Abstract
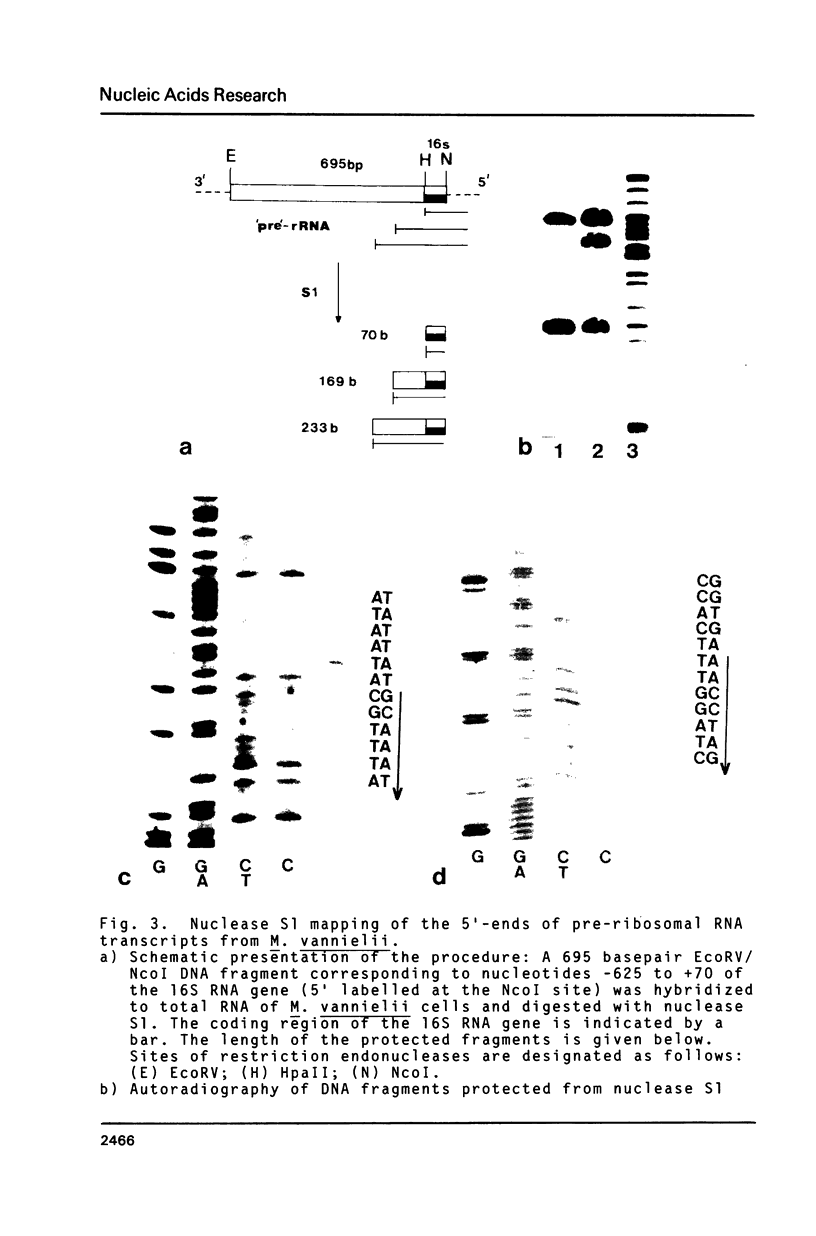
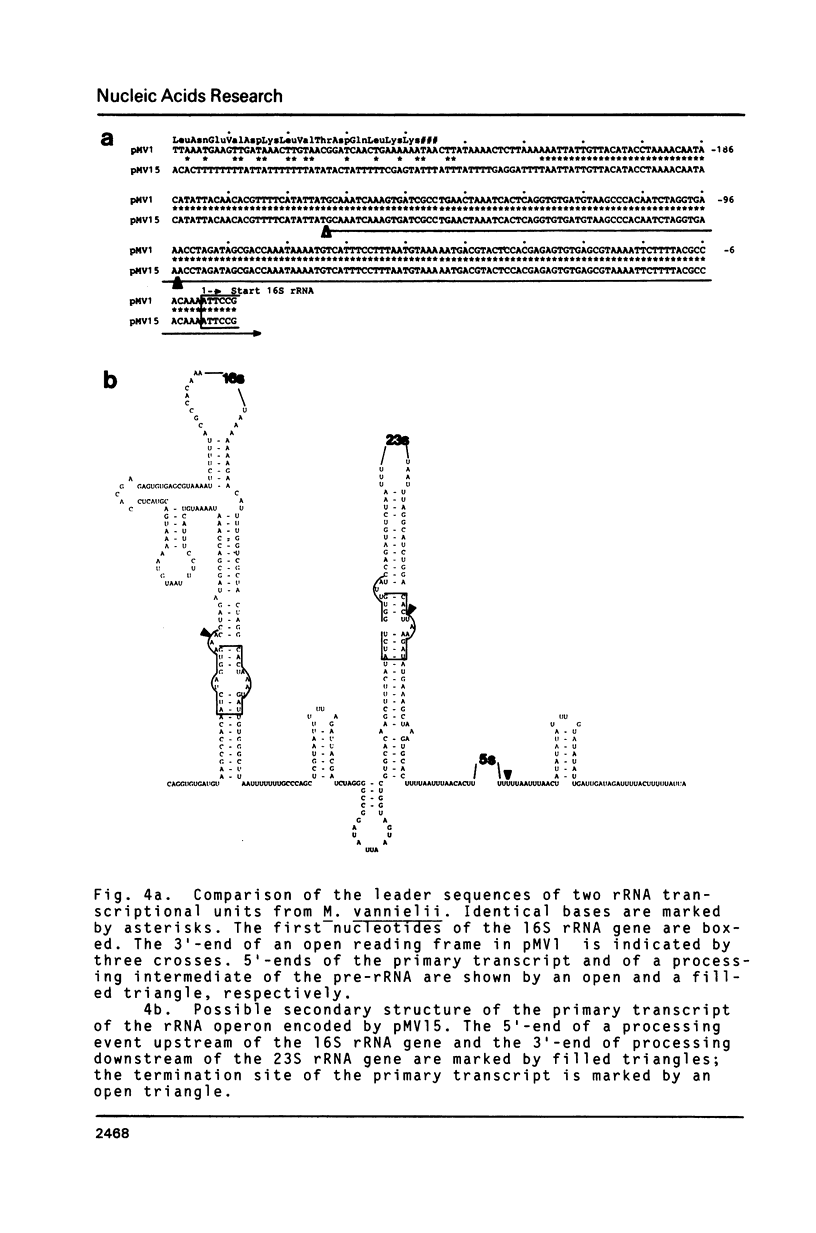
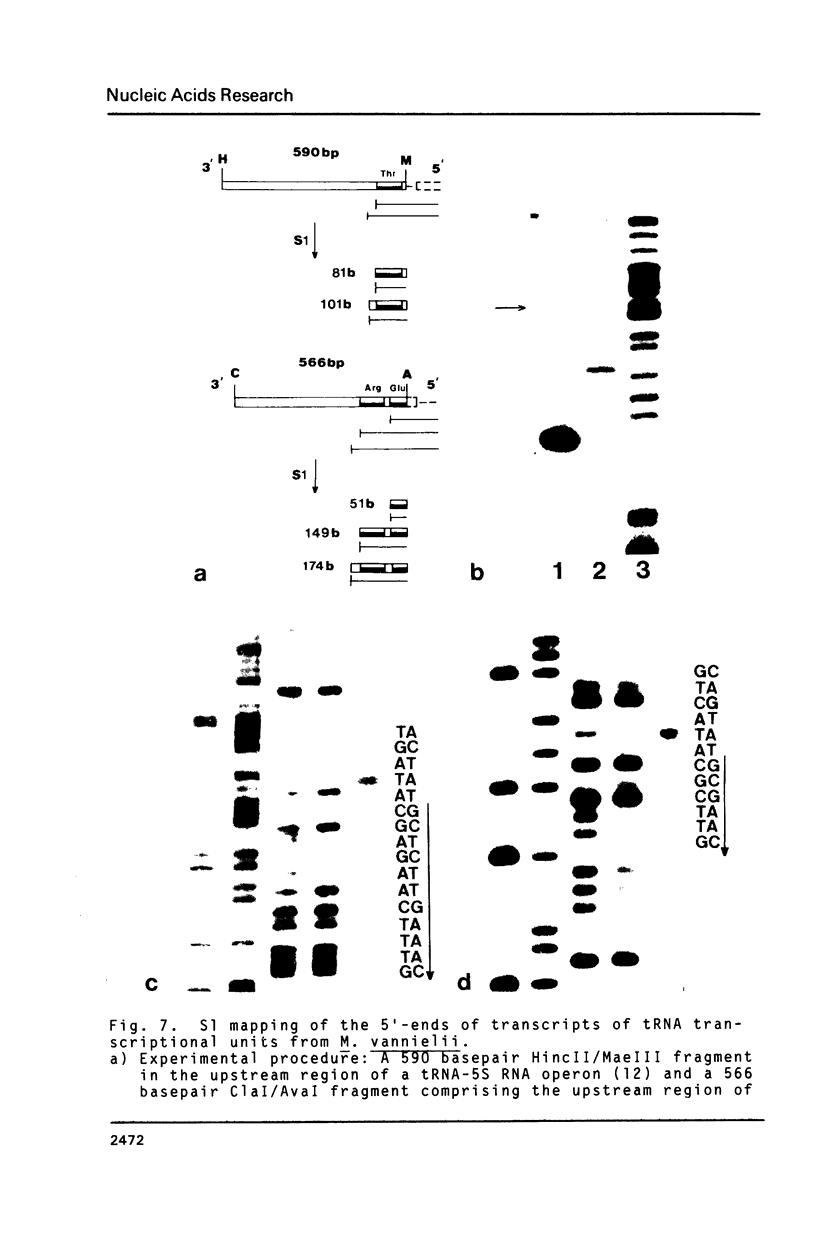
A previous survey of upstream sequences of tRNA genes from the archaebacterium Methanococcus vannielii has revealed that there are two boxes of sequence homology: A box "A" of about 20 conserved nucleotides at a distance of 30 to 49 basepairs upstream from the gene and a box "B" 18 to 19 nucleotides downstream from box "A" (Wich, G., Sibold, L., and Böck, A. (1985) System. Appl. Microbiol. (in press). Nuclease S1 mapping experiments were carried out with two of these tRNA transcriptional units and with a ribosomal RNA operon, to determine whether these consensus sequences have a function in the initiation of transcription. Use was made of the fact that cells from Methanococcus accumulate primary transcript and processing intermediates of ribosomal RNA under conditions of protein synthesis inhibition. The following results were obtained: (i) Transcription in all three systems starts at the G within the conserved trinucleotide TGC of box "B". Since the box "B" motif, 5'TGCaagT3', also occurs at the site of transcription initiation of protein encoding genes, both in methanogenic and halophilic organisms, it appears to constitute a frequently used transcription start signal within these archaebacterial groups. (ii) The box "A" motif occurs with constant spacing, relative to box "B", in all 10 tRNA and ribosomal RNA transcriptional units investigated from Methanococcus. Since it is not present in the leader region of genes coding for proteins, it seems to function as a specific element which is required for the expression of genes for stable RNA. (iii) Termination of transcription of the ribosomal RNA operon from Methanococcus occurs at a distinct T within an oligo-T stretch immediately downstream from the 3'-terminal 5S RNA gene. This signal occurs in all 3'-flanking regions of transcriptional units for stable RNA from the Methanococcus strains studied. Termination signals for stable RNA genes in Methanococcus appear to be similar with those of stable RNA genes in eukaryotes. (iv) By nuclease S1 mapping a recognition site was identified for a processing enzyme involved in the maturation of preribosomal RNA.
