Abstract
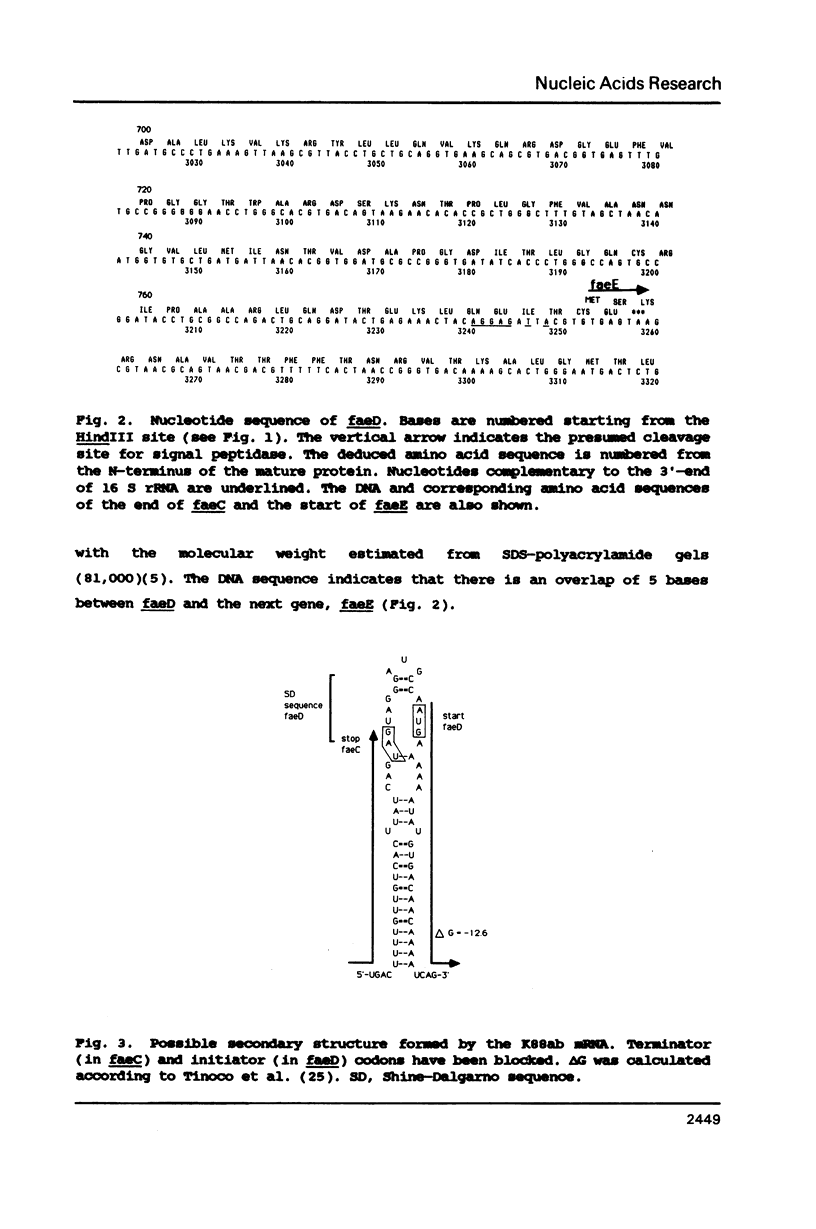
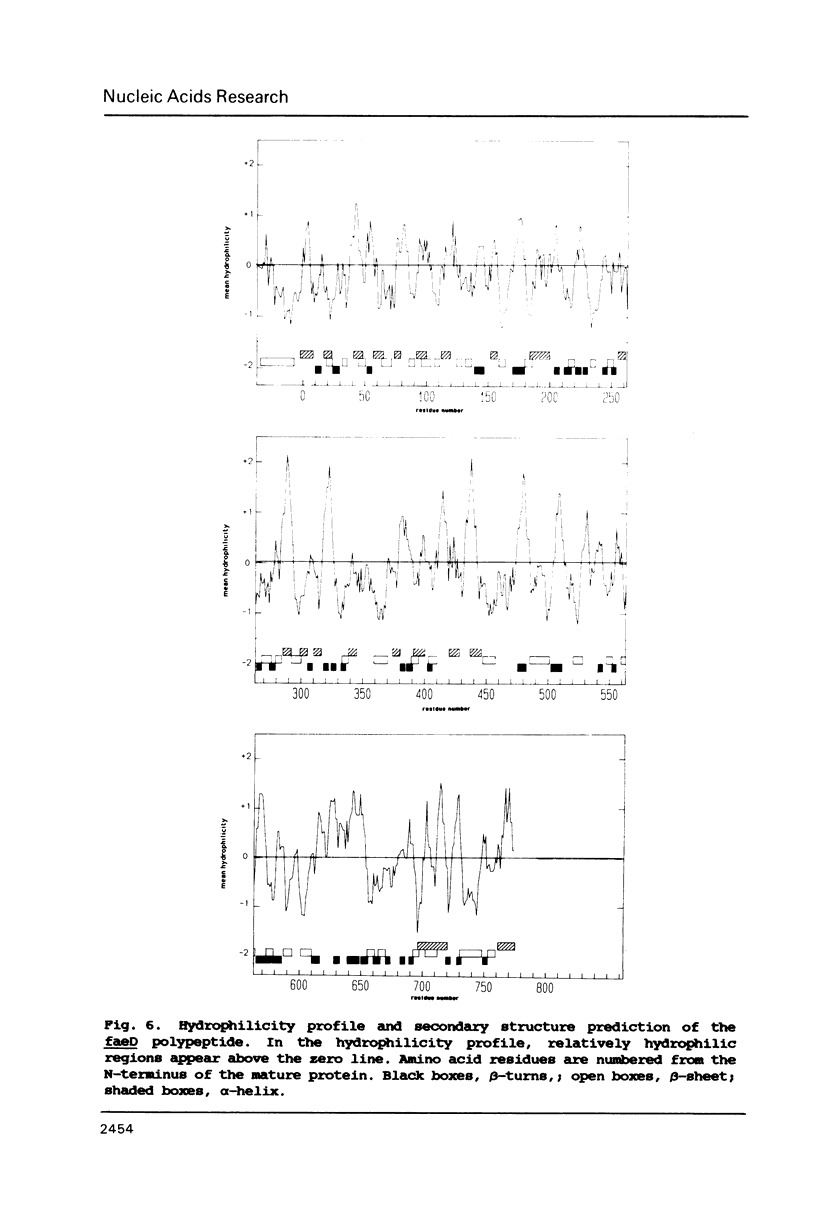
The nucleotide sequence of the faeD gene of Escherichia coli and the amino acid sequence of its product is presented. The faeD product is an outer membrane protein required for transport of K88ab fimbrial subunits across the outer membrane. The protein is synthesized as a precursor containing a signal peptide, and the tentative mature protein comprises 777 amino acid residues. The distribution of amino acids in the faeD protein is similar to that of other outer membrane proteins; showing a fairly even distribution of charged residues and the absence of extensive hydrophobic stretches. Secondary structure predictions revealed a region of 250 amino acid residues which might be embedded in the outer membrane. The 5'-end of faeD is located within a region showing dyad symmetry. This region serves to couple translation of faeD to the translation of the gene preceding it (faeC). The 3'-end of faeD shows an overlap of 5 bases with the next gene (faeE).
