Abstract
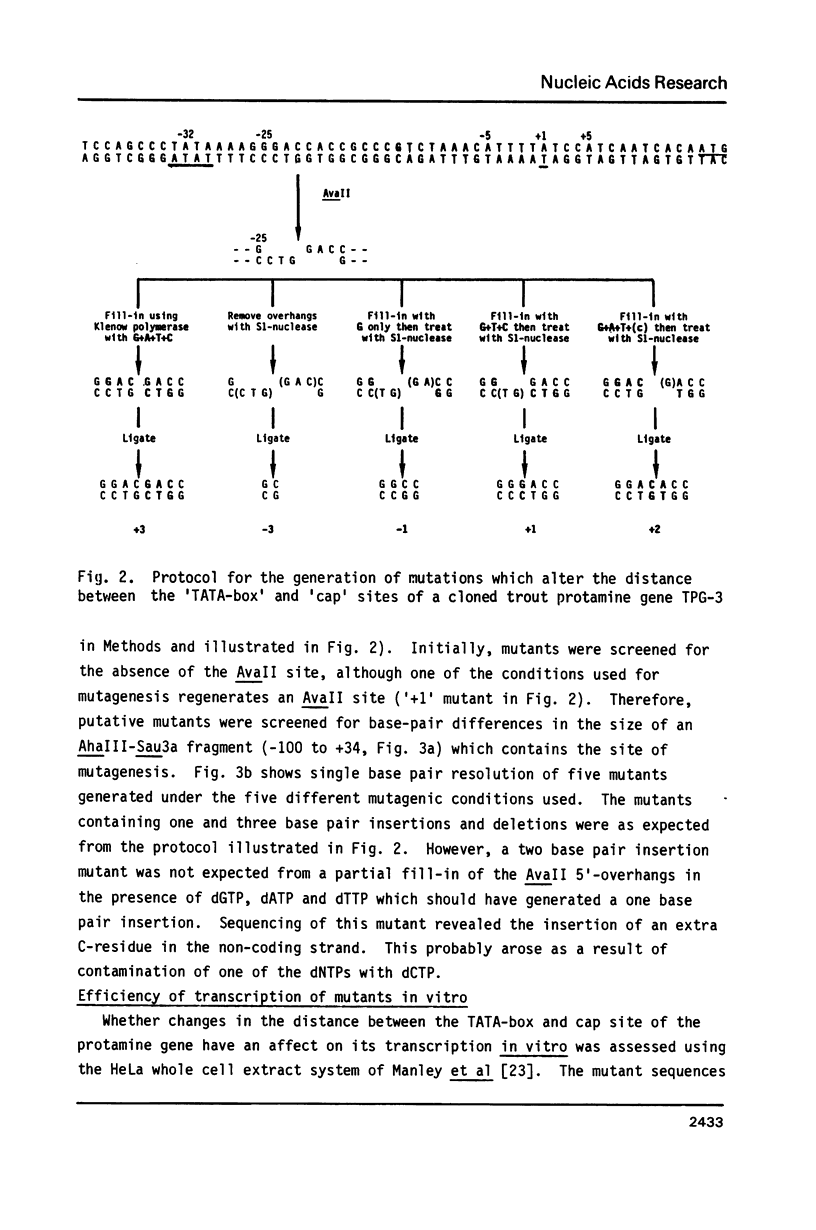
We have studied how small changes in the distance between the TATA-box and cap site affect transcription of a eukaryotic gene in vitro and in vivo. The trout protamine gene TPG-3 [Gregory et al. (1982) Nucl. Acids Res. 10, 7581-7592] is a good model for such a study as it has (i) a consensus TATA-box 32 base pairs (bp) upstream from an A-residue which is the natural cap site (designated +1) (ii) two further A-residues at -5 and +5, providing alternative transcriptional start sites which are in significantly different sequence environments and (iii) a unique AvaII restriction site immediately downstream from the TATA-box which is ideal for the insertion or deletion of up to 3bp. Transcripts of the wild type and mutant genes were generated in vitro using a HeLa whole cell extract or 'in vivo' by transient expression following their transfection into HeLa cells. These 'spacer' mutations did not affect the efficiency of transcription of the gene in vitro but they did affect the selection of transcriptional start site both in vitro and 'in vivo'. Analysis of 5'-ends by S1-mapping and primer extension showed that the A-residue(s) selected are those which, by insertion or deletion, come to lie on the same face of the DNA double helix as the TATA-box, although the DNA sequence in the immediate vicinity of the potential start sites influences their utilisation. Comparison of the TPG-3 wild type transcripts in these experimental systems with natural mRNA suggests that cap site selection is more stringent in the developing trout testis.