Full text
PDF

















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abelson J. RNA processing and the intervening sequence problem. Annu Rev Biochem. 1979;48:1035–1069. doi: 10.1146/annurev.bi.48.070179.005131. [DOI] [PubMed] [Google Scholar]
- Beauclerk A. A., Hummel H., Holmes D. J., Böck A., Cundliffe E. Studies of the GTPase domain of archaebacterial ribosomes. Eur J Biochem. 1985 Sep 2;151(2):245–255. doi: 10.1111/j.1432-1033.1985.tb09095.x. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Betlach M., Friedman J., Boyer H. W., Pfeifer F. Characterization of a halobacterial gene affecting bacterio-opsin gene expression. Nucleic Acids Res. 1984 Oct 25;12(20):7949–7959. doi: 10.1093/nar/12.20.7949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollschweiler C., Kühn R., Klein A. Non-repetitive AT-rich sequences are found in intergenic regions of Methanococcus voltae DNA. EMBO J. 1985 Mar;4(3):805–809. doi: 10.1002/j.1460-2075.1985.tb03701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cue D., Beckler G. S., Reeve J. N., Konisky J. Structure and sequence divergence of two archaebacterial genes. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4207–4211. doi: 10.1073/pnas.82.12.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassarma S., Rajbhandary U. L., Khorana H. G. Bacterio-opsin mRNA in wild-type and bacterio-opsin-deficient Halobacterium halobium strains. Proc Natl Acad Sci U S A. 1984 Jan;81(1):125–129. doi: 10.1073/pnas.81.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diver W. P., Grinsted J., Fritzinger D. C., Brown N. L., Altenbuchner J., Rogowsky P., Schmitt R. DNA sequences of and complementation by the tnpR genes of Tn21, Tn501 and Tn1721. Mol Gen Genet. 1983;191(2):189–193. doi: 10.1007/BF00334812. [DOI] [PubMed] [Google Scholar]
