Abstract
A VpreB surrogate light (SL) chain was identified for the first time in a marsupial, the opossum Monodelphis domestica. Comparing the opossum VpreB to homologues from eutherian (placental mammals) and avian species supported the marsupial gene being VpreB3. VpreB3 is a protein that is not known to traffic to the cell surface as part of the pre-B cell receptor. Rather, VpreB3 associates with nascent immunoglobulin (Ig) chains in the endoplasmic reticulum. Homologues of other known SL chains VpreB1, VpreB2, and λ5, which are found in eutherian mammals, were not found in the opossum genome, nor have they been identified in the genomes of non-mammals. VpreB3 likely evolved from earlier gene duplication, independent of that which generated VpreB1 and VpreB2 in eutherians. The apparent absence of VpreB1, VpreB2, and λ5 in marsupials suggests that an extra-cellular pre-B cell receptor containing SL chains, as it has been defined in humans and mice, may be unique to eutherian mammals. In contrast, the conservation of VpreB3 in marsupials and its presence in non-mammals is consistent with previous hypotheses that it is playing a more primordial role in B cell development.
Keywords: VpreB, Evolution, Marsupial, B cell ontogeny
B-lymphocytes progress through distinct stages of gene expression and gene rearrangement during their ontogeny (Hardy et al. 1991; Li et al. 1993). Two defining steps are the sequential rearrangement of Ig heavy (H) and light (L) chain genes through the process of RAG dependent V(D)J recombination (Schatz 2004). Associated with the rearrangement of the Ig H genes and expression of H chain proteins is the expression of the SL chains VpreB and λ5, which precedes the rearrangement and expression of Ig L chain genes. These SL chains associate with the IgM H (μH) chain, which along with the CD79a and CD79b (Igα and Igβ) signaling molecules form the extra-cellular pre-B cell receptor (BCR) (Kudo and Melchers 1987; Karasuyama et al. 1990; Tsubata and Reth 1990; Shirasawa et al. 1993; Mårtensson et al. 2007). Successful expression of the pre-BCR is critical for signaling developmental progression in B cells.
Three VpreB gene lineages, VpreB1, VpreB2 and VpreB3, have been identified. Mice have all three genes, whereas humans only have VpreB1 and VpreB3. VpreB1 and VpreB2 (VpreB1/2) are closely related and used on the cell-surface as part of the pre-BCR in early developing B cells and, although both genes are present, in mice VpreB1 appears to play a dominant role (Tsubata and Reth 1990; Ohnishi and Takemori 1994; Mundt et al. 2005; Mårtensson et al. 2007). VpreB3 is a more distantly related gene whose function is less well studied. VpreB3 has been shown to associate with nascent μH chains and λ5 in the endoplasmic reticulum (ER) in mice, but not traffic to the cell surface (Ohnishi and Takemori 1994). VpreB3 has also been found in a non-mammalian vertebrate, the chicken, where it appears to participate in the retention of free Ig L chains in the ER to prevent their secretion independent of H chains (Rosnet et al. 2004). The presence of VpreB3 in birds as well as mammals have lead some investigators to speculate that it is the more ancient of the SL chains, whose primordial function may be to associate with Ig chains intra-cellularly (Vettermann and Jäck 2010).
Here we describe a VpreB homologue in a marsupial, the gray short-tailed opossum Monodelphis domestica. Marsupials are a mammalian lineage that diverged from eutherians at least 147 million years ago, and are noteworthy for giving birth to highly altricial young (reviewed in Old and Deane 2000; Bininda-Emonds et al. 2007). In general, newborn marsupials are unable to initiate B and T cell dependent immune responses until they are greater than a week old, which correlates well with the appearance of lymphocyte specific markers and gene expression in many cases (Kalmutz 1962; LaPlante et al. 1969; Rowlands et al. 1972; Parra et al. 2009; Duncan et al. 2010). As part of an ongoing study of postnatal ontogeny of the opossum immune system, we wished to identify markers of B cell development including the surrogate light chains; however, surrogate light chains have not been described previously for any marsupial species. Using the available opossum whole genome sequence (GenBank accession no. AAFR03000000; Mikkelsen et al. 2006) the identification of genes homologous to VpreB and λ5 was attempted using in silico screening methods (Altschul et al. 1990). Using mouse VpreB1, VpreB2, and λ5 was unsuccessful, although these sequences did identify previously annotated opossum Vλ and Cλ genes known to be used in the conventional Igλ repertoire (Wang et al. 2009). Mouse VpreB3, however, matched a partial gene among the unassembled opossum genome sequences (Scaffold Un.55000001-60000000). The partial VpreB gene contained a 329 bp gap at the 5’ end of the gene, which was filled by sequencing a PCR fragment spanning the gap that was amplified directly from genomic M. domestica DNA (Fig. 1; The complete gene sequence was deposited in GenBank as accession number JN863116).
