Abstract
During Epithelial to Mesenchymal Transition (EMT), cells modulate expression of proteins resulting in loss of apical-basal polarity. Effectors of this EMT switch target the polarity protein Crumbs3a, a small transmembrane protein that is essential for generation of the apical membrane and tight junctions of mammalian epithelial cells. We previously showed that the Crumbs3 gene is a direct target of transcriptional regulation by Snail, a potent inducer of EMT. However, Snail has also been shown to have multiple non-transcriptional roles, including regulation of cell adhesion, proliferation and survival. Using SNAP-tag labeling, we determined that cell surface Crumbs3a has a half-life of approximately 3 hours and that this cell surface half-life is significantly reduced when EMT is induced by Snail. We further observe that Snail induces differential glycosylation of Crumbs3a, including sialylation, suggesting a mechanism by which Crumbs3a may be destabilized. These results indicate that Crumbs3a is a post-translational target of Snail, in addition to being a transcriptional target. We conclude that Snail’s ability to post-translationally modify and destabilize Crumbs3a augments the depolarizing process of EMT.
Keywords: Crumbs, Snail, EMT, apical-basal, epithelial, polarity, glycosylation, sialylation
Introduction
Properly functioning epithelial tissues require that cells establish and maintain segregated plasma membrane regions (1, 2). These regions include an apical or luminal membrane, and a basolateral membrane separated by the gating function of tight junctions in vertebrates. The resulting apical-basal polarity allows asymmetric trafficking of receptors, positioning of channels and release of secretagogues, which is the basis for mature epithelial cell function. Yet there is intrinsic fluidity in the control of this cell polarity program, illustrated by dismantling of apical-basal polarity, cell migration, and re-establishment of cell apical-basal polarity during normal development or tissue healing following injury in a process known as Epithelial to Mesenchymal Transition (EMT) (3, 4). Control of the cell polarity program can also be maladaptively usurped, as occurs in malignancy (5–8).
Establishing proper epithelial cell apical-basal polarity and function requires Crumbs, an evolutionarily conserved apical transmembrane protein. Crumbs expression is tightly regulated such that too little or too much Crumbs both result in disruption of apical-basal cell polarity and tissue architecture (9–14). Excessive Crumbs induces expansion of the apical membrane and cell proliferation leading to organ enlargement. In agreement with these results, Crumbs was recently shown to be an upstream effector of the Hippo-YAP pathway, involved in cell proliferation and organ size (15). Conversely, lack of Crumbs results in the absence of epithelial organ formation in Drosophila and zebrafish, as well as in the loss of tight junction and primary cilia formation in mammalian epithelial cells in culture (16). Additionally, loss of Crumbs disrupts tight junction formation, apical-basal polarity, and contact-inhibited growth in an in vivo tumorigenic mouse model (17). Thus, understanding how epithelial cells control trafficking and localization of Crumbs is central to understanding control of apical-basal polarity.
The dominant Crumbs homolog in mammalian epithelial cells is Crumbs3 (10, 18). Alternate splicing of the mammalian Crumbs3 transcript results in production of two Crumbs3 isoforms, Crumbs3a and Crumbs3b, which encode divergent intracellular C-termini of lengths 20 and 23 amino acids respectively (19). Crumbs3a contains the canonical, evolutionarily conserved intracellular region shared by most Crumbs homologs. Crumbs3a interacts with PALS1 via the former’s C-terminal PDZ binding domain, resulting in formation of the apical polarity complex Crumbs3a-PALS1-PATJ, which localizes to apical membranes and tight junctions. This protein complex interacts with a second key polarity complex, Par6-Par3-atypical PKC. The direct interaction of these two protein complexes is crucial for generation and maintenance of tight junctions and apical-basal polarity in epithelial cells (2). These complexes also interact with a third basolateral polarity complex, Lgl-Dlg-Scribble (20–22). Mechanisms directed at destabilizing these interactions are necessary for EMT (23–25).
Most investigations into mechanisms controlling the epithelial cell polarity program in mammalian cells have focused on transcriptional regulation of proteins (26). Snail is a well-described and potent transcriptional suppressor of several apical-basal polarity genes, including E-cadherin and Crumbs3 (27–29). However, such an all-or-none approach to control of polarity protein expression inadequately explains the spectrum of partial and transient depolarization seen in processes such as tubulogenesis and wound healing (30, 31). Moreover, expression of both Snail and Crb3a is detected in breast cancer cell lines MCF7 and SKBR3 (29, 32–34) suggesting that non-transcriptional mechanisms are also involved in regulating the cell polarity program. As Snail has recently been shown to have multiple non-transcriptional roles including those as an inducer of cell survival and cell movement (35–37), we investigated whether Snail has additional roles in regulating Crumbs beyond transcriptional suppression.
We previously showed that Snail potently suppresses transcription of the Crumbs3 gene (29). However, transcriptional repression of Crumbs3 gene cannot be the sole mechanism of control of this apical polarity protein, since there are cell lines which co-express Crumbs3a and Snail (29, 32–34). Indeed, relatively little is known about how cells control polarity during partial EMT required during processes such as wound healing.
Here, we extend our earlier work with a new study that reveals a novel, post-transcriptional relationship between Snail and Crumbs3a. Namely, we show that when Crumbs3a is exogenously expressed in the presence of Snail, the gel mobility of Crumbs3a is altered suggesting that Snail exerts post-transcriptional effects on Crumbs3a as well. In this set of experiments, we explore the etiology of this mobility shift and examine the effect of Snail on intracellular trafficking of exogenously expressed Crumbs3a. We show that although Crumbs3a still traffics correctly to the apical cell surface, Snail significantly destabilizes cell surface Crumbs3a and alters its glycosylation. We propose that Snail induces post-translational modification and accelerates the degradation of the Crumbs3a polypeptide. In conjunction with its established role as a direct repressor of Crumbs transcription, the modulation of Crumbs3a post-translational modifications by Snail seems to further contribute to the loss of apical-basal polarity observed during EMT.
Results
Crumbs3a dynamics in polarized MDCK monolayers
We first characterized the trafficking and stability of Crumbs3a (Crb3a) in polarized Madin-Darby canine kidney (MDCK) cells at steady state. Attempts to study trafficking of endogenous Crb3a using conventional techniques were hampered by two factors. First, Crb3a exhibits minimal biotinylation likely due to its small extracellular domain, which contains only two lysines. Second, the entire mature protein contains no cysteines or methionines, the latter due to cleavage of the N-terminal signal peptide region. Thus, endogenous Crb3a is not amenable to traditional trafficking methods, including cell surface biotinylation and 35S-methionine/cysteine metabolic labeling.
These limitations necessitated our use of exogenously expressed epitope-tagged Crb3a for trafficking experiments. By using mammalian expression vectors with minimal promoters, we achieved protein expression levels where proper apical-basal polarity is maintained. Our laboratory’s previous experience studying trafficking of Crb3a during the genesis of a central lumen in 3D cyst assay suggests similar trafficking for endogenous Crb3a and exogenously expressed GFP-Crb3a (38). In our experience the presence of an extracellular epitope near the N-terminus of the mature protein does not seem to qualitatively alter Crb3a’s trafficking. Thus, we posit that the cell dynamics of extracellular epitope-tagged Crb3a expressed stably at near-endogenous levels reflects those of endogenous Crb3a in polarized MDCK cells at steady state.
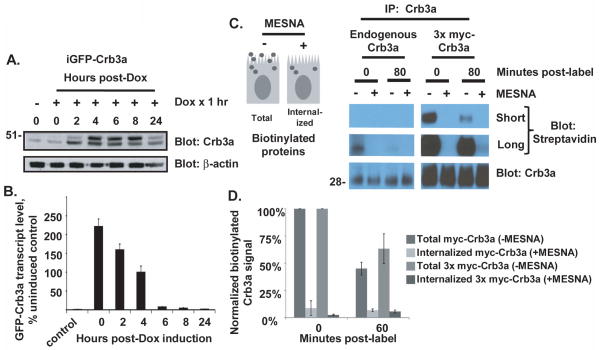
We first set out to determine the half-life of total cell Crb3a by performing a nonradioactive pulse-chase experiment using inducible GFP-Crb3a (iGFP-Crb3a) as shown in Figures 1A and 1B. After 1 hour of treatment with 50 ng/ml of doxycycline, transcript levels for iGFP-Crb3a were maximally induced and declined following doxycycline washout. The induced protein was maximally expressed between 6–8 hours following doxycycline pulse but fell to near uninduced level by 24 hours indicating that several half-lives had occurred within this interval.
Figure 1. Crumbs3a dynamics in polarized MDCK monolayers.