- Dunn R., McCoy J., Simsek M., Majumdar A., Chang S. H., Rajbhandary U. L., Khorana H. G. The bacteriorhodopsin gene. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6744–6748. doi: 10.1073/pnas.78.11.6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant J. A. Stringent control in E. coli. Annu Rev Genet. 1979;13:393–415. doi: 10.1146/annurev.ge.13.120179.002141. [DOI] [PubMed] [Google Scholar]
- Gegenheimer P., Watson N., Apirion D. Multiple pathways for primary processing of ribosomal RNA in Escherichia coli. J Biol Chem. 1977 May 10;252(9):3064–3073. [PubMed] [Google Scholar]
- Gray C. P., Sommer R., Polke C., Beck E., Schaller H. Structure of the orgin of DNA replication of bacteriophage fd. Proc Natl Acad Sci U S A. 1978 Jan;75(1):50–53. doi: 10.1073/pnas.75.1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M. W., Doolittle W. F. Has the endosymbiont hypothesis been proven? Microbiol Rev. 1982 Mar;46(1):1–42. doi: 10.1128/mr.46.1.1-42.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton P. T., Reeve J. N. Structure of genes and an insertion element in the methane producing archaebacterium Methanobrevibacter smithii. Mol Gen Genet. 1985;200(1):47–59. doi: 10.1007/BF00383311. [DOI] [PubMed] [Google Scholar]
- Hecht N. B., Woese C. R. Separation of bacterial ribosomal ribonucleic acid from its macromolecular precursors by polyacrylamide gel electrophoresis. J Bacteriol. 1968 Mar;95(3):986–990. doi: 10.1128/jb.95.3.986-990.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel C., Irminger J. C., Bucher P., Birnstiel M. L. Sea urchin histone mRNA termini are located in gene regions downstream from putative regulatory sequences. Nature. 1980 May 15;285(5761):147–151. doi: 10.1038/285147a0. [DOI] [PubMed] [Google Scholar]
- Huet J., Schnabel R., Sentenac A., Zillig W. Archaebacteria and eukaryotes possess DNA-dependent RNA polymerases of a common type. EMBO J. 1983;2(8):1291–1294. doi: 10.1002/j.1460-2075.1983.tb01583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui I., Dennis P. P. Characterization of the ribosomal RNA gene clusters in Halobacterium cutirubrum. J Biol Chem. 1985 Jan 25;260(2):899–906. [PubMed] [Google Scholar]