Figure 1.
Alignment of deduced amino acid sequences of opossum VpreB3 with mouse VpreB1, 2, and 3, human VpreB1 and 2, rabbit VpreB3, and chicken VpreB3. Leader peptide, and the regions that correspond to FR1 through 3 and CDR1 and 2 in Ig V domains are indicated above the alignment. Residues identical to the opossum sequence are indicated by dashes; dots indicate insertions necessary for generating the alignment. Conserved cysteines are shaded while the conserved HXAC motif is highlighted in black. The mouse VpreB1, VpreB2, and VpreB3 sequences that were used to perform an in silico search of the opossum whole genome using the BLAST algorithm were Genbank accession nos. NM_016982, BC141459, and NM_009514). To complete the partial opossum VpreB gene identified, primers were designed to flank a 329 bp gap at the 5’ end of the gene (5’-AGGAGGGCCTTCTCAGGA and 5’-GCTCCTGCTCCTCTTCATTG) and a product that covered the gap was cloned and sequenced.
The VpreB gene in eutherian mammals consists of two exons that encode the leader peptide and extracellular V domain, respectively. Based on sequence similarity between the opossum gene and mouse and human VpreB, the presumptive opossum VpreB exons were identified along with the predicted mRNA splice sites (not shown). PCR primers located within the two exons were used to amplify a cDNA clone from splenic mRNA from an eight-week-old opossum. When compared to the genomic sequence, the cDNA sequence confirmed the opossum VpreB gene structure predicted from the alignments. When compared to VpreB genes from eutherians and birds the opossum gene clustered with other VpreB3 genes in a phylogenetic analysis, consistent with the opossum gene being a VpreB3 homologue (Figs. 1 and 2).
Figure 2.
Phylogenetic tree based on nucleotide alignments of VpreB1, 2, and 3 along with Vλ and Vκ genes from the species indicated. The opossum VpreB3 is bolded and boxed. Analyses were performed on nucleotide alignments using the neighbor-joining and minimal evolution methods in MEGA4 with similar results; the minimal evolution tree is shown (Tamura et al. 2007). Amino acid translations were first aligned to establish gap position and then converted back to nucleotide using the BioEdit program (Hall 1994). The GenBank accession numbers of the sequences used in the phylogenetic analysis were: Opossum VpreB3, JN863116; Human VpreB1, CR456609; Human VpreB3, NM_013378; Human VL5b, BAA20017; Human VL4c, CAA80218; Human VL9a, CAP74528; Human VL1a, BAA20004; Human VL6a, AAB33217; Chimpanzee VpreB1, NW_003458635; Chimpanzee VpreB3, NW_003458643; Rhesus Monkey VL5, AM056012; Mouse VpreB2, BC141459; Mouse VpreB1, NM_016982; Mouse VpreB3, NM_009514; Mouse VLX, AAA39169; Rat VpreB2, NM_001134788; Rat VpreB1, NM_001108845; Rat VK, EDL82784; Hamster VK, AAA82732; Rabbit VpreB2, AY351268; Rabbit VpreB3, XM_002724010; Rabbit VpreB1, AY351269; Rabbit VL2, PS0055; Rabbit VL3, PS0056; Sheep VK, S33161; Cow VpreB3, NW_003104461; Cow VpreB1, NW_003104461; Horse VK1, CAA53283; Pig VpreB3, NW_003611976; Pig VpreB1, NW_003611976; Elephant 490, NW_003573490 (1148141-1148468); Elephant 490b, NW_003573490 (3338560-3338236); Elephant 560, NW_003573560; Dog VpreB1, NC_0066083; Dog Vi930, XM_003639930; Dog Vi107, NW_003726107; Opossum VK, XP_003339882; Opossum VKII, XP_003339837; Possum VKI, AY074425; Possum VLIV, AAM09967; Opossum VLI, AF049774; Echidna VL167, AAM76525; Platypus VL1, AAO16067; Platypus VL97, AAO16074; Chicken VpreB3, XM_415223; Zebra finch VpreB3, NW_002197395; Turkey VpreB3, NW_003436164; Chicken VL1, I51216.