Full text
PDF












Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aoyama T., Takanami M. Essential structure of E. coli promoter II. Effect of the sequences around the RNA start point on promoter function. Nucleic Acids Res. 1985 Jun 11;13(11):4085–4096. doi: 10.1093/nar/13.11.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoist C., Chambon P. In vivo sequence requirements of the SV40 early promotor region. Nature. 1981 Mar 26;290(5804):304–310. doi: 10.1038/290304a0. [DOI] [PubMed] [Google Scholar]
- Chambon P., Dierich A., Gaub M. P., Jakowlev S., Jongstra J., Krust A., LePennec J. P., Oudet P., Reudelhuber T. Promoter elements of genes coding for proteins and modulation of transcription by estrogens and progesterone. Recent Prog Horm Res. 1984;40:1–42. doi: 10.1016/b978-0-12-571140-1.50005-0. [DOI] [PubMed] [Google Scholar]
- Concino M. F., Lee R. F., Merryweather J. P., Weinmann R. The adenovirus major late promoter TATA box and initiation site are both necessary for transcription in vitro. Nucleic Acids Res. 1984 Oct 11;12(19):7423–7433. doi: 10.1093/nar/12.19.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corden J., Wasylyk B., Buchwalder A., Sassone-Corsi P., Kedinger C., Chambon P. Promoter sequences of eukaryotic protein-coding genes. Science. 1980 Sep 19;209(4463):1406–1414. doi: 10.1126/science.6251548. [DOI] [PubMed] [Google Scholar]
- Davison B. L., Egly J. M., Mulvihill E. R., Chambon P. Formation of stable preinitiation complexes between eukaryotic class B transcription factors and promoter sequences. Nature. 1983 Feb 24;301(5902):680–686. doi: 10.1038/301680a0. [DOI] [PubMed] [Google Scholar]
- Dierks P., van Ooyen A., Cochran M. D., Dobkin C., Reiser J., Weissmann C. Three regions upstream from the cap site are required for efficient and accurate transcription of the rabbit beta-globin gene in mouse 3T6 cells. Cell. 1983 Mar;32(3):695–706. doi: 10.1016/0092-8674(83)90055-7. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. Control of eukaryotic messenger RNA synthesis by sequence-specific DNA-binding proteins. 1985 Aug 29-Sep 4Nature. 316(6031):774–778. doi: 10.1038/316774a0. [DOI] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Fire A., Samuels M., Sharp P. A. Interactions between RNA polymerase II, factors, and template leading to accurate transcription. J Biol Chem. 1984 Feb 25;259(4):2509–2516. [PubMed] [Google Scholar]
- Ghosh P. K., Reddy V. B., Piatak M., Lebowitz P., Weissman S. M. Determination of RNA sequences by primer directed synthesis and sequencing of their cDNA transcripts. Methods Enzymol. 1980;65(1):580–595. doi: 10.1016/s0076-6879(80)65061-7. [DOI] [PubMed] [Google Scholar]
- Green M. R., Roeder R. G. Definition of a novel promoter for the major adenovirus-associated virus mRNA. Cell. 1980 Nov;22(1 Pt 1):231–242. doi: 10.1016/0092-8674(80)90171-3. [DOI] [PubMed] [Google Scholar]