A) Time course showing induced GFP-Crb3a protein levels in MDCK cells following treatment with 50 ng/ml doxycycline for 1 hour. Cell lysates were produced at the indicated times post-treatment and subjected to immunoblot with the indicated antibodies. The left lane shows uninduced expression; actin is included as a loading control. B) Time course for doxycycline-induced GFP-Crb3a mRNA levels harvested from cells treated in parallel to (A), as detected by qRT-PCR. Transcript levels are normalized to GAPDH and shown relative to uninduced control levels. The experiment was performed in triplicate; averaged levels are shown with standard deviations. C) and D) Cell surface Crb3a internalization assays using cell surface biotinylation (see Methods). Residual cell surface biotin is cleaved by treatment with MESNA prior to cell lysis to reveal internalized pool of biotinylated proteins (see schematic in C). C) Endogenous Crb3a and 3x myc-Crb3a were isolated from parental (left immunoblot) and 3x myc-Crb3a (right immunoblot) cells at indicated time points and assayed for biotinylation. D) Graph of Crb3a internalization assay using 1x myc-Crb3a and 3x myc-Crb3a cells, performed as in (C) except that the biotinylated Crb3a signal intensities were normalized to myc immunoblot signal. Averaged levels obtained from two experiments are shown with standard deviations.
We then investigated the resident time of Crb3a at the apical cell surface of polarized MDCK cells at steady state. We performed cell surface biotinylation and uptake studies on both endogenous and epitope-tagged Crumbs3a as shown in Figures 1C and 1D. Endogenous Crb3a was poorly biotinylated, presumably related to the few potential biotinylation sites within a small extracellular region. Addition of a 1x-myc and a 3x-myc extracellular tag increased the number of extracellular lysine residues potentially available for conjugation with biotin from 2 to 4 and 8, respectively. As shown in Figure 1C, 80 minutes following cell surface biotinylation, the residual pools of both biotinylated cell surface (−MESNA) endogenous and 3x myc-Crb3a were markedly reduced. Further, minimal internalized biotinylated protein was detected by immunoblot following stripping of residual cell surface biotin (+MESNA). We repeated the experiment at an earlier time point to detect internalized Crb3a, with both 1x myc-Crb3a and 3x myc-Crb3a as shown in Figure 1D. At 60 minutes, 45% (SD±5%) of myc-Crb3a and 63% (SD±13%) of 3x myc-Crb3a remain, suggesting a half-life of cell surface Crb3a on the order of one hour. Yet, minimal internalized myc-tagged Crb3a is detected. These results suggest that both endogenous and exogenous Crb3a exhibit similar cell surface kinetics; both rapidly disappear from the cell surface and are metabolized.
Characterization of SNAP-Crumb3a MDCK cells
In order to more accurately characterize Crb3a’s dynamics at the apical cell surface, we developed a system to quantitatively analyze Crb3a trafficking. We produced an MDCK II cell line that stably expresses Crb3a fused with an extracellular SNAP-tag (SNAP-Crb3a, depicted in Figure 2A) at near endogenous Crb3a levels. We assert that there are at least two reasons why the SNAP-tag labeling approach may be more representative of endogenous Crb3a trafficking. The first is that the SNAP-tag labeling reaction results in a single covalent modification as opposed to biotin labeling of several lysines, which may or may not be available for labeling at any given time. The second is that detection of SNAP-Crb3a involves direct quantification of fluorescence labeling of protein present in cell lysates, as opposed to indirect quantification following potentially inefficient immunoprecipitation of Crumbs3a as performed in biotinylation experiments. Thus, we produced an MDCK II cell pool stably expressing SNAP-Crb3a.
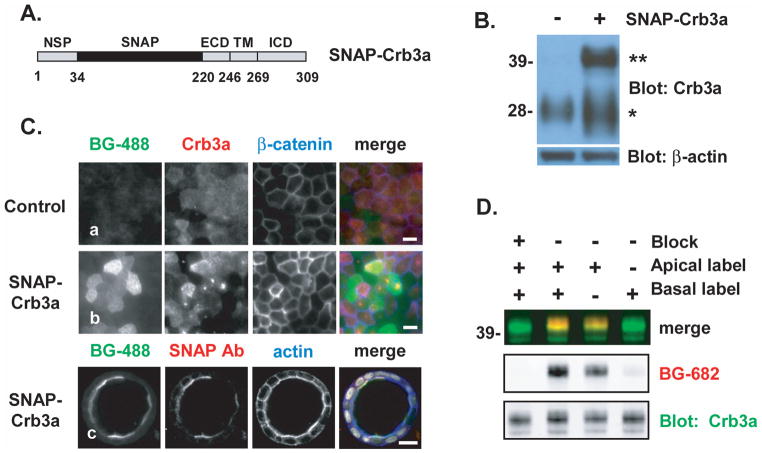
Figure 2. Characterization of SNAP-Crumb3a MDCK cells.
A) Schematic representation of the SNAP-Crb3a construct; the SNAP-tag lies within the proximal native extracellular domain (ECD) just distal to the natural signal peptide (NSP); the transmembrane domain (TM) and intracellular domain (ICD) are also labeled. Numbers refer to residues. B) Immunoblot showing relative expression levels of endogenous Crb3a (indicated by *, left lane) in parental MDCK cells versus stably expressed exogenous SNAP-Crb3a (indicated by **, right lane) in a FACS-enriched MDCK cell pool. Actin is included as a loading control. C) Immunofluorescence imaging of MDCK cells in 2D monolayers (rows a and b, epifluorescence, white bars 10 μm) and 3D cyst (row c, confocal, white bar 20 μm) revealing localization and specificity of in-culture labeling of SNAP-Crb3a with BG-488. Cells were fixed following labeling, permeabilized and immunostained with indicated antibodies and phalloidin (to indicate F-actin). D) Immunoblot of in-culture labeled cell surface SNAP-Crb3a in polarized MDCK cells grown on transwell filters to allow selective exposure to BG-682 on the apical or basal cell surfaces. Cell surface SNAP block was performed prior to labeling with BG-682 as indicated; cells were lysed immediately following labeling. The experiment was performed twice in duplicate and representative images are shown.
We then created a pool enriched for cells expressing SNAP-Crb3a near endogenous Crb3a levels. We labeled cell surface SNAP-Crb3a with a fluorescently tagged cell-impermeable SNAP-tag substrate (benzylguanine-488, BG-488), and subjected this labeled cell pool to fluorescence-activated cell sorting (FACS). Immunoblotting of cell lysates from the enriched pool with anti-Crb3a antibody (Figure 2B) confirmed that these cells express total cell SNAP-Crb3a at levels comparable to endogenous Crb3a. The SNAP-Crb3a fusion protein (marked as **) migrates around 42 kDa on SDS-PAGE, higher than the anticipated mass of 33 kDa. Endogenous Crb3a (marked as *) migrates around 28 kDa while the calculated protein mass is predicted to be 13 kDa; we previously demonstrated that this difference is due to glycosylation of the endogenous polypeptide (10). The lower bands visible in the SNAP-Crb3a lane are not visible by immunoblotting with anti-SNAP antibody (not shown), suggesting breakdown products.
We confirmed proper trafficking of SNAP-Crb3a by labeling cells grown in 2-dimensional (2D) and 3-dimensional (3D) culture with the SNAP-tag reagent prior to fixation. As shown in Figure 2C, control cells (panel a) exposed to BG-488 in culture show minimal background fluorescent signal, while SNAP-Crb3a cells exhibit co-localization of the 488 signal with antibodies recognizing Crb3a and SNAP epitopes at the apical cell surface in both 2D monolayer (panel b) and 3D cysts (panel c). The difference seen in 2D between BG-488 and Crb3a immunostaining reflects BG-488’s specific labeling of cell surface SNAP-tagged Crb3a exclusively, as opposed to Crb3a immunostaining which detects both intracellular and cell surface Crb3a. The mid-cell punctate Crb3a staining in 2D corresponds to primary cilia, also visualized with SNAP-tag labeling but not seen in these images. These results confirm both proper trafficking of SNAP-Crb3a to the apical cell surface and specificity of SNAP-Crb3a labeling. The ability to label SNAP-Crb3a at the apical surface of MDCK cells in 3D cysts in culture with benzylguanine reagents is seen only in developing cysts. By day 7, in-culture labeling with these reagents is unsuccessful despite the confirmed presence of apical SNAP-Crb3a by antibody staining after permeabilization, presumably due to exclusion by mature lateral and tight junctions.
Specificity of in-culture labeling of SNAP-Crb3a and localization of SNAP-Crb3a to the apical surface in polarized MDCK cells were further evaluated by exposing either the apical or basolateral surface of cells grown on transwell filters to BG-682. By immunoblotting cell lysates produced immediately following in-culture SNAP-tag labeling, we show that the vast majority of labeled SNAP-Crb3a is located on the apical rather than the basolateral cell surface as seen in Figure 2D. Furthermore, pre-incubating cells with a cell impermeable SNAP-blocking reagent abrogates SNAP-tag labeling. In combination, these results both confirm proper apical localization of SNAP-Crb3a and specificity of the in-culture cell surface SNAP-tag labeling reaction. This novel SNAP-Crb3a MDCK cell pool was then used for subsequent trafficking experiments.