The translated sequence was aligned to VpreB protein sequence from other species and many key residues are conserved including cysteines (Cys) in predicted β-strands B and F, and the canonical WF/YQQ in the region corresponding to framework region (FR)-2 (Fig. 1; Williams and Barclay 1988). Also present is a conserved HXAC motif was evident in all VpreB3, providing an unusual unpaired Cys (Cys-99), which is absent from VpreB1/2 (Fig. 1). Chicken VpreB3 has been shown to form covalent heterodimers with Ig L chains, however these are unaffected by mutation of Cys-99 (Rosnet et al. 2004). Which of the three Cys in VpreB3 is forming the covalent bond with L chain in chicken has not been determined, nor has been the role of Cys-99 in any species. Such covalent binding has not been reported for mammalian VpreB3 where the interactions with μH chain and λ5 may be transient early in B cell ontogeny (Ohnishi and Takemori 1994).
The predicted opossum VpreB3 protein, like VpreB3 from other species, lacked the extended carboxy-end tail (non-immunoglobulin domain) sometimes referred to as the unique region (UR) found in VpreB1/2 (Fig. 1). In VpreB1/2 the UR does not form β-strand G, as would the region corresponding to FR4 in conventional V domains (Bankovich et al. 2007; Williams and Barclay 1988). Rather an extended UR at the amino terminus of λ5 replaces the G-strand in VpreB1/2 in the complete pre-BCR (Bankovich et al. 2007). VpreB3 in mouse also interacts with μH and of λ5 presumably in a similar interaction (Ohnishi and Takemori 1994). Although a VpreB3 crystal structure is unavailable based on the sequence it appears that this molecule would also lack β-strand G, as the protein only extends four residues past the last Cys (Fig. 1). However opossum, like chicken, appears to lack λ5; therefore it is not clear what structural interaction the VpreB3 has with other pre-BCR components.
The UR of VpreB1/2 extends beyond the Ig domain and interacts with the complementarity determining region (CDR)-3 of the μH chain V domain (Bankovich et al. 2007). Intact Pre-BCR has been shown to form dimers and this cross-linking is dependent on the UR of VpreB1/2 and λ5 (Bankovich et al. 2007). The absence of an extended UR in VpreB3 suggests it would be incapable of performing this role on the cell surface, consistent with an intracellular role, early in the trafficking of the pre-BCR (Ohnishi and Takemori 1994; Rosnet et al. 2004). This looks to be as true for opossum VpreB3 as for chicken and mouse. One of the roles of the Ig domain part of SL, independent of an extended UR, may be to interact with nascent μH chain to evaluate its compatibility with L chains (Smith and Roman 2010). It is possible that this is a primary role for VpreB3 as well.
Phylogenetic analyses of representative VpreB genes from a variety of mammalian and non-mammalian species, that included Vλ and Vκ genes, was performed (Fig. 2). Also included was VpreB3 from turkey and zebra finch, which were identified in the available whole genome assemblies for these species (Dalloul et al. 2010; Warren et al. 2010). The results of these analyses support the origin of VpreB3 independently of VpreB1/VpreB2 from different ancestral Vλ genes (Fig. 2). An early duplication gave rise to the more ancient VpreB3, which is present in birds, opossum, and eutherians. A second duplication appears to be specific to eutherians and gave rise to the VpreB1 and VpreB2 lineages. In other words, the extant VpreB genes are not monophyletic. VpreB1 and VpreB2 share 97% amino acid identity, while VpreB3 shares only 37% amino acid identity with VpreB1 and 2 (Mårtensson et al. 2007). VpreB1 and VpreB2 appear to be recent duplications occurring independently in different species (Fig. 2). Alternatively gene conversion (concerted evolution) may be homogenizing VpreB1 and VpreB2 in a species-specific manner in different eutherian lineages, creating the pattern of clades found in mice, rats and rabbits in Fig. 2. A VpreB-like gene reported from an agnathan, the sea lamprey, was also included in the initial analyses (Cannon et al. 2005). However, in phylogenetic analyses this gene fell outside the clade containing VpreB, Vλ and Vκ, and is likely not directly ancestral to the VpreB genes found in amniotes (not shown).