- Gregory S. P., Dillon N. O., Butterworth P. H. The localisation of the 5'-termini of in vivo transcripts of a cloned rainbow trout protamine gene. Nucleic Acids Res. 1982 Dec 11;10(23):7581–7592. doi: 10.1093/nar/10.23.7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory S. P., Dillon N. O., Butterworth P. H. The localisation of the 5'-termini of in vivo transcripts of a cloned rainbow trout protamine gene. Nucleic Acids Res. 1982 Dec 11;10(23):7581–7592. doi: 10.1093/nar/10.23.7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosschedl R., Birnstiel M. L. Identification of regulatory sequences in the prelude sequences of an H2A histone gene by the study of specific deletion mutants in vivo. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1432–1436. doi: 10.1073/pnas.77.3.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosveld G. C., Shewmaker C. K., Jat P., Flavell R. A. Localization of DNA sequences necessary for transcription of the rabbit beta-globin gene in vitro. Cell. 1981 Jul;25(1):215–226. doi: 10.1016/0092-8674(81)90246-4. [DOI] [PubMed] [Google Scholar]
- Grosveld G. C., de Boer E., Shewmaker C. K., Flavell R. A. DNA sequences necessary for transcription of the rabbit beta-globin gene in vivo. Nature. 1982 Jan 14;295(5845):120–126. doi: 10.1038/295120a0. [DOI] [PubMed] [Google Scholar]
- Haynes J., Weissmann C. Constitutive, long-term production of human interferons by hamster cells containing multiple copies of a cloned interferon gene. Nucleic Acids Res. 1983 Feb 11;11(3):687–706. doi: 10.1093/nar/11.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel C., Irminger J. C., Bucher P., Birnstiel M. L. Sea urchin histone mRNA termini are located in gene regions downstream from putative regulatory sequences. Nature. 1980 May 15;285(5761):147–151. doi: 10.1038/285147a0. [DOI] [PubMed] [Google Scholar]
- Hirose S., Takeuchi K., Hori H., Hirose T., Inayama S., Suzuki Y. Contact points between transcription machinery and the fibroin gene promoter deduced by functional tests of single-base substitution mutants. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1394–1397. doi: 10.1073/pnas.81.5.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose S., Takeuchi K., Suzuki Y. In vitro characterization of the fibroin gene promoter by the use of single-base substitution mutants. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7258–7262. doi: 10.1073/pnas.79.23.7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer J., Shen C. K., Maniatis T. The chromosomal arrangement of human alpha-like globin genes: sequence homology and alpha-globin gene deletions. Cell. 1980 May;20(1):119–130. doi: 10.1016/0092-8674(80)90240-8. [DOI] [PubMed] [Google Scholar]
- Lusky M., Botchan M. Inhibition of SV40 replication in simian cells by specific pBR322 DNA sequences. Nature. 1981 Sep 3;293(5827):79–81. doi: 10.1038/293079a0. [DOI] [PubMed] [Google Scholar]
- Manley J. L., Fire A., Cano A., Sharp P. A., Gefter M. L. DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3855–3859. doi: 10.1073/pnas.77.7.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Stefano J. E., Gralla J. D. Spacer mutations in the lac ps promoter. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1069–1072. doi: 10.1073/pnas.79.4.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga K., Hirose S., Suzuki Y. In monkey COS cells only the TATA box and the cap site region are required for faithful and efficient initiation of the fibroin gene transcription. Nucleic Acids Res. 1984 Feb 10;12(3):1543–1558. doi: 10.1093/nar/12.3.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twigg A. J., Sherratt D. Trans-complementable copy-number mutants of plasmid ColE1. Nature. 1980 Jan 10;283(5743):216–218. doi: 10.1038/283216a0. [DOI] [PubMed] [Google Scholar]