Determination of SNAP-Crumbs3a cell surface kinetics
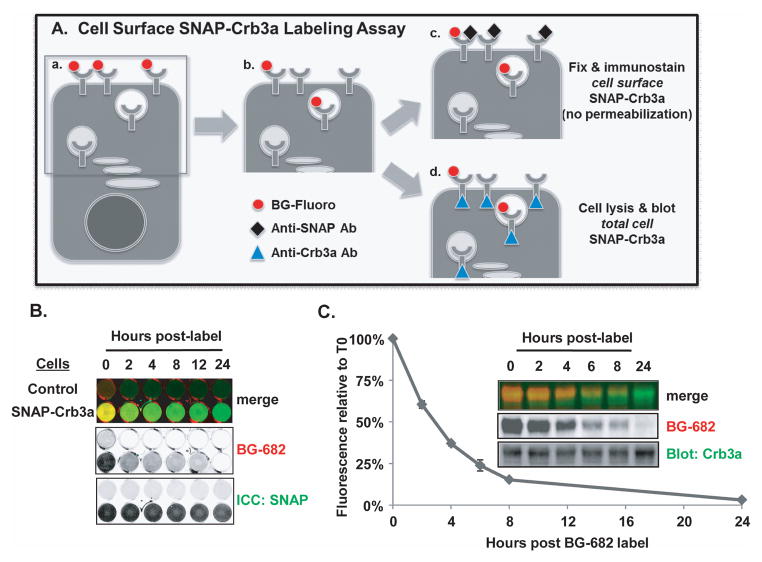
We determined the kinetics of cell surface SNAP-Crb3a by performing nonradioactive pulse-chase labeling assays in polarized MDCK cells at steady state. As depicted in Figure 3A, cell surface SNAP-Crb3a is labeled with cell-impermeable benzylguanine (BG), a SNAP-tag substrate, and then chased for the times indicated and processed as described in the figure legend. Figure 3B shows a cell surface labeling experiment, performed as illustrated in Figure 3A(c). Cells were grown in 24 well tissue culture dishes, labeled with BG-682 at time 0, and fixed at the indicated time points. Immunocytochemistry of non-permeabilized cells with anti-SNAP antibody to label total cell surface SNAP-Crb3a was performed and cells were imaged in situ. By 2 hours post-labeling, the fluorescent signal of residual labeled SNAP-Crb3a is significantly decreased relative to total cell surface SNAP-Crb3a. These results suggest that the half-life of cell surface SNAP-Crb3a is similar to the results obtained by surface biotinylation shown in Figure 1C.
Figure 3. Determination of SNAP-Crumbs3a cell surface kinetics.
A) Schematic representation of cell surface SNAP-Crb3a labeling time course assay performed in 24 well tissue culture plates: (a) cell surface SNAP-Crb3a is labeled in culture with cell impermeable benzylguanine (BG) tethered to a fluorescent label; (b) cells are incubated for indicated durations, and processed by either (c) fixation and immunostaining (no permeabilization) with anti-SNAP antibody to assess residual quantity of BG-labeled SNAP-Crb3a relative to cell surface SNAP-Crb3a (labeled plus newly delivered unlabeled), or (d) cell lysis and immunoblot with anti-Crb3a antibody to assess residual quantity of BG-labeled SNAP-Crb3a relative to total cell SNAP-Crb3a. B) In situ immunocytochemistry (ICC) showing half-life of cell surface SNAP-Crb3a relative to cell surface SNAP-Crb3a as described in 3A(c). Cells expressing control vector (top row) or SNAP-Crb3a were grown in 24 well tissue culture plates, labeled in culture with BG-682 then fixed and immunostained with anti-SNAP antibody without permeabilization. Fluorescence images of cells in wells were taken in situ. C) Quantitative half-life of cell surface SNAP-Crb3a assessed by immunoblot of cell lysates produced as described in 3A(d). Residual labeled SNAP-Crb3a fluorescence is shown relative to total cell SNAP-Crb3a as detected by immunoblot with anti-Crb3a antibody. The experiment was performed four times with representative images shown; the graph shows mean experimental time points with standard errors.
In Figure 3C, we quantitatively characterize the half-life of cell surface SNAP-Crb3a relative to the total cell pool of SNAP-Crb3a by immunoblot, as illustrated in Figure 3A(d). Cell surface SNAP-Crb3a was labeled with BG-682 at time 0, and cell were lysed at the indicated time points and subjected to standard protein electrophoresis. The BG-682 fluorescence signal of SNAP-Crb3a was normalized to Crb3a signal. Quantitative analysis reveals a mean half-life for cell surface SNAP-Crb3a of 2.78 hours (SEM±0.12). The mean half-life was statistically equivalent for cells grown on transwell filters, 2.87 hours (SEM±0.21).
Closer examination of the immunoblots in Figure 3C reveals that the SNAP-Crb3a migrates as a doublet, which is also seen with immunoblots of endogenous and exogenous GFP- and myc-tagged Crb3a. It is possible that these bands represent species of Crb3a with different post-translational modifications (PTMs) resulting in different intracellular trafficking pathways or stability. However, when we analyzed the bands separately, we found that both bands of the doublet are labeled equally in cell surface labeling experiments and maintain a consistent ratio relative to the total cell protein level, indicating that the cell surface half-life of each is the same (data not shown).
Cell surface Crumbs3a is metabolized via the endosomal-lysosomal pathway
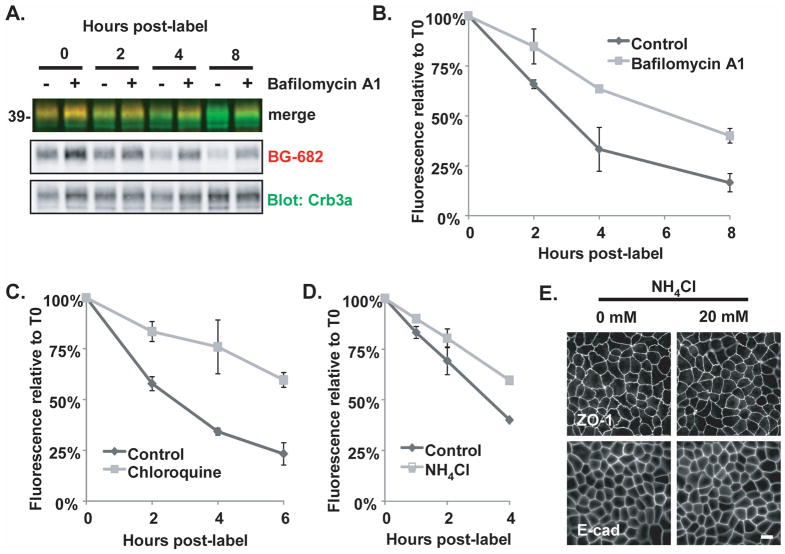
Next, we explored the pathway by which cell surface Crb3a is metabolized in polarized MDCK cells at steady state. We performed the same cell surface SNAP-Crb3a trafficking experiments described in Figure 3A(d), but treated cells with inhibitors of the endosomal-lysosomal pathway following cell surface labeling. Bafilomycin A1 specifically inhibits vacuolar-type H+-ATPase, while chloroquine and NH4Cl inhibit lysosomal acidification (39). Treatment of cells with 100 nM bafilomycin A1 doubled the half-life of labeled cell-surface SNAP-Crb3a as seen in Figure 4A and 4B; the mean half-life increased from 2.70 hours (SD±0.30) in control-treated cells to 6.01 hours (SD±0.19) in bafilomycin A1-treated cells. Similar results were seen following treatment of cells with 250 μg/ml chloroquine (CQ) and 20 mM NH4Cl as seen in Figures 4C and 4D. Treatment with CQ increased the mean half-life of SNAP-Crb3a from 2.70 hours (SD±0.09) in control-treated cells to 8.37 hours (SD±0.36), while NH4Cl increased the mean half-life from 3.18 hours (SD±0.15) in control-treated cells to 5.47 hours (SD±0.29). Because NH4Cl has been shown to have a dose and treatment duration effect on tight and adherens junctions (40), we checked the integrity of the junctions for the relatively short duration and dose of NH4Cl used in this trafficking experiment. Tight and adherens junction formation remained intact during treatment with 20 mM NH4Cl over 4 hours as assessed by immunofluorescence for ZO-1 and E-cadherin as seen in Figure 4E.
Figure 4. Cell surface Crumbs3a is metabolized via the endosomal-lysosomal pathway.
A) SNAP-Crb3a cells were cell surface labeled in-culture with BG-682 and incubated for times indicated in the presence of 100 nM bafilomycin A1. Quantification of cell surface labeling experiments performed in the presence of B) 100 nM bafilomycin A1 as in (A), C) 250 μg/ml chloroquine, or D) 20 mM NH4Cl, and appropriate vehicle controls. Cell lysates were immunoblotted with anti-Crb3a and relative residual fluorescence was determined relative to time 0. All experiments were performed twice, in duplicate; graphs show experimental averages with standard deviations. E) Immunofluorescence images of SNAP-Crb3a cells grown on transwell filters and incubated in the absence (0 mM) or presence (20 mM) of NH4Cl for 4 hours, then fixed, permeabilized, and immunostained with indicated antibodies.