The VpreB genes appear to have evolved from Vλ genes and the mouse VpreB1 and VpreB2 and human VpreB1 genes remain linked to the Igλ locus (Kudo and Melchers 1987). Chicken and human VpreB3 genes are also linked to the Igλ locus, however the mouse VpreB3 gene is on chromosome 10, non-syntenic to Igλ (Rosnet et al. 1999; Rosnet et al. 2004). A search of the region on opossum chromosome 3, where Igλ is located (Deakin et al. 2006), did not uncover any genes resembling SL chain homologues (not shown). Unfortunately the VpreB3 gene was found amongst the unassembled sequences in the opossum genome database and, therefore, it is not currently known where VpreB3 is physically located the opossum, although it is clear from the mouse genome that its linkage to Igλ is not required for its function. As with opossum and chicken there was no evidence of VpreB1 or VpreB2 in the turkey and zebra finch genomes (not shown).
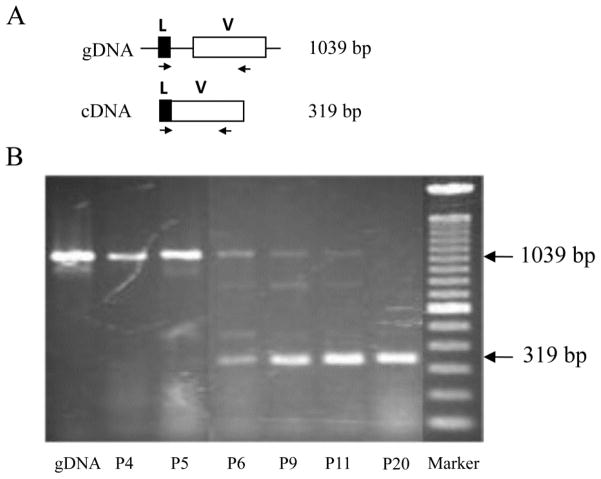
To investigate transcription of the VpreB3 gene in postnatal opossums, RT-PCR was performed using RNA isolated from neonatal animals. The earliest age which VpreB3 transcripts could be detected was postnatal day 6 (Fig. 3). This is following the detection of CD79a, CD79b, and IgH transcription in newborn opossums, but preceding Ig L chain gene rearrangement and transcription (not shown; XW and RDM manuscript in review). These results are consistent with marsupials not having mature B cells capable of generating antibody responses until greater than one week of age (Kalmutz 1962; Rowlands et al. 1972; Old and Deane 2000; Duncan et al. 2010). Furthermore, the expression of VpreB3 just prior to Ig light chain gene rearrangement and transcription in the opossum would be consistent with its apparent function in chickens, where it associates with nascent light chains in the ER.
Figure 3.
A, Diagram of the VpreB3 gene (top) and cDNA (bottom). Leader region and V domain are represented by solid and open boxes, respectively. Location of the primers used for RT-PCR, complementary to leader exon and V exon are indicated as arrows. B, Detection of VpreB3 transcripts by RT-PCR in postnatal opossums. To investigate the timing of transcription of the opossum VpreB3 gene RT-PCR was performed on total RNA isolated from newborn opossums at postnatal (P) days 4, 5, 6, 9, 11, and 20. gDNA is a control for the 1039 bp product produced when contaminating genomic DNA is present. Primers used were 5’-AGGAGGGCCTTCTCAGGA and 5-GCTCCTGCTCCTCTTCATTG.
