
ΔSTM1485with pFPV25.1 (used in confocal studies) |
This study |
| STM1485::6xHis |
S. Typhimurium expressing the1485::6xHis fusion protein |
This study |
| STMΔssaV |
SPI-2 null mutant strain |
15 |
|
Plasmid |
Description |
|
| pQE601485 |
S. Typhimurium 1485 complementing vector (carbr) |
This study |
| pKD46 |
Plasmid expressing λ red recombinase (carbr) |
23 |
| pKD3 |
Plasmid with FRT-flanked chloramphenicol resistance gene |
23 |
| pKD4 |
Plasmid with FRT-flanked kanamycin resistance gene |
23 |
| pFPV25.1 |
Plasmid expressing GFP |
45 |
| pHG86 |
Promoterless reporter plasmid (carbr) |
46 |
| pHG86sseJ-His | Plasmid expressing 6X His tagged SseJ | This study |
Growth study of ΔSTM1485 strain was done in different media such as LB (pH 7.5), minimal media (pH 7.5), F-media (pH 5.0) and ISM media (pH 5.0). For growth kinetics at low pH, cultures were grown in LB or LPM buffered at different pH values (pH 4.5, 4.0 and 3.5) by sodium citrate (0.05 M). Overnight grown cultures were sub-cultured (equal number of bacteria) into the respective media and growth was monitored by measurement of optical density at 600 nm every 1 h. Acid tolerance response of Salmonella was done as described previously with some modifications.22
For ATR analysis, cultures were grown at 37°C in LPM (pH 7.0) to an OD600 of 1.5 for the stationary phase ATR assay and to an OD600 of 1.5 for log-phase assay. For acid adaptation, cells were resuspended in fresh LPM at pH 4.5 and incubated for 2 h. Unadapted and adapted cultures were inoculated into fresh LPM (pH 3.0) at approximately 1 × 108 cells/ml. Cultures were incubated for 2 h at pH 3.0 and viable bacteria were calculated by plating aliquots of serially diluted cultures onto the LB (pH 7.5) plates at 0 h and 2 h. The number of viable bacteria was expressed as % survival after 2 h incubation at pH 3.0.
Eukaryotic cell lines and growth conditions
RAW264.7 cells were a kind gift of Prof. Anjali Karande (Department of Biochemistry, Indian Institute of Science, India). INT-407, HeLa and CaCo-2 cells were obtained from National Center for Cell Science (NCCS), Pune, India. The cells were grown in Dulbecco’s modified Eagle’s medium (DMEM; Sigma) or RPMI 1640, both supplemented with 10% fetal calf serum (Sigma). Elicited mouse macrophages were harvested from BALB/c mice by peritoneal lavage with RPMI 1640 medium (Sigma) 5 d after intraperitoneal injection of 5 ml of brewer’s thioglycolate medium (Hi-media). All cells were maintained at 37°C and 5% CO2. For Caco-2 cells, 1% non-essential amino acids was added to the medium.
Construction of the STM1485 mutant in Salmonella Typhimurium
ΔSTM1485 strain was constructed by one-step gene deletion strategy.23 The knockout was selected for chloramphenicol resistance and confirmed by colony PCR using specific confirmatory primers (Table S1).
Complementation of the ΔSTM1485 strain
First, STM1485 gene was amplified by polymerase chain reaction (PCR) using primers listed in Table S1 and cloned into pQE60 vector. Plasmid containing STM1485 gene was confirmed by restriction digestion and using vector specific primers (Table S1). The resulting plasmid DNA was electroporated into the ΔSTM1485 strain to get pQE601485 strain.