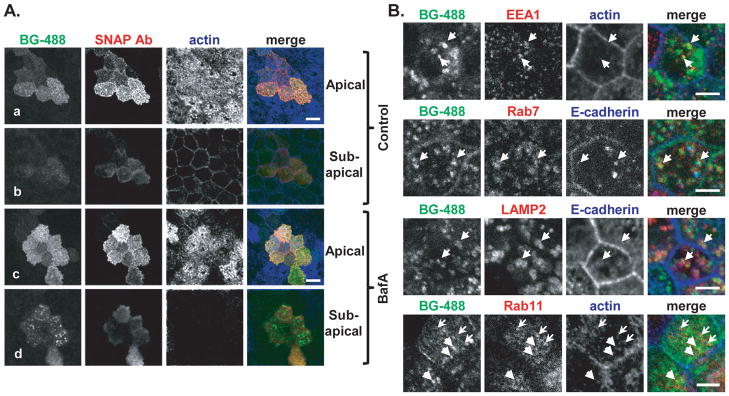
Next we examined the fate of cell surface-labeled SNAP-Crb3a following treatment with inhibitors of the endosomal-lysosomal pathway. As seen in Figure 5A, confocal images of cells treated with bafilomycin A1 (BafA, panel c) and vehicle control (Control, panel a) both show residual labeled SNAP-Crb3a at the cell surface co-localizing with anti-SNAP antibody in non-permeabilized cells. However, puncta of labeled SNAP-Crb3a that do not co-localize with anti-SNAP antibody are also seen (panels b and d), indicating internalized pools of SNAP-Crb3a from the cell surface. This was observed to a much larger extent with bafilomycin treatment (panel d) as compared to control treatment (panel b).
Figure 5. Crumbs3a accumulates intracellularly in the presence of inhibition of the endosomal-lysosomal pathway.
A) Confocal immunofluorescence images of SNAP-Crb3a cells grown on transwell filters, labeled in culture with BG-488, treated for 8 hours with either 100 nM bafilomycin A1 (BafA, panels c and d) or vehicle control (panels a and b), then fixed. Cells were immunostained (not permeabilized) with anti-SNAP antibody and phalloidin. Z-stack images captured at the apical and sub-apical regions (relative to actin-laden microvilli) are shown. White bars, 10 μm. B) Confocal immunofluorescence images of cells treated with bafilomycin A1 as in 5A, but cells were permeabilized and immunostained with indicated antibodies. In the top three rows, arrows highlight regions of co-localization of internalized, labeled SNAP-Crb3a and antibody of interest. In the bottom row, heavy arrows (pointing to right) highlight internalized, labeled SNAP-Crb3a from the cell surface while the light arrows (pointing to left) highlight Rab11-positive vesicles. White bars, 5 μm.
To further characterize the vesicles containing internalized cell surface-labeled SNAP-Crb3a following treatment with bafilomycin A1, we co-stained with several antibodies recognizing proteins in the endosomal-lysosomal pathway as shown in Figure 5B. We found that under these conditions, cell surface-labeled SNAP-Crb3a partially co-localizes with EEA1, Rab7 and LAMP2, suggesting that Crb3a traffics from the cell surface through the endosomal-lysosomal pathway. Conversely, we were unable to show significant colocalization of accumulated cell surface-labeled SNAP-Crb3a with Rab11, suggesting that in polarized cells at steady state, recycling of cell surface Crb3a through the apical recycling endosome is less prominent. Together, these results indicate that at steady state, cell surface SNAP-Crb3a is internalized and metabolized through the endosomal-lysosomal pathway.
Endogenous Crumbs3a and exogenous Snail expression correlate inversely
We next turned our attention to conditions that are known to affect apical-basal polarity and asked whether Crb3a stability is affected by these conditions. Snail is a potent destabilizer of apical-basal polarity. Although Snail is known to suppress transcription of both Crumbs3 and E-cadherin genes, its effect on protein stability and membrane retention is less clear. Since multiple proteins involved in apical-basal polarity have relatively long half-lives, it has been suggested that post-transcriptional mechanisms are also employed to control expression levels (41). We previously showed that endogenous Crb3a expression decreases in the presence of Snail (29), and here we explore the temporal relationship of expression of these two proteins.
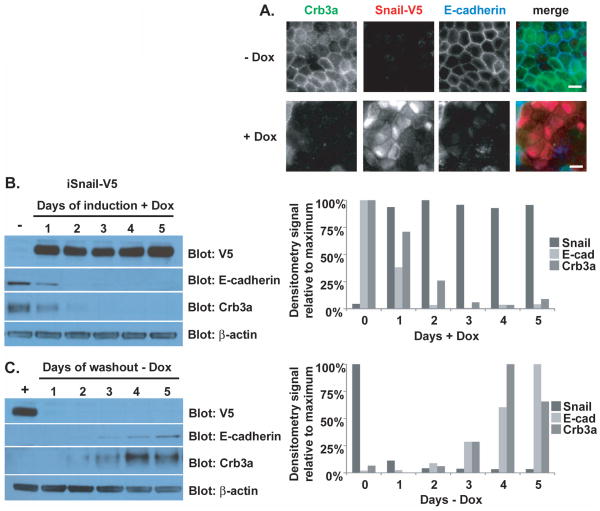
We produced MDCK cells stably expressing doxycycline-inducible Snail-V5. As shown in Figure 6A, uninduced cells (−Dox) exhibit normal-appearing adherens (E-cadherin) and tight (Crb3a) junctions as detected by immunofluorescence. Following induction of Snail-V5 (+Dox), cells lose endogenous Crb3a and E-cadherin expression and dismantle their intercellular junctions as we have seen with stable Snail expression. We then analyzed protein expression levels in these cells as a function of time following doxycycline induction or washout. As shown in Figure 6B, endogenous Crb3a expression is almost undetectable within two days of induction of Snail expression in parallel with E-cadherin. In Figure 6C, Crb3a expression resumes within two days of doxycycline washout similarly to E-cadherin. Graphs depicting the relative densitometry signal of each protein during the time course accompany the immunoblots. The parallel pattern of disappearance and reappearance of Crb3a and E-cadherin suggests common mechanisms controlling protein expression.
Figure 6. Endogenous Crumbs3a and exogenous Snail expression correlate inversely.
A) Epifluorescent images showing loss of tight (Crb3a) and adherens (E-cadherin) junctions after induction of Snail. Inducible Snail-V5 MDCK cells were grown on transwell filters in the absence (−Dox) or presence (+Dox) of 100 ng/ml doxycycline for 4 days, fixed and immunostained with Crb3a, V5 and E-cadherin antibodies. White bars, 10 μm. B) The same cells were lysed after growth in absence (−) or presence (+) of 100 ng/ml doxycycline for the indicated number of days, or C) for the indicated number of days following doxycycline washout (following initial induction for 7 days). Cell lysates were immunoblotted with indicated antibodies. Graphs (right of each figure) show quantification of immunoblot band intensities as determined by densitometry (see Methods), normalized to β-actin, and reported relative to the maximum protein expression level particular to that experiment.
Snail destabilizes Crumbs3a at the cell surface
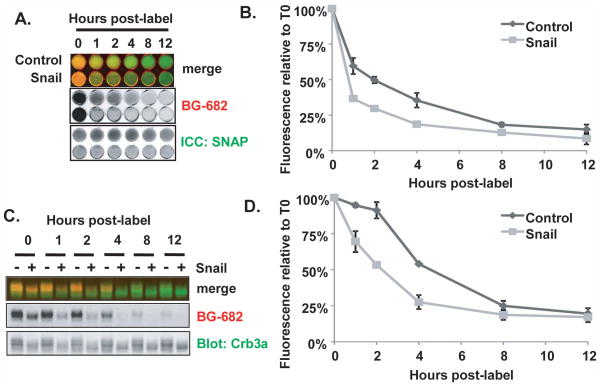
To investigate whether Snail affects Crb3a stability, we produced MDCK cells stably co-expressing Snail-FLAG and SNAP-Crb3a as seen in Supplementary Figure S1A. We performed cell surface SNAP-Crb3a labeling experiments and compared the half-life of SNAP-Crb3a in cells expressing Snail versus vector control as assessed by immunofluorescence in situ as illustrated in Figure 3A(c). As shown in Figure 7A and 7B, the half-life of cell surface SNAP-Crb3a is decreased by 50% in the presence of Snail when compared to control cells; the half-life is decreased from 1.83 hours (SD±0.19) in control cells, to 0.92 hours (SD±0.03) in Snail-coexpressing cells.
Figure 7. Snail destabilizes Crumbs3a at the cell surface.