Homologues of the VpreB1, VpreB2, and λ5 genes were not identified in the opossum genome and, to our knowledge, have only been found in eutherian mammals. VpreB1 and/or VpreB2, and λ5, therefore, may be specific to eutherians; marsupials, birds, and other lineages may use an alternative pre-BCR form that lacks these surrogate light chains on the cell surface. If this is the case then the cell-surface pre-BCR, as it has been defined in eutherian mammals such as mice and humans, is both a unique and significant adaptation given its requirement for normal B cell development. In mice pre-BCR containing H chain and the CD79a and CD79b signaling molecules, but lacking one or both SL or conventional L chains can be expressed, signal phosphorylation and Ca2+ release, and mediate allelic exclusion (Shimizu et al. 2002; Schuh et al. 2003; Su et al. 2003). There is, however, severe impairment of B cell development in SL knockout mice, demonstrating the clear important role these chains play in eutherian B cell development (Kitamura et al. 1992; Ehlich et al. 1993; Pelanda et al.1996).
One hypothesis is that a functional cell-surface pre-BCR lacking SL or conventional L chains is present in birds and non-eutherian mammals and may be an ancestral form of the pre-BCR. However, there is the possibility of alternatives to the conventional SL. Rearrangement of the Igκ L chain genes prior to IgH rearrangement and expression has been proposed as an explanation for the development of some B cells in SL knockout mice (Ehlich et al. 1993; Pelanda et al 1996). Germline Vk and JCk transcripts encoding proteins that functionally substitute for VpreB1/2 and λ5 have been described in mouse and may be a partial explanation for why B cell development is not completely blocked in VpreB1/2 and λ5 deficient mice (Francés et al. 1994; Rangel et al. 2005). Indeed, an Igκ transgene, under some circumstances, can rescue impaired B cell development in SL knockout mice (Pelanda et al. 1996). Opossum does have the Igκ L chain, however the presence of germline Igκ transcripts has yet to be explored in this species (Miller et al. 1999).
In conclusion, VpreB3 is an ancient gene whose function in B cell development has been retained at least among the amniotes (Rosnet et al. 2004; Vettermann and Jäck 2010). VpreB3 is the only of the Igλ derived SL that may be universally present in the amniotes. Further structural analyses of VpreB3 interactions with other chains in the BCR complex are needed.
Acknowledgments
This article was made possible in part by support from a National Institutes of Health grant no. IP20RR18754 from the Institutional Development Award program and National Science Foundation award IOS-0641382.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bankovich AJ, Raunser S, Juo ZS, Walz T, Davis MM, Garcia KC. Structural insight into pre-B cell receptor function. Science. 2007;316:291–294. doi: 10.1126/science.1139412. [DOI] [PubMed] [Google Scholar]
- Bininda-Emonds OR, Cardillo M, Jones KE, MacPhee RD, Beck RM, Grenyer R, Price SA, Vos RA, Gittleman JL, Purvis A. The delayed rise of present-day mammals. Nature. 2007;446:507–512. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- Cannon JP, Haire RN, Pancer Z, Mueller MG, Skapura D, Cooper MD, Litman GW. Variable domains and a VpreB-like molecule are present in a jawless vertebrate. Immunogenetics. 2005;56:924–929. doi: 10.1007/s00251-004-0766-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalloul RA, et al. Multi-platform next-generation sequencing of the domestic turkey (Meleagris gallopavo): genome assembly and analysis. PLoS Biol. 2010;8:e1000475. doi: 10.1371/journal.pbio.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deakin JE, Olp JJ, Graves JAM, Miller RD. Physical mapping of immunoglobulin loci IGH@, IGK@, and IGL@in the opossum (Monodelphis domestica) Cytogen Genome Res. 2006;114:94H. doi: 10.1159/000091942. [DOI] [PubMed] [Google Scholar]