Construction of His-tagged SseJ
To generate His-tagged fusion protein, the sseJ ORF along with its promoter was amplified using primers listed in Table S1. Plasmid pHG86 and the PCR product were digested with EcoRI and BamHI and ligated together, yielding His-tagged construct. In this strategy, reverse primer was modified to carry a sequence coding for 6X histidine residues at 5′ end followed by a stop codon so as to obtain only a SseJ tagged with 6X His at C-terminal end. The His-tagged SseJ construct was introduced into STM-WT strain, ΔSTM1485 strain and ΔssaV mutant strain by electrporation.
Construction of the Salmonella strain expressing the chromosomal STM1485::6xHis fusion protein
The Salmonella strain expressing the STM1485::6xHis fusion protein was engineered in serovar Typhimurium by a modified method from the one-step deletion strategy as described previously.39 Briefly, electrocompetent cells from serovar Typhimurium transformants, carrying a red helper plasmid (pKD46), were prepared. In this strategy, the forward knockout primer was modified to carry a sequence coding for six histidine residues at 5′ end. This histidine tag containing primer was targeted against the region immediately downstream of the STM1485 gene (including the STM1485 stop codon). The knockout primer set effectively knocked out the 270 bp of intergenic region downstream of STM1485 while concomitantly tagging STM1485 on its C-terminal with the sequence coding for 6 histidine residues. This was followed by another stop codon so as to obtain only a STM1485:: 6xHis fusion protein. PCR product containing the kanamycin resistance gene (from pKD4 plasmid) flanked by sequences upstream and downstream of the STM1485 gene was prepared. This DNA was then electroporated in Salmonella Typhimurium carrying pKD46. The mutants were selected by kanamycin resistance and confirmed by PCR using the kanamycin internal forward and STM1485 cloning forward primers. In the knock-in strain, a band corresponding to 850 bp was amplified whereas in the WT strain, no such product could be detected. We also confirmed the chromosomal knock-in strain in the bacterial lysate by immune-blot using anti-His antibody staining. A 10 kDa single band corresponding to the STM1485:: 6xHis fusion protein was observed.
Intracellular proliferation assay
Different cell lines were seeded in 24-well plate at a concentration of 1 × 105 to 2 × 105 per well 24 h prior to infection. For the infection of RAW264.7 cells, the S. Typhimurium strains were grown to stationary phase in LB with the appropriate antibiotic. For infection of epithelial cells, stationary-phase cultures were diluted 1:33 in LB and grown for 3 h (late-exponential phase) prior to infection to induce Salmonella pathogenicity island 1(SPI-1) encoded genes required for invasion of non-phagocytic cells.40 The gentamicin protection assay was done as described previously.41 The fold intracellular replication was calculated by dividing the intracellular bacterial load at 16 h by the bacterial load at 2 h. Alternatively, GFP latex beads and heat killed bacteria (at a ratio of 10 to 1) were added to the cells for 20 min and followed further as in the case of infection. To inhibit the vacuolar acidification, inhibitors like bafilomycin A1 (BAF) (10–100 nM), folimycin (100 nM) and NH4Cl (100 mM) (purchased from Calbiochem) were used. Cells were pretreated with these inhibitors for 30 min followed by the infection and gentamicin protection assay.
Bacterial RNA extraction from infected cells and in vitro grown bacteria
This was done as described previously.17 After 6 h of infection, cells were lysed on ice for 30 min in 1 ml of 0.1% SDS, 1% acidic phenol, and 19% ethanol in water. Salmonella were isolated from the lysate by centrifugation and RNA was isolated using TRI reagent (Sigma) according to manufacturer’s protocol. RNA from different in vitro-grown bacterial strains (WT and ΔSTM1485) was obtained by growing bacteria in LB at neutral pH values (pH 7.0 and 4.5) at 37°C to mid-log phase using TRI reagent (Sigma). Bacteria grown under static condition at 37°C to mid-log phase in DMEM/5% CO2 were used as control. These conditions mimicked those used for the cell-infection experiments.