SNAP-Crb3a cells co-expressing Snail or vector control were cell surface labeled with BG-682 and either fixed (A) or lysed (B) at the indicated time points. A) in situ immunocytochemistry showing half-life of cell surface SNAP-Crb3a relative to cell surface SNAP-Crb3a as described in 3A(c), in the presence of Snail-FLAG (Snail) or vector control. B) Quantitation of fluorescence images from (A) showing relative residual fluorescence signal at the specified time points, normalized to the total anti-SNAP antibody signal in each well. C) Immunoblot of cell lysates labeled in parallel to those in 7A with D) quantification of residual fluorescence signal of cell surface SNAP-Crb3a normalized to total cell SNAP-Crb3a performed as described in Figure 3A(d). Experiments were performed twice in duplicate; graphs show averaged experimental time points with standard deviations.
In Figure 7A, cell surface SNAP-Crb3a expression appeared to be less than control cells, possibly as a result of decreased production or destabilization of SNAP-Crb3a transcript in the presence of Snail. Accordingly, we enriched for cell surface SNAP-Crb3a expression in Snail cells using FACS. We confirmed that both the protein and transcript levels of SNAP-Crb3a in this enriched pool were at least that of control cells as seen in Supplementary Figure S1A and S1B. We then performed cell surface SNAP-Crb3a labeling experiments with these enriched pools and assessed SNAP-Crb3a half-life by immunoblotting as illustrated in Figure 3A(d). As seen in Figure 7C and 7D, there is also an ~50% decrease in half-life of cell surface SNAP-Crb3a in the presence of Snail, consistent with the results in Figure 7A. The half-life is decreased from a mean of 4.56 hours (SD±0.11) in control cells, to 2.13 hours (SD±0.26). We note that the half-life of the control cells in this experiment is longer than the half-life for SNAP-Crb3a reported in Figure 3C. We attribute this to altered inter-experimental conditions including examination of cells at earlier time points post-labeling increasing duration of exposure of cells to room conditions, and the presence of two expression vectors requiring treatment with two selection antibiotics.
Because MDCK cells expressing Snail do not form adherens or tight junctions, we explored whether cell surface Crb3a is destabilized when cells lack lateral junctions. We cultured MDCK cells expressing SNAP-Crb3a (from Figure 2) in low calcium media overnight and then performed cell surface trafficking experiments. We confirmed that at the time of initiation of the cell surface trafficking experiment, intercellular junctions were abolished as assessed by immunofluorescence imaging (Supplementary Figure S2); the expected subapical accumulation of SNAP-Crb3a characteristic of MDCK cells cultured in low calcium media was seen under these conditions. As shown in Supplementary Figures S2B and S2C, there is no difference in half-life of cell surface labeled SNAP-Crb3a in the presence or absence of intercellular junctions. This result suggests that destabilization of cell surface Crb3a by Snail cannot be attributed to the absence of cell junctions. Taken together, these results indicate that cell surface Crb3a is significantly destabilized in the presence of Snail.
Snail induces differential post-translational modifications of Crumbs3a
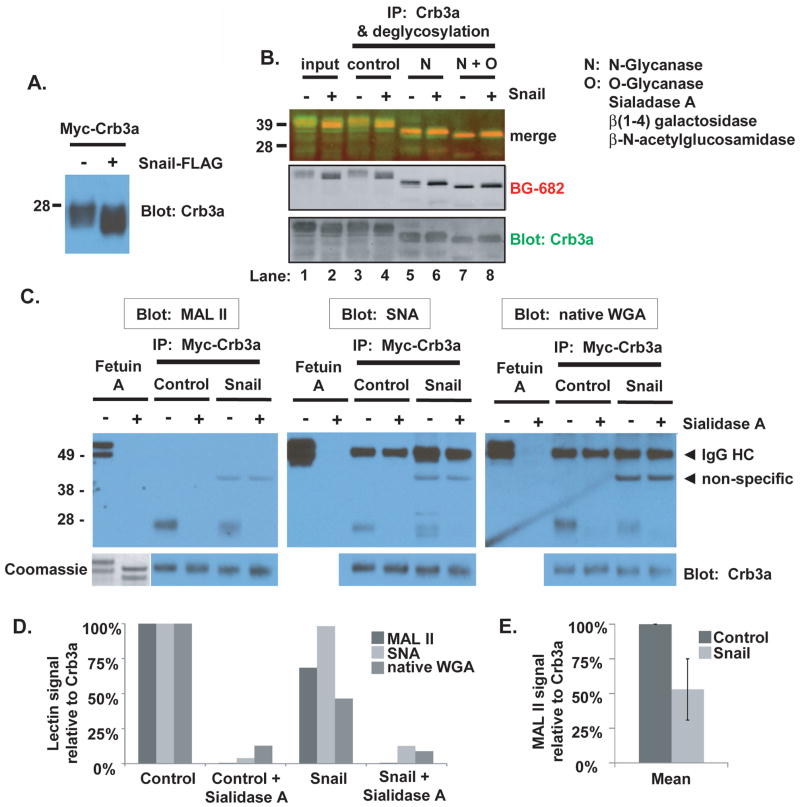
We consistently noticed that the apparent mass of exogenously expressed Crb3a on SDS-PAGE was altered in the presence of Snail (Supplementary Figure S1A and Figure 8A). We pursued the etiology of the mobility shift that we observed for SNAP-Crb3a and myc-Crb3a induced by the presence of Snail. Since PTMs are a common mechanism to alter protein trafficking and stability (42), we performed deglycosylation experiments following labeling of cell surface SNAP-Crb3a in the absence or presence of Snail. Crumbs3a contains a single potential N-glycosylation site and multiple potential serine and threonine sites for O-glycosylation. N-deglycosylation was performed with N-Glycanase (PNGase F), which removes virtually all N-linked oligosaccharides. O-deglycosylation was performed with O-Glycanase (endo-α-N-acetylgalactosaminidase) in combination with Sialidase A (α-2(3,6,8,9)-neuraminidase), β(1–4) galactosidase and β-N-acetylglucosamidase. Together, these exoglycosidases sequentially remove most terminal monosaccharides to allow access to the disaccharide galactose β(1–3)-N-acetylgalactosamine (Gal β(1–3)GalNAc) core for removal by O-Glycanase. Specifically, these enzyme treatments cleave all non-reducing terminal branched and unbranched sialic acids, catalyze the hydrolysis of β1–4 linked D-galactopyranosyl residues, and catalyze the hydrolysis of terminal, non-reducing β-N-acetylglucosamine residues. These enzyme treatments do not remove oligosaccharides containing α-linked galactose or fucose, or directly O-linked N-acetylglucosamine, N-acetylgalactosamine, fucose or mannose.
Figure 8. Snail induces differential post-translational modifications of Crumbs3a.
A) Mobility of myc-Crb3a is altered in the presence of Snail as revealed by Crb3a immunoblot of lysates from cells co-expressing myc-Crb3a and either Snail-FLAG (+) or vector control (−). B) Glycosylation pattern analysis of SNAP-Crb3a in the presence or absence of Snail. Following cell surface in-culture labeling with BG-682, SNAP-Crb3a was immunoprecipitated from cell lysates with anti-Crb3a antibody and subjected to N- and O-deglycosylation. SNAP-Crb3a was detected with affinity purified anti-Crb3a antibody. C) Sialylation pattern analysis of SNAP-Crb3a in the presence of absence or Snail by lectin blot. Myc-Crb3a was immunopurified using myc antibody-conjugated beads, denatured and incubated with Sialidase A or control. Following standard PAGE and protein transfer, membranes were blotted with indicated lectins. All membranes were stripped and reprobed with anti-Crb3a antibody. Fetuin A was included as a glycoprotein control; protein presence following desialylation was confirmed in gel by Coomassie staining (lower left corner). D) Graph showing the relative band intensities of the lectin blots in (C), analyzed by densitometry as described in Methods, normalized to Crb3a signal, and reported relative to untreated control cells. E) Graph of average MAL II lectin blot signal intensity as in (C) and (D) obtained from two separate experiments, reported with standard deviations (bars).
Figure 8B shows that following N-deglycosylation, the difference in SNAP-Crb3a’s mobility in the presence of Snail is almost eliminated (compare lanes 5 and 6 to 3 and 4). This result suggests that Snail induces differential terminal N-glycosylation of Crb3a. Following subsequent O-deglycosylation, SNAP-Crb3a migrates faster in the absence (lanes 7) rather than the presence of Snail (lane 8). Indeed, following N- and O-deglycosylation, the mobility was slower in the presence of Snail (compare lane 8 to 7) although SNAP-Crb3a mobility was initially faster in the presence of Snail (compare lane 2 to 1). These results indicate that differential terminal N- and O-glycosylation of Crb3a occur in the presence of Snail. Moreover, subtly altered mobility of SNAP-Crb3a persists in the presence of Snail following both N- and O-deglycosylation (compare lane 8 to 7) suggesting that Snail induces additional PTM(s) not identified by these deglycosylation methods. Another significant observation is discovered when comparing the blots of cell surface-labeled Crb3a (BG-682, middle panel) to total cell Crb3a (lower panel). The identical banding patterns indicate that Crb3a still traffics to the cell surface despite altered terminal glycosylations induced by Snail.