Full text
PDF












Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- Hall M. N., Gabay J., Débarbouillé M., Schwartz M. A role for mRNA secondary structure in the control of translation initiation. Nature. 1982 Feb 18;295(5850):616–618. doi: 10.1038/295616a0. [DOI] [PubMed] [Google Scholar]
- Kadonaga J. T., Gautier A. E., Straus D. R., Charles A. D., Edge M. D., Knowles J. R. The role of the beta-lactamase signal sequence in the secretion of proteins by Escherichia coli. J Biol Chem. 1984 Feb 25;259(4):2149–2154. [PubMed] [Google Scholar]
- Levitt M. A simplified representation of protein conformations for rapid simulation of protein folding. J Mol Biol. 1976 Jun 14;104(1):59–107. doi: 10.1016/0022-2836(76)90004-8. [DOI] [PubMed] [Google Scholar]
- McKenney K., Shimatake H., Court D., Schmeissner U., Brady C., Rosenberg M. A system to study promoter and terminator signals recognized by Escherichia coli RNA polymerase. Gene Amplif Anal. 1981;2:383–415. [PubMed] [Google Scholar]
- Mooi F. R., Harms N., Bakker D., de Graaf F. K. Organization and expression of genes involved in the production of the K88ab antigen. Infect Immun. 1981 Jun;32(3):1155–1163. doi: 10.1128/iai.32.3.1155-1163.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooi F. R., Wijfjes A., de Graaf F. K. Identification and characterization of precursors in the biosynthesis of the K88ab fimbria of Escherichia coli. J Bacteriol. 1983 Apr;154(1):41–49. doi: 10.1128/jb.154.1.41-49.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooi F. R., Wouters C., Wijfjes A., de Graaf F. K. Construction and characterization of mutants impaired in the biosynthesis of the K88ab antigen. J Bacteriol. 1982 May;150(2):512–521. doi: 10.1128/jb.150.2.512-521.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooi F. R., de Graaf F. K. Molecular biology of fimbriae of enterotoxigenic Escherichia coli. Curr Top Microbiol Immunol. 1985;118:119–138. doi: 10.1007/978-3-642-70586-1_7. [DOI] [PubMed] [Google Scholar]
- Mooi F. R., van Buuren M., Koopman G., Roosendaal B., de Graaf F. K. K88ab gene of Escherichia coli encodes a fimbria-like protein distinct from the K88ab fimbrial adhesin. J Bacteriol. 1984 Aug;159(2):482–487. doi: 10.1128/jb.159.2.482-487.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H., Vaara M. Molecular basis of bacterial outer membrane permeability. Microbiol Rev. 1985 Mar;49(1):1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Callaghan C. H., Morris A., Kirby S. M., Shingler A. H. Novel method for detection of beta-lactamases by using a chromogenic cephalosporin substrate. Antimicrob Agents Chemother. 1972 Apr;1(4):283–288. doi: 10.1128/aac.1.4.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheim D. S., Yanofsky C. Translational coupling during expression of the tryptophan operon of Escherichia coli. Genetics. 1980 Aug;95(4):785–795. doi: 10.1093/genetics/95.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudega B., Mooi F. R., de Graaf F. K. Excretion of proteins by gram-negative bacteria: export of bacteriocins and fimbrial proteins by Escherichia coli. Antonie Van Leeuwenhoek. 1984;50(5-6):569–584. doi: 10.1007/BF02386227. [DOI] [PubMed] [Google Scholar]
- Paul C., Rosenbusch J. P. Folding patterns of porin and bacteriorhodopsin. EMBO J. 1985 Jun;4(6):1593–1597. doi: 10.1002/j.1460-2075.1985.tb03822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roggenkamp R., Dargatz H., Hollenberg C. P. Precursor of beta-lactamase is enzymatically inactive. Accumulation of the preprotein in Saccharomyces cerevisiae. J Biol Chem. 1985 Feb 10;260(3):1508–1512. [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schümperli D., McKenney K., Sobieski D. A., Rosenberg M. Translational coupling at an intercistronic boundary of the Escherichia coli galactose operon. Cell. 1982 Oct;30(3):865–871. doi: 10.1016/0092-8674(82)90291-4. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Uhlin B. E., Båga M., Göransson M., Lindberg F. P., Lund B., Norgren M., Normark S. Genes determining adhesin formation in uropathogenic Escherichia coli. Curr Top Microbiol Immunol. 1985;118:163–178. doi: 10.1007/978-3-642-70586-1_9. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem. 1983 Jun 1;133(1):17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]