RT-PCR
Bacterial RNA (extracted either from infected cells, bacteria grown in DMEM medium or from bacteria grown in LB medium) was reverse transcribed using reverse transcription system (Fermentas) for STM1485 gene and control 16S rRNA gene with primers mentioned in (Table S1). Samples were pretreated with DNase I to remove the genomic DNA. To check the expression level of specific gene, cDNA was amplified for 35 cycles using gene specific primers (Table S1). Control reactions were done without reverse transcription to verify the absence of DNA in RNA preparations.
Antibodies and confocal laser scanning microscopy (CLSM)
Antisera raised in rabbits against recombinant SseB, and SseD have been described before by Nikolaus et al.18 RAW264.7 cells were seeded on a coverslip at 1 × 105 cells/well in 24-well plates and infection was done as described above. Infected cells were fixed with 3.5% paraformaldehyde in PBS (pH 7.4) (Sigma) for 15 min at indicated time points and then cells were washed thrice with PBS. Cells were incubated with specific antibody [LPS (HyTest Ltd.); SseD (kind gift from Michael Hensel)] diluted in blocking buffer [0.1% saponin, 2% BSA and 2% goat serum (all from Sigma) in PBS] for 1 h. Cells were then washed twice with PBS and incubated with appropriate fluorescent secondary antibodies (Dynova or Jackson Laboratory) diluted in the blocking buffer (SseB: Cy3-conjugated anti-rabbit IgG). In some experiments, Salmonella and heat killed bacteria were stained for LPS using the monoclonal anti-Salmonella Typhimurium LPS antibody (HyTest) raised in mouse, as the primary antibody and FITC-conjugated goat anti-mouse IgG antibody (Dianova) as the secondary antibody. The cells were washed thrice with PBS and coverslips were mounted on a glass slide. After staining images were taken using a confocal laser scanning microscope (LSM Meta, Zeiss).
For detection of SseD in vitro, bacteria expressing GFP were grown in ISM media (pH 5.0) overnight at 37°C. Bacteria were recovered by centrifugation at 5,000 g for 10 min and fixed for 20 min in 3.5% PFA in PBS. Subsequently bacteria were washed twice with PBS very gently. Antibodies were diluted 1:100 in PBS containing 2% BSA and 2% goat serum and added to the fixed bacteria. Antibody staining was performed under shaking conditions for 2 h followed by Cy3-conjugated anti-rabbit IgG for 1 h. After 3–4 washes with PBS, the final pellet was resuspended in 50% glycerol in PBS. Aliquot of stained bacteria were mounted on glass slides, covered and sealed and analyzed for CLSM. Adobe Photoshop 7 was used to adjust the contrast and brightness of the images.
Texas Red-Ovalbumin (TROv) pulse chasing to load the lysosomes
RAW264.7 cells were seeded on glass coverslips and were labeled by pulsing with 50 µg/ml of TROv (Molecular Probes) for 30 min at 37°C in 5% CO2.34 The labeling media was replaced with fresh DMEM medium after three washes and incubated further for 30 min. After 30 min of incubation, cells were infected as described above. Infected cells were fixed at respective time points and analyzed. In some experiments, cells were incubated with fluorescent yellow-green latex beads of 1 µm size (L4655, Sigma-Aldrich). Images were obtained as projections of Z-stacks and co-localization analysis was done using FCS-CLSM META software.
Preparation of cell fraction and protein gel blot analysis
Bacterial strains WT and ΔSTM1485 were grown overnight in ISM media at pH 5.0. Bacterial lysate, surface and secretary proteins were isolated from the culture as described by Beuzon et al. with slight modifications.30
Aliquots containing equal amount of protein were loaded onto 12% SDS-PAGE gel and transferred to nitrocellulose membrane (Millipore) using semi dry gel transfer apparatus. The membranes were treated with 5% skimmed milk for 1 h to block non-specific binding and incubated with primary antibody raised in rabbit against SseB (kind gift from Michael Hensel) (1:2,000) for 2 h and followed by washes. The blots were further stained with goat anti-rabbit IgG-horseradish peroxidase conjugate (AP-Biotech, 1:2,000) for 2 h. The immune complex on the blots were detected with enhanced chemiluminescence substrate (Perkin Elmer) and exposed to Eastman Kodak XAR X-ray film.