- Duncan L, Webster K, Gupta V, Nair S, Deane E. Molecular characterisation of the CD79a and CD79b subunits of the B cell receptor complex in the gray short-tailed opossum (Monodelphis domestica) and tammar wallaby (Macropus eugenii): Delayed B cell immunocompetence in marsupial neonates. Vet Immunol Immunopath. 2010;136:235–247. doi: 10.1016/j.vetimm.2010.03.013. [DOI] [PubMed] [Google Scholar]
- Ehlich A, Schaal S, Gu H, Kitamura D, Müller W, Rajewsky K. Immunoglobulin heavy and light chain genes rearrange independently at early stages of B cell development. Cell. 1993;72:695–704. doi: 10.1016/0092-8674(93)90398-a. [DOI] [PubMed] [Google Scholar]
- Francés V, Pandrau-Garcia D, Guret C, Ho S, Wang Z, Duvert V, Saeland S, Martinez-Valdez H. A surrogate 15 kDa JC kappa protein is expressed in combination with mu heavy chain by human B cell precursors. EMBO J. 1994;13:5937–5943. doi: 10.1002/j.1460-2075.1994.tb06939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nuc Acids Symp Ser. 1994;41:95–98. [Google Scholar]
- Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med. 1991;173:1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmutz SE. Antibody production in the opossum embryo. Nature. 1962;193:851–853. doi: 10.1038/193851a0. [DOI] [PubMed] [Google Scholar]
- Karasuyama H, Kudo A, Melchers F. The proteins encoded by the VpreB and lambda 5 pre-B cell-specific genes can associate with each other and with mu heavy chain. J Exp Med. 1990;172:969–972. doi: 10.1084/jem.172.3.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura D, Kudo A, Schaal S, Müller W, Melchers F, Rajewsky K. A critical role of lambda 5 protein in B cell development. Cell. 1992;69:823–831. doi: 10.1016/0092-8674(92)90293-l. [DOI] [PubMed] [Google Scholar]
- Kudo A, Melchers F. A second gene, VpreB in the lambda 5 locus of the mouse, which appears to be selectively expressed in pre-B lymphocytes. EMBO J. 1987;6:2267–2272. doi: 10.1002/j.1460-2075.1987.tb02500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPlante ES, Burrell R, Watne AL, Taylor DL, Zimmermann B. Skin allograft studies in the pouch young of the opossum. Transplantation. 1969;7:67–72. doi: 10.1097/00007890-196901000-00006. [DOI] [PubMed] [Google Scholar]
- Li YS, Hayakawa K, Hardy RR. The regulated expression of B lineage associated genes during B cell differentiation in bone marrow and fetal liver. J Exp Med. 1993;178:951–960. doi: 10.1084/jem.178.3.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mårtensson I-L, Keenan R, Licence S. The pre-B-cell receptor. Curr Opin Immunol. 2007;19:137–142. doi: 10.1016/j.coi.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TJ, Wakefield MJ, Aken B, Amemiya CT, Chang JL, Duke S, Garber M, Gentles AJ, Goodstadt L, Heger A, et al. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature. 2007;447:167–177. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- Miller RD, Bergemann ER, Rosenberg GH. Marsupial light chains: Ig kappa with four V families in the opossum Monodelphis domestica. Immunogenetics. 1999;50:329–335. doi: 10.1007/s002510050609. [DOI] [PubMed] [Google Scholar]
- Mundt C, Licence S, Maxwell G, Melchers F, Mårtensson I-L. Only VpreB1, but not VpreB2, is expressed at levels which allow normal development of B cells. Int Immunol. 2006;18:163–172. doi: 10.1093/intimm/dxh359. [DOI] [PubMed] [Google Scholar]
- Ohnishi K, Takemori TJ. Molecular components and assembly of μ•surrogate light chain complexes in pre-B cell lines. J Biol Chem. 1994;269:28347–28353. [PubMed] [Google Scholar]
- Old JM, Deane EM. Development of the immune system and immunological protection in marsupial pouch young. Dev Comp Immunol. 2000;24:445–454. doi: 10.1016/s0145-305x(00)00008-2. [DOI] [PubMed] [Google Scholar]