To further explore terminal glycosylation events of Crb3a seen in the presence of Snail, we analyzed glycosylation patterning of Crumbs3a by lectin blotting. We chose to analyze for the presence of sialic acid modifications since sialylation is the most common modification of the Gal β(1–3)GalNAc core and the addition of sialic acid (N-acetylneuraminic acid) is known to affect mass and charge of glycoproteins. Moreover, altered sialylation of glycoproteins is well described in malignant transformation and affects the half-lives of circulating glycoproteins (43, 44).
We used Maackia amurensis lectin II (MAL II), Sambucus nigra bark (SNA) and native wheat germ agglutinin (WGA) to identify sialic acid modifications of Crb3a. MAL II preferentially binds sialic acid residues with (α-2,3) linkages, while SNA has higher affinity for sialic acid residues with (α-2,6) linkages. Native WGA interacts specifically with non-reducing terminal N-acetylneuraminic acid (sialic acid) and N-acetylglucosamine (GlcNAc) residues. Myc-Crb3a was immunopurified from lysates of cells expressing Snail or control vector using myc antibody-conjugated sepharose beads. To confirm specificity of lectin binding, we desialylated immunopurified myc-Crb3a with Sialidase A; this exoglycosidase cleaves all non-reducing terminal and branched sialic acid residues from N- and O-glycans. Each of these lectins recognizes sialic acid residues of the glycoprotein fetuin A so this was used as a positive control (45). Lectin blotting following protein electrophoresis of myc-Crb3a shows that Crb3a is sialylated as seen in Figure 8C. MAL II (left panel) and native WGA (right panel) both recognize myc-Crb3a but the mean relative signal intensity is decreased in the presence of Snail suggesting a decrease in α-2,3 sialylation. The relative band intensities of the lectin immunoblots are shown graphically in Figure 8D. Figure 8E shows that the MAL II lectin blotting results are reproducible, and that the mean signal intensity decreases to 53% (SD±22%) of control in the presence of Snail. Although the signal intensity of SNA binding to myc-Crb3a is similar in the presence or absence of Snail, altered mobility of myc-Crb3a is clearly apparent and a doublet is observed suggesting the presence of other PTM(s). Together, these results indicate that Crb3a is sialylated via both α-2,3 and α-2,6 linkages, and that α-2,3 sialylation of Crb3a is decreased in the presence of Snail.
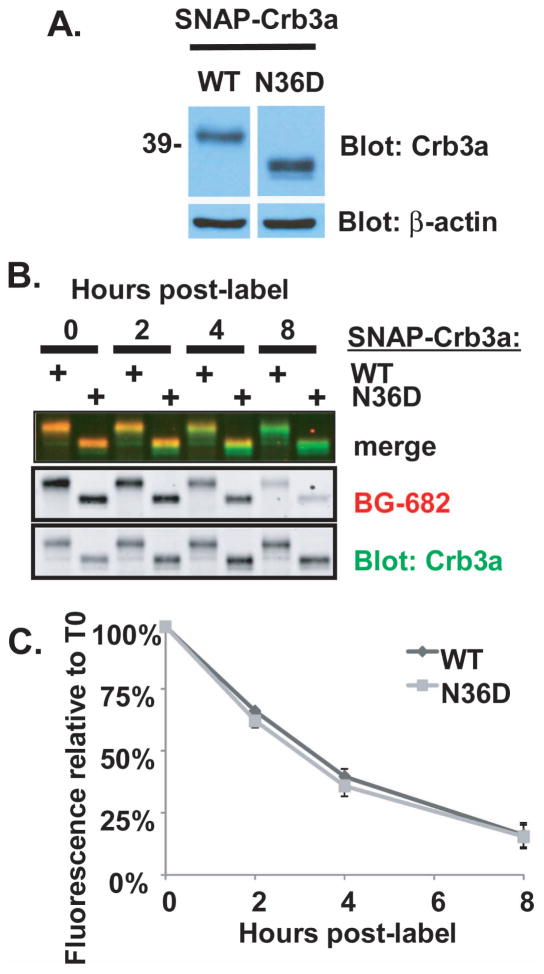
Since differential sialylation could be due to modifications at either O- or N-glycans, we explored the effect of N-glycosylation on stability of Crb3a at the cell surface. Earlier studies in our laboratory revealed that mutation of the only extracellular asparagine of wild type Crb3a to aspartate (N36D) induced a significant mobility shift (10) indicating that this is the only N-glycosylation site of Crb3a. We produced an expression vector with the analogous mutation in SNAP-Crb3a (SNAP-Crb3a-N36D) and created a stable MDCKII cell line co-expressing SNAP-Crb3a-N36D as shown in Figure 9A. We then performed cell surface SNAP-Crb3a trafficking experiments. Figures 9B and 9C show that the stability of cell surface SNAP-Crb3a-N36D is no different than wild type SNAP-Crb3a, with a mean half-life of 2.84 hours (SD±0.05) versus 3.06 hours (SD±0.17) respectively. These results indicate that complete removal of N-glycosylation does not alter Crb3a trafficking to or stability at the cell surface.
Figure 9. N-glycosylation of Crumbs3a is not critical for trafficking to or stability at the apical cell surface.
A) Lysates from cells expressing wild type SNAP-Crb3a (WT) or SNAP-Crb3a-N36D (N36D) immunoblotted with indicated antibodies. B) Comparison of the half-lives of cell surface WT or N36D SNAP-Crb3a. Cells were labeled in-culture with BG-682, incubated for indicated time points, lysed and immunoblotted with anti-Crb3a antibody. B) Graph showing residual fluorescence of cell surface labeled SNAP-Crb3a normalized to total SNAP-Crb3a signal and expressed relative to the 0 time point. The experiment was performed twice in duplicate; a representative immunoblot is shown. The graph shows averaged experimental time points with standard deviations.
Discussion
In this report, we characterize the cell membrane trafficking of Crumbs3, the Crumbs ortholog expressed in mammalian epithelial cells. Specifically, Crumbs3a is the most prevalent splice isoform present in these cells and is the focus of this study. In summary, our novel findings are: 1) cell surface Crb3a has a half-life of 2.8 hours, 2) Crb3a is internalized from the cell surface and metabolized via the endosomal-lysosomal pathway, 3) Snail expression substantially decreases the cell surface stability of exogenously expressed Crb3a, and 4) Snail expression modulates the PTMs (glycosylation and sialylation) of exogenously expressed Crb3a.
Our data reveal that in polarized MDCK cells at steady state, the half-life of SNAP-Crb3a at the cell surface is 2.8 hours as quantitatively assessed by SNAP-tag labeling. We further found that the half-life of SNAP-Crb3a was unaffected by dissolution of the intercellular junctions, suggesting that the stability of Crb3a at the cell surface is similar whether it is located within the apical membrane or tight junctions. Once internalized from the cell surface, SNAP-Crb3a appears to be rapidly metabolized via the endosomal-lysosomal pathway. Together with data obtained in Drosophila (11), our data reinforce the critical role of the endocytic pathway in regulation of accumulation of Crumbs at the cell surface. We previously showed that Crb3a traffics through Rab11-positive endosomes during cystogenesis (38). However, we were unable to show in the present study that a significant amount of Crb3a traffics through the Rab11-positive recycling endosome pathway once internalized from the cell surface, also consistent with results seen in Drosophila (11). A plausible scenario is that Crb3a traffics differently at various points along the polarization spectrum. Indeed, recently published studies reveal that the retromer complex is involved in Crumbs trafficking, suggesting a mechanism by which Crumbs may be either recycled to the cell surface or sent to the lysosome for degradation (46–48).
Generalization of such a mechanism as a means to centrally control the cell polarity program, however, is challenged by the breadth of different trafficking and metabolic pathways of transmembrane proteins involved in cell polarity. While the approximately 3 hour half-life of cell surface Crb3a is similar to the 2 hour half-life reported for podocalyxin/gp135 (49), it is shorter than the 5 hour half-life reported for E-cadherin (50). The finding that Crb3a is rapidly metabolized via the endosomal-lysosomal pathway also contrasts with pathways determined for other transmembrane polarity proteins. While E-cadherin recycles to the cell surface through Rab11-positive recycling endosomes (51, 52), occludin recycles to the cell surface via a Rab13-mediated process (53). Further, degradation of each involves a different ubiquitin ligase (54, 55). The disparate modes of trafficking and degradation among these adherens and tight junction transmembrane proteins make it challenging to envision a single unifying mechanism to control epithelial polarization.
However, a potential unifying mechanism is suggested by our discovery that Snail destabilizes cell surface Crb3a. While Snail is a downstream target of many different signaling pathways including EGF, Wnt, FGF, Notch, integrins and TGFβ, it is also an upstream effector of cell proliferation, survival, polarity and migration (36). This central position makes Snail an attractive candidate as a potent and multifaceted regulator of the cell polarity program. Snail family members are frequently described as inducers of partial or full EMT, and Snail’s roles beyond transcriptional regulation are currently being elucidated.