For the expression of STM1485::6xHis fusion protein, both the strains were inoculated in LB broth and epithelial cells were infected separately with each strain. Equal amount of total protein from the whole cell lysate (purified bacteria either from infected cells or from those grown in DMEM or LB medium) was processed for immunoblotting.
Protein secretion analysis
STM-WT strain, ΔSTM1485 strain and ΔssaV mutant carrying the His-tagged SseJ construct were grown in F media, pH 5.0 (SPI-2 inducing conditions). Bacterial lysate and secretary proteins were isolated from the culture as described by Freman et al.42 with slight modifications.
STM1485::6xHis and SseJ::6xHis fusion proteins were probed using HRP-conjugated anti-His antibody (1:20,000, Sigma). As loading control anti-RRF (ribosome recycling factor) antibody was used (a kind gift of Professor Umesh Varshney, Indian Institute of Science, India. Polyclonal, 1:2,000). The WT Salmonella was used as a control to study the expression of His-tagged protein in Salmonella strain expressing STM1485::6xHis fusion protein.
Competitive index (CI)
Competitive index assay was performed as described previously.43 The competitive index (CI) was calculated by dividing the number of mutant bacteria recovered from infected animals by the number of wild-type bacteria recovered.44 This value was then corrected by the initial ratio of mutant to wild-type bacteria used to infect each animal. Values are mean ± SD for ten mice from two experiments.
Statistical analysis and software
For analyzing data Student's t-test was employed using commercially available software (GraphPad Prism). Statistical significance was defined as follows: **, p < 0.005; *, p < 0.05. Densitometric analyses performed using Fuji Multi Gauge V2.3 software.
Supplementary Material
Acknowledgments
We would like to thank Preeti for her critical suggestions. This work was supported by the grant, Provision (2A) Tenth Plan (191/MCB) from the Director of Indian Institute of Science, Bangalore, India and Department of Biotechnology (DBT 197 and DBT 172) to D.C. Infrastructure support from ICMR (Center for Advanced Study in Molecular Medicine), DST (FIST) and UGC (special assistance) is acknowledged. U.S.A. acknowledges UGC for fellowship. We thank Central Animal facility for providing us with the animals.
Acknowledgments
Supplemental materials may be found here: www.landesbioscience.com/journals/virulence/article/19029
Glossary
Abbreviations:
- STM
Salmonella Typhimurium
- SCV
Salmonella containing vacuole
- SPI-2
Salmonella pathogenicity island-2
- T3SS
type three secretion system
- ATR
acid tolerance response
- iNOS
inducible nitric oxide synthase
- NADPH
nicotinamide adenine dinucleotide phosphate-oxidase
- BAF
bafilomycin A1
- WT
wild-type
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Footnotes
Previously published online: www.landesbioscience.com/journals/virulence/article/19029
References
- 1.Sabbagh SC, Forest CG, Lepage C, Leclerc JM, Daigle F. So similar, yet so different: uncovering distinctive features in the genomes of Salmonella enterica serovars Typhimurium and Typhi. FEMS Microbiol Lett. 2010;305:1–13. doi: 10.1111/j.1574-6968.2010.01904.x. [DOI] [PubMed] [Google Scholar]