- Mankin A. S., Teterina N. L., Rubtsov P. M., Baratova L. A., Kagramanova V. K. Putative promoter region of rRNA operon from archaebacterium Halobacterium halobium. Nucleic Acids Res. 1984 Aug 24;12(16):6537–6546. doi: 10.1093/nar/12.16.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Moss B. 5' end labeling of RNA with capping and methylating enzymes. Gene Amplif Anal. 1981;2:253–266. [PubMed] [Google Scholar]
- Müller B., Allmansberger R., Klein A. Termination of a transcription unit comprising highly expressed genes in the archaebacterium Methanococcus voltae. Nucleic Acids Res. 1985 Sep 25;13(18):6439–6445. doi: 10.1093/nar/13.18.6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace N. R. Structure and synthesis of the ribosomal ribonucleic acid of prokaryotes. Bacteriol Rev. 1973 Dec;37(4):562–603. doi: 10.1128/br.37.4.562-603.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Venkatesan S., Moss B. In vitro transcription of the inverted terminal repetition of the vaccinia virus genome: correspondence of initiation and cap sites. J Virol. 1981 Feb;37(2):738–747. doi: 10.1128/jvi.37.2.738-747.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver R. F., Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979 Nov 10;7(5):1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wich G., Jarsch M., Böck A. Apparent operon for a 5S ribosomal RNA gene and for tRNA genes in the archaebacterium Methanococcus vannielii. Mol Gen Genet. 1984;196(1):146–151. doi: 10.1007/BF00334107. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Fox G. E. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci U S A. 1977 Nov;74(11):5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer H. A., Gilbert S. F., Nomura M. DNA sequences of promoter regions for rRNA operons rrnE and rrnA in E. coli. Cell. 1979 May;17(1):201–209. doi: 10.1016/0092-8674(79)90308-8. [DOI] [PubMed] [Google Scholar]