- Parra ZE, Baker ML, Lopez AM, Trujillo J, Volpe JM, Miller RD. TCR mu recombination and transcription relative to the conventional TCR during postnatal development in opossums. J Immunol. 2009;182:154–163. doi: 10.4049/jimmunol.182.1.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelanda R, Schaal S, Torres R, Rajewsky K. A prematurely expressed Ig kappa transgene, but not a V kappa J kappa gene segment targeted into the Ig kappa locus, can rescue B cell development in lambda 5-deficient mice. Immunity. 1996;5:229–239. doi: 10.1016/s1074-7613(00)80318-0. [DOI] [PubMed] [Google Scholar]
- Rangel R, McKeller MR, Sims-Mourtada J, Kashi C, Cain K, Wieder ED, Molldrem JJ, Pham LV, Ford RJ, Yotnda P, Guret C, Francés V, Martinez-Valdez H. Assembly of the kappa preB receptor requires a V kappa-like protein encoded by a germline transcript. J Biol Chem. 2005;280:17807–17814. doi: 10.1074/jbc.M409479200. [DOI] [PubMed] [Google Scholar]
- Rosnet O, Blanco-Betancourt C, Grivel K, Richter K, Schiff C. Binding of free immunoglobulin light chains to VpreB3 inhibits their maturation and secretion in chicken B cells. J Biol Chem. 2004;279:10228–10236. doi: 10.1074/jbc.M312169-A200. [DOI] [PubMed] [Google Scholar]
- Rosnet O, Mattei M, Delattre O, Schiff C. VPREB3: cDNA characterization and expression in human and chromosome mapping in human and mouse. Cytogenet Cell Genet. 1999;87:205–208. doi: 10.1159/000015468. [DOI] [PubMed] [Google Scholar]
- Rowlands DT, Blakeslee D, Lin HH. The early immune response and immunoglobulins of opossum embryos. J Immunol. 1972;108:941–946. [PubMed] [Google Scholar]
- Schatz DG. V(D)J recombination. Immunol Rev. 2004;200:5–11. doi: 10.1111/j.0105-2896.2004.00173.x. [DOI] [PubMed] [Google Scholar]
- Schuh W, Meister S, Roth E, Jäck H-M. Cutting edge: signaling and cell surface expression of a mu H chain in the absence of lambda 5: a paradigm revisited. J Immunol. 2003;171:3343–3347. doi: 10.4049/jimmunol.171.7.3343. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Mundt C, Licence S, Melchers F, Mårtensson I-L. VpreB1/VpreB2/λ5 Triple-Deficient Mice Show Impaired B Cell Development but Functional Allelic Exclusion of the IgH Locus. J Immunol. 2002;168:6286–6293. doi: 10.4049/jimmunol.168.12.6286. [DOI] [PubMed] [Google Scholar]
- Shirasawa T, Ohnishi K, Hagiwara S, Shigemoto K, Takebe Y, Rajewsky K, Takemori T. A novel gene product associated with mu chains in immature B cells. EMBO J. 1993;12:1827–1834. doi: 10.1002/j.1460-2075.1993.tb05831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BP, Roman CAJ. The unique and immunoglobulin-like regions of surrogate light chain component λ5 differentially interrogate immunoglobulin heavy-chain structure. Mol Immunol. 2010;47:1195–1206. doi: 10.1016/j.molimm.2010.01.002. [DOI] [PubMed] [Google Scholar]
- Su YW, Flemming A, Wossning T, Hobeika E, Reth M, Jumaa H. Identification of a pre-BCR lacking surrogate light chain. J Exp Med. 2003;198:1699–1706. doi: 10.1084/jem.20031428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tsubata T, Reth M. The products of pre-B cell-specific genes (lambda 5 and VpreB) and the immunoglobulin mu chain form a complex that is transported onto the cell surface. J Exp Med. 1990;172:973–976. doi: 10.1084/jem.172.3.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Olp JJ, Miller RD. On the genomics of immunoglobulins in the gray, short-tailed opossum Monodelphis domestica. Immunogenetics. 2009;61:581–596. doi: 10.1007/s00251-009-0385-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren WC, et al. The genome of a songbird. Nature. 2010;464:757–762. doi: 10.1038/nature08819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AF, Barclay AN. The immunoglobulin superfamily-domains for cell surface recognition. Ann Rev Immunol. 1988;6:381–405. doi: 10.1146/annurev.iy.06.040188.002121. [DOI] [PubMed] [Google Scholar]
- Vettermann C, Jäck H-M. The pre-B cell receptor: turning autoreactivity into self-defense. Trends Immunol. 2010;31:176–183. doi: 10.1016/j.it.2010.02.004. [DOI] [PubMed] [Google Scholar]