- 2.Andrews-Polymenis HL, Baumler AJ, McCormick BA, Fang FC. Taming the elephant: Salmonella biology, pathogenesis, and prevention. Infect Immun. 2010;78:2356–69. doi: 10.1128/IAI.00096-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rychlik I, Barrow PA. Salmonella stress management and its relevance to behaviour during intestinal colonisation and infection. FEMS Microbiol Rev. 2005;29:1021–40. doi: 10.1016/j.femsre.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 4.Foster JW, Hall HK. Adaptive acidification tolerance response of Salmonella typhimurium. J Bacteriol. 1990;172:771–8. doi: 10.1128/jb.172.2.771-778.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foster JW. Low pH adaptation and the acid tolerance response of Salmonella typhimurium. Crit Rev Microbiol. 1995;21:215–37. doi: 10.3109/10408419509113541. [DOI] [PubMed] [Google Scholar]
- 6.Foster JW. The acid tolerance response of Salmonella typhimurium involves transient synthesis of key acid shock proteins. J Bacteriol. 1993;175:1981–7. doi: 10.1128/jb.175.7.1981-1987.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rathman M, Sjaastad MD, Falkow S. Acidification of phagosomes containing Salmonella typhimurium in murine macrophages. Infect Immun. 1996;64:2765–73. doi: 10.1128/iai.64.7.2765-2773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schneider B, Gross R, Haas A. Phagosome acidification has opposite effects on intracellular survival of Bordetella pertussis and B. bronchiseptica. Infect Immun. 2000;68:7039–48. doi: 10.1128/IAI.68.12.7039-7048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huynh KK, Grinstein S. Regulation of vacuolar pH and its modulation by some microbial species. Microbiol Mol Biol Rev. 2007;71:452–62. doi: 10.1128/MMBR.00003-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chakravortty D, Hansen-Wester I, Hensel M. Salmonella pathogenicity island 2 mediates protection of intracellular Salmonella from reactive nitrogen intermediates. J Exp Med. 2002;195:1155–66. doi: 10.1084/jem.20011547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallois A, Klein JR, Allen LA, Jones BD, Nauseef WM. Salmonella pathogenicity island 2-encoded type III secretion system mediates exclusion of NADPH oxidase assembly from the phagosomal membrane. J Immunol. 2001;166:5741–8. doi: 10.4049/jimmunol.166.9.5741. [DOI] [PubMed] [Google Scholar]
- 12.Brumell JH, Grinstein S. Salmonella redirects phagosomal maturation. Curr Opin Microbiol. 2004;7:78–84. doi: 10.1016/j.mib.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 13.Groisman EA. The pleiotropic two-component regulatory system PhoP-PhoQ. J Bacteriol. 2001;183:1835–42. doi: 10.1128/JB.183.6.1835-1842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rappl C, Deiwick J, Hensel M. Acidic pH is required for the functional assembly of the type III secretion system encoded by Salmonella pathogenicity island 2. FEMS Microbiol Lett. 2003;226:363–72. doi: 10.1016/S0378-1097(03)00638-4. [DOI] [PubMed] [Google Scholar]
- 15.Chakravortty D, Rohde M, Jager L, Deiwick J, Hensel M. Formation of a novel surface structure encoded by Salmonella Pathogenicity Island 2. EMBO J. 2005;24:2043–52. doi: 10.1038/sj.emboj.7600676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu XJ, McGourty K, Liu M, Unsworth KE, Holden DW. pH sensing by intracellular Salmonella induces effector translocation. Science. 2010;328:1040–3. doi: 10.1126/science.1189000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol. 2003;47:103–18. doi: 10.1046/j.1365-2958.2003.03313.x. [DOI] [PubMed] [Google Scholar]