Indeed, our results support the concept that Snail is involved in complex regulation of cell polarity proteins. First, we showed that endogenous Crb3a and E-cadherin expression do not recover to steady state levels until 4–5 days following the disappearance of Snail in our inducible expression system. Whether this is an epigenetic regulatory effect of Snail (56) or a post-transcriptional effect is unclear. We then showed that Snail expression (and ensuing EMT) substantially decreases the cell surface stability of exogenously expressed Crb3a, findings that cannot be merely attributed to destabilization of intercellular junctions. We provide evidence that Snail alters PTMs of exogenously expressed Crb3a, and we propose that these PTMs may underlie the destabilization of cell surface Crb3a.
Our experiments reveal that these altered PTMs affect terminal N- and O-glycosylation and sialylation of Crb3a (Figure 8B and 8C). Although extracellular N-glycosylation has been suggested as a mechanism involved in protein trafficking to the apical membrane, our analysis of a Crb3a point mutant (N36D) suggests that N-glycosylation does not affect trafficking to or stability at the cell surface (Figure 9). This finding is consistent with previously published data that N-glycosylation does not alter delivery of other integral membrane proteins involved in apical-basal polarity, including gp135 and E-cadherin (49, 50). However, it is still plausible that modification of Crb3a’s N-glycan affects Crb3a’s stability at the cell surface, which could arise by altering Crb3a’s binding partners that hasten Crb3a’s metabolism. In this case, the N36D mutant would be anticipated to exhibit the “default” trafficking characteristics of wild-type Crb3a. Such a scenario is suggested by a recent study showing that alterations of terminal N-glycan modifications affect TOLL-like receptor 4 (TLR4)’s activity at the cell surface. Removal of terminal α-2,3-linked sialic acid residues by neuraminidase 1 (NEU1) was proposed to be necessary to overcome steric hindrance and allow TLR4 dimerization and activation (57). Alternatively, other non-N-glycan-related PTMs may be involved in modulating Crb3a’s cell surface stability and metabolism.
The differential sialylation of Crb3a induced by Snail could be involved in destabilization of Crb3a at the cell surface. Indeed, altered sialylation is known to affect stability of circulating glycoproteins (58, 59). Sialic acid modifications are produced by transfer of sialic acid residues to acceptor carbohydrates, reactions catalyzed by members of the sialyltransferase (ST) family which each create glycoprotein specific linkages. Altered ST activity is described in multiple malignancies, including members of the α-2,3-sialyltransferase (ST3Gal) family (44, 60, 61). Interestingly, while increased expression of several ST3Gal enzymes and increased levels of α–2,3-linked sialic residues detected in tissue correlates with increased risk for metastatic potential in several human carcinomas (62, 63), we observe decreased α-2,3 sialylation of Crb3a in the presence of Snail. We speculate that this difference may be related to the net negative charge that sialic acid modifications impart on proteins that reside within different membrane regions of the epithelial cell surface. For lumen-forming epithelial cells, a negative charge on proteins at the apical surface may provide a repulsive cue to opposing apical membranes, thus supporting the maintenance of a central lumen. A decrease in sialic acid modifications of these proteins may then be expected to decrease inhibition of cell-cell contact at the apical membrane. Conversely, for cell surface proteins involved in cell-cell contact at the basolateral surface, such a decrease in sialic acid modifications may promote cell-cell adhesion while an increase in sialic acid modifications may alter cell-cell contacts and adhesion. Thus, we anticipate different effects on epithelial cell polarity depending on the proteins modified by the various STs, and the STs’ relative activity at any point. Alternatively, there could be upregulation or alteration of associated sialidases, or a relative increase in α-2,6 sialylation which could halt further terminal glycosylation opportunities. Such alterations are described in human malignancies including colon carcinoma (64–66).
Another interesting implication for differential sialylation involving a potential role in regulation of polarity ties back to our finding that Crb3a is internalized and metabolized through the endosomal-lysosomal pathway (Figures 4 and 5). Since human sialidases NEU1 and NEU4 are found in lysosomes (64), internalization of Crb3a from the cell surface into lysosomes could provide an opportunity to post-translationally regulate Crb3a’s cell surface stability and cell polarization status.
In the current study, we find that cell surface Crb3a is substantially destabilized in the setting of Snail-induced EMT. We coincidentally observe that Snail induces differential glycosylation and sialylation of Crb3a. Although it is unclear whether it is these or other changes that are responsible for the observed destabilization of cell surface Crb3a, the role of PTMs in modulating protein trafficking and stability is well established, and altered glycosylation patterns are commonly found in human malignancy (67–70). Future studies will focus on whether modified glycosylation or other factors such as destabilization of Crb3a binding partners is the reason for Crb3a’s reduced cell surface half-life in EMT.
Materials and Methods
Antibodies and chemicals
Primary antibodies: rabbit anti-Crumbs3a (10), mouse anti-β-actin (Sigma Aldrich, St. Louis, MO), rat anti-E-cadherin clone DECMA1 (Sigma Aldrich), mouse anti-β-catenin (BD Biosciences, Franklin Lakes, NJ), mouse anti-EEA1 (BD Biosciences), rabbit anti-Rab7 (Cell Signaling Technology, Danvers, MA), rabbit-anti-(canine) LAMP2 was a generous gift from Enrique Rodriguez-Boulan (Weill Cornell Medical College, New York, NY), mouse anti-Rab11 (BD Biosciences), mouse anti-gp135 was a generous gift from George Ojakian (State University of New York), rabbit-anti-V5 (Bethyl Laboratories, Montgomery, TX), rabbit anti-SNAP (New England Biolabs, Ipswich, MA), rabbit anti-GFP (Invitrogen, Carlsbad, CA), goat-anti-Snail (R&D Systems, Minneapolis, MN). Alexa-Fluor conjugated secondary antibodies, phalloidin-647 and DAPI were obtained from Invitrogen. Horseradish peroxidase-conjugated Streptavidin and secondary antibodies were obtained from BioLegend (San Diego, CA) and GE Healthcare (Piscataway, NJ) respectively. Benzylguanine-488, Benzylguanine-682, SNAP-Surface Block were obtained from New England Biolabs. Bafilomycin A1 was obtained from EMD/Calbiochem (Gibbstown, NJ). Doxycycline was obtained from Clontech (Mountain View, CA). NH4Cl and biotinylated (native) wheat germ agglutinin were from Sigma-Aldrich. Biotinylated Maackia amurensis lectin II and Sambucus nigra lectin were obtained from Vector Laboratories (Burlingame, CA).
DNA constructs and stable cell lines
A custom mammalian minimal-expression vector was produced by replacing the CMV enhancer and promoter of pcDNA3.1/Zeo(+) (Invitrogen) with the minimal CMV promoter from pTRE-Tight (Clontech). Myc-(human) Crb3a: cDNA was PCR amplified from a previously established expression vector (16, 29) and subcloned into the multiple cloning site of the new expression vector. 3x myc-Crb3a: oligonucleotides corresponding to the 3x myc epitope were annealed and subcloned in to replace the single myc tag. SNAP-Crb3a: SNAP tag was PCR amplified from pSNAP(m)-tag vector (New England Biolabs) and subcloned into replace the single myc tag. SNAP-Crb3a retroviral expression vector: SNAP-Crb3a was amplified from the previous vector and subcloned into pRev-TRE (Clontech). SNAP-Crb3a-N36D: site directed mutagenesis was performed on SNAP-Crb3a retroviral expression vector using forward primer 5′-GGACTTCTGCAAATGAGgATAGCACTGTTTTGCCT-3′ and corresponding reverse complement primer. These vectors were introduced into stable MDCK cell lines expressing Snail-FLAG or corresponding empty vector (29) without a Tet regulatory vector, so that SNAP-Crb3a is constitutively expressed at low level. Inducible GFP-Crumbs3a: cDNA was PCR amplified from mGFP-Crb3a (38) and subcloned into pRetroX-Tight-Puro (Clontech). Inducible Snail-V5: cDNA was PCR amplified from Snail-FLAG (29) using primers that introduced a C-terminal V5 tag and subcloned into pRetroX-Tight-Puro. Inducible retroviral protein expression vectors were transduced into MDCKII cells containing stably incorporated pTet-On Advanced (Clontech). Transfection of mammalian expression vectors into MDCKII cells was performed using Lipofectamine 2000 (Invitrogen) according to manufacturer’s directions. Retroviral-mediated gene transfer was performed as previously described (71). Cells incorporating desired elements were stably selected by culturing cells in growth media supplemented with 200 μg/ml Zeocin (Invitrogen), 100 ng/ml Hygromycin B (Invitrogen) or 5 μg/ml Puromycin (InvivoGen, San Diego, CA) as appropriate.