- 18.Nikolaus T, Deiwick J, Rappl C, Freeman JA, Schroder W, Miller SI, et al. SseBCD proteins are secreted by the type III secretion system of Salmonella pathogenicity island 2 and function as a translocon. J Bacteriol. 2001;183:6036–45. doi: 10.1128/JB.183.20.6036-6045.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faucher SP, Porwollik S, Dozois CM, McClelland M, Daigle F. Transcriptome of Salmonella enterica serovar Typhi within macrophages revealed through the selective capture of transcribed sequences. Proc Natl Acad Sci USA. 2006;103:1906–11. doi: 10.1073/pnas.0509183103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goetz SM, Stuck AE, Hirschi A, Gillmann G, Dapp U, Nikolaus T, et al. [Test-retest reliability of a German language multidimensional assessment instrument in elderly probands] Z Gerontol Geriatr. 2001;34:196–206. doi: 10.1007/s003910170064. [DOI] [PubMed] [Google Scholar]
- 21.Lindemann B, Nikolaus T. Outcomes of percutaneous endoscopic gastrostomy in dementia patients. J Am Geriatr Soc. 2001;49:838–9. doi: 10.1046/j.1532-5415.2001.49170.x. [DOI] [PubMed] [Google Scholar]
- 22.Seputiene V, Motiejunas D, Suziedelis K, Tomenius H, Normark S, Melefors O, et al. Molecular characterization of the acid-inducible asr gene of Escherichia coli and its role in acid stress response. J Bacteriol. 2003;185:2475–84. doi: 10.1128/JB.185.8.2475-2484.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–5. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suziedeliené E, Suziedelis K, Garbenciute V, Normark S. The acid-inducible asr gene in Escherichia coli: transcriptional control by the phoBR operon. J Bacteriol. 1999;181:2084–93. doi: 10.1128/jb.181.7.2084-2093.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshimori T, Yamamoto A, Moriyama Y, Futai M, Tashiro Y. Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J Biol Chem. 1991;266:17707–12. [PubMed] [Google Scholar]
- 26.Cirillo DM, Valdivia RH, Monack DM, Falkow S. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol Microbiol. 1998;30:175–88. doi: 10.1046/j.1365-2958.1998.01048.x. [DOI] [PubMed] [Google Scholar]
- 27.García-del Portillo F. Salmonella intracellular proliferation: where, when and how? Microbes Infect. 2001;3:1305–11. doi: 10.1016/S1286-4579(01)01491-5. [DOI] [PubMed] [Google Scholar]
- 28.Puiac S, Negrea A, Richter-Dahlfors A, Plant L, Rhen M. Omeprazole antagonizes virulence and inflammation in Salmonella enterica-infected RAW264.7 cells. Antimicrob Agents Chemother. 2009;53:2402–9. doi: 10.1128/AAC.01483-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoon H, McDermott JE, Porwollik S, McClelland M, Heffron F. Coordinated regulation of virulence during systemic infection of Salmonella enterica serovar Typhimurium. PLoS Pathog. 2009;5:e1000306. doi: 10.1371/journal.ppat.1000306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beuzón CR, Banks G, Deiwick J, Hensel M, Holden DW. pH-dependent secretion of SseB, a product of the SPI-2 type III secretion system of Salmonella typhimurium. Mol Microbiol. 1999;33:806–16. doi: 10.1046/j.1365-2958.1999.01527.x. [DOI] [PubMed] [Google Scholar]
- 31.Tobar JA, Carreno LJ, Bueno SM, Gonzalez PA, Mora JE, Quezada SA, et al. Virulent Salmonella enterica serovar typhimurium evades adaptive immunity by preventing dendritic cells from activating T cells. Infect Immun. 2006;74:6438–48. doi: 10.1128/IAI.00063-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCollister BD, Bourret TJ, Gill R, Jones-Carson J, Vazquez-Torres A. Repression of SPI2 transcription by nitric oxide-producing, IFNgamma-activated macrophages promotes maturation of Salmonella phagosomes. J Exp Med. 2005;202:625–35. doi: 10.1084/jem.20050246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uchiya K, Barbieri MA, Funato K, Shah AH, Stahl PD, Groisman EA. A Salmonella virulence protein that inhibits cellular trafficking. EMBO J. 1999;18:3924–33. doi: 10.1093/emboj/18.14.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garvis SG, Beuzon CR, Holden DW. A role for the PhoP/Q regulon in inhibition of fusion between lysosomes and Salmonella-containing vacuoles in macrophages. Cell Microbiol. 2001;3:731–44. doi: 10.1046/j.1462-5822.2001.00153.x. [DOI] [PubMed] [Google Scholar]
- 35.Audia JP, Webb CC, Foster JW. Breaking through the acid barrier: an orchestrated response to proton stress by enteric bacteria. Int J Med Microbiol. 2001;291:97–106. doi: 10.1078/1438-4221-00106. [DOI] [PubMed] [Google Scholar]
- 36.Bearson S, Bearson B, Foster JW. Acid stress responses in enterobacteria. FEMS Microbiol Lett. 1997;147:173–80. doi: 10.1111/j.1574-6968.1997.tb10238.x. [DOI] [PubMed] [Google Scholar]
- 37.Miki T, Okada N, Danbara H. Two periplasmic disulfide oxidoreductases, DsbA and SrgA, target outer membrane protein SpiA, a component of the Salmonella pathogenicity island 2 type III secretion system. J Biol Chem. 2004;279:34631–42. doi: 10.1074/jbc.M402760200. [DOI] [PubMed] [Google Scholar]
- 38.Fardini Y, Trotereau J, Bottreau E, Souchard C, Velge P, Virlogeux-Payant I. Investigation of the role of the BAM complex and SurA chaperone in outer-membrane protein biogenesis and type III secretion system expression in Salmonella. Microbiology. 2009;155:1613–22. doi: 10.1099/mic.0.025155-0. [DOI] [PubMed] [Google Scholar]
- 39.Basler HD, Bloem R, Casser HR, Gerbershagen HU, Griessinger N, Hankemeier U, et al. [A structured pain interview for geriatric patients] Schmerz. 2001;15:164–71. doi: 10.1007/s004820170018. [DOI] [PubMed] [Google Scholar]
- 40.Steele-Mortimer O. Infection of epithelial cells with Salmonella enterica. Methods Mol Biol. 2008;431:201–11. doi: 10.1007/978-1-60327-032-8_16. [DOI] [PubMed] [Google Scholar]
- 41.Eswarappa SM, Panguluri KK, Hensel M, Chakravortty D. The yejABEF operon of Salmonella confers resistance to antimicrobial peptides and contributes to its virulence. Microbiology. 2008;154:666–78. doi: 10.1099/mic.0.2007/011114-0. [DOI] [PubMed] [Google Scholar]
- 42.Freeman JA, Rappl C, Kuhle V, Hensel M, Miller SI. SpiC is required for translocation of Salmonella pathogenicity island 2 effectors and secretion of translocon proteins SseB and SseC. J Bacteriol. 2002;184:4971–80. doi: 10.1128/JB.184.18.4971-4980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nikolaus T, Baethe M. [Influenza, pneumococci, tetanus: the most important vaccinations in old age] MMW Fortschr Med. 2001;143:39–41. [PubMed] [Google Scholar]
- 44.Beuzón CR, Holden DW. Use of mixed infections with Salmonella strains to study virulence genes and their interactions in vivo. Microbes Infect. 2001;3:1345–52. doi: 10.1016/S1286-4579(01)01496-4. [DOI] [PubMed] [Google Scholar]
- 45.Valdivia RH, Falkow S. Bacterial genetics by flow cytometry: rapid isolation of Salmonella typhimurium acid-inducible promoters by differential fluorescence induction. Mol Microbiol. 1996;22:367–78. doi: 10.1046/j.1365-2958.1996.00120.x. [DOI] [PubMed] [Google Scholar]
- 46.Das P, Lahiri A, Chakravortty D. Novel role of the nitrite transporter NirC in Salmonella pathogenesis: SPI2-dependent suppression of inducible nitric oxide synthase in activated macrophages. Microbiology. 2009;155:2476–89. doi: 10.1099/mic.0.029611-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.