Tissue culture, immunofluorescence and microscopy
Culture conditions for all MDCKII cell lines were performed as previously described (29, 38). IF using indicated antibodies and imaging of cells by epifluorescent and confocal microscopy of 2D monolayers and 3D cysts were performed as previously described (38).
Protein immunopurification and immunoblotting
Cells were lysed in SDS denaturing lysis buffer (1% SDS, 50 mM Tris pH 7.4, 5 mM EDTA, mammalian protease inhibitor cocktail (Sigma Aldrich)). For immunoprecipitation (IP), lysates were diluted 10-fold in ice-cold IP buffer (50 mM HEPES, 22.5 mM NaOH, 100 mM NaCl, 1% Triton X-100). IP was performed using TrueBlot anti-rabbit Ig IP beads (eBioscience, San Diego, CA) according to manufacturer’s directions. For myc-Crb3a pulldown using myc-antibody conjugated beads, cells were lysed in RIPA buffer (137 mM NaCl, 20 mM Tris-Cl pH 7.4, 10% glycerol, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 2 mM EDTA). Affinity purified rabbit c-myc antibody was coupled to Sepharose G beads using 20 mM dimethyl pimelimidate dihydrochloride in 0.2 M sodium borate pH 9.0 buffer; the reaction was stopped by washing with 0.2 M ethanolamine pH 8.0, and non-covalently bound antibodies eluted with glycine. Beads were incubated with myc-Crb3a lysate, washed with RIPA, high salt buffer (20 mM Tris pH 7.4, 500 mM NaCl) and 0.2% CHAPS in TBS. Immunopurified protein was eluted in lithium dodecyl sulfate buffer with 50 mM DTT.
Cell lysates and immunopurified proteins were run on SDS-PAGE gel using standard technique. Proteins were transferred to PVDF (Millipore, Billerica, MA) and blotted with antibodies or lectins. For chemiluminescent detection, immunoblotting and detection were performed as previously described (38), except that membranes incubated with lectins were blocked with Carbo-Free blocking solution (Vector Laboratories). Densitometric measurements of signal intensity of bands observed on immunoblot (IB) films were performed by creating a Photoshop document (600 dpi) of the film using a Canon 8800F image scanner, and measuring average signal intensity within regions of interest using MetaMorph; signal intensities were corrected for background and normalized to corresponding Crb3a, myc, or β-actin signal, as indicated. For fluorescence detection, IB and detection were performed according to LI-COR (Lincoln, NE) Odyssey manual using Protein Loading Buffer (LI-COR) and IRDye 800CW goat anti-rabbit IgG (H+L) secondary antibody (LI-COR). For fluorescence detection using ECL-Plus, images were obtained with Typhoon 9400 (GE Healthcare) scanner and data quantified using ImageQuant TL software. For infrared fluorescence detection, images were obtained with Licor Odyssey Imager and data quantified using Odyssey infrared imager 2.0 software.
Induction of GFP-Crumbs3a and Snail-V5
For modified pulse chase assays, iGFP-Crb3a cells were grown post-confluence on tissue culture dishes. Cells were pulse-treated with 50 ng/ml doxycycline for 1 hour at 37°C, copiously washed three times with complete growth media and then returned to incubator following addition of fresh media containing tetracycline-free fetal bovine serum (Clontech). For Snail-V5 induction, media containing 100 ng/ml of doxycycline was replaced daily. Washout was performed as described earlier. Control cells were treated with vehicle only.
Quantitative Real-time PCR (qRT-PCR)
mRNA was isolated from confluent cells, reverse transcribed into cDNA and qRT-PCR performed as previously described (72). Primers were designed using PerlPrimer software (73) and are as follows: GFP-Crb3a forward 5′-TTCAAGATCCGCCACAACAT-3′ and reverse 5′-GAGGGAGAAGACCACGATGA-3′; SNAP-Crb3a forward 5′-CTCAACGCCTACTTTCACC-3′ and reverse 5′-GCTGATGACCTCTCCGAA-3′. C. familiaris Crb3a and GAPDH primers were already described (29). Data were analyzed by normalizing to GAPDH and levels reported relative to SNAP-Crb3a or Crb3a.
Cell surface biotinylation assay
Parental MDCKII cells and cells stably expressing 1x or 3x myc-Crb3a were grown 3 days post-confluent on tissue culture dishes. Cell surface biotinylation was performed with EZ-Link Sulfo-NHS-SS-Biotin using the Pierce Cell Surface Protein Isolation Kit (Thermo Fisher Scientific Inc., Rockford, IL) according to manufacturer’s directions. Following labeling and washing, time 0 cell lysates were prepared by either lysing immediately or treating with MESNA to strip (residual) cell surface biotin and then lysed. For later time points, cells were returned to 37°C incubator for indicated duration and then either lysed or treated with MESNA and lysed. 500 μg of protein for each lysate sample was subjected to IP using anti-Crb3a antibody in combination with Protein A-Sepharose 4B beads (GE Healthcare). IP’d proteins were run on SDS-PAGE as described above, and membranes were immunoblotted with Streptavidin-HRP to detect biotinylated proteins, and anti-Crb3a or anti-myc antibody to confirm Crb3a IP. Quantification of biotinylated Crb3a signal intensities was performed by densitometry (see Protein Purification and Immunoblotting), normalized to the Crb3a or myc immunoblot signal and reported relative to the corresponding time 0 signal intensity without MESNA treatment.
SNAP-Crumbs3a cell surface fluorescence labeling and trafficking assay
SNAP-Crb3a cells were grown 3 days post-confluence on either 24 well flat-bottom tissue culture plates (Corning) or 2.4 cm polyester transwell filters (0.4 μm pore size, Corning). Growth media was pre-incubated overnight. Media was replaced with pre-incubated media containing BG-488, BG-682 or SNAP-Surface Block. Cells were incubated for 30 minutes at 37°C, and then washed with growth media. For time course experiments, pre-incubated media was added back to cells, cells returned to incubator, then fixed for IF or lysed for IB at desired time points. For trafficking experiments in low calcium media, cells were cultured on transwell filters in normal growth media for 3 days post-confluence, and washed copiously with PBS. Media containing 5 μM Ca2+ media-10% dialyzed fetal bovine serum (Invitrogen) was added to cells and incubated overnight before proceeding with subsequent studies. Labeling and subsequent time course incubations were also performed in low calcium media. For IF analysis of cell surface trafficking, cells were fixed in 4% paraformaldehyde in PBS, rinsed with PBS, blocked with PBS plus 2% goat serum, and incubated with anti-SNAP antibody without permeabilization, followed by anti-rabbit IRDye800CW. Cells were imaged using LI-COR Odyssey imager, data quantified using Odyssey 2.0 software and analyzed using Microsoft Excel software. Quantifications of residual BG-fluorescence labeling were corrected for background, normalized to SNAP-antibody signal for each sample and reported as percent residual fluorescence relative to time 0. Note that because there was significant background labeling of cells, results are reported only as relative change in half-life as opposed to absolute change in half-life. For IB analysis, data were quantified and analyzed as described with IF. Quantifications of residual BG-fluorescence labeling were corrected for background, normalized to Crb3a signal for each sample and reported as percent residual fluorescence relative to time 0. Half-lives for SNAP-Crb3a were calculated for each data set by using the equation generated by Microsoft Excel software best-fit exponential curve (based on R2 values). The half-life values were then pooled for each experiment and statistically analyzed by Excel’s descriptive statistics. In Figure 7, data points from hours 0–4 were used to generate the equations in order to best fit the data given the short half-life of SNAP-Crb3a in the presence of Snail.
Deglycosylation assay
Cells expressing SNAP-Crb3a or SNAP-Crb3a-N36D were labeled with BG-682 in culture as described above, then immediately lysed. IP was performed by adding anti-Crb3a antibody to lysate containing 500 μg total protein. Isolated protein was then treated with the GlycoPro Enzymatic Deglycosylation Kit and prO-LINK Extender Kit (ProZyme, Hayward, CA) according to manufacturer’s directions and analyzed by IB as described earlier.
Supplementary Material
Acknowledgments
We thank Subramanium Pennathur and Anuradha Vivekanandan-Giri for helpful discussions and advice. We also thank Shuling Fan, Marc Schlüter, and Toby Hurd for valuable discussions and sharing of reagents. This work was supported by National Institutes of Health (NIH) grants R01DK069605 and R01DK058208 to BM, National Kidney Foundation Postdoctoral Research Fellowship and NIH Institutional Postdoctoral Kirschstein National Research Service Award T32DK007378 to JLH, and NIH Individual Postdoctoral Kirschstein National Research Service Award F32GM079906 to ELW. This work used the Imaging Core of the Michigan Diabetes Research and Training Center funded by DK020572 from the National Institute of Diabetes and Digestive and Kidney Diseases.
References
- 1.Assémat E, Bazellières E, Pallesi-Pocachard E, Le Bivic A, Massey-Harroche D. Polarity complex proteins. Biochim Biophys Acta. 2008;1778(3):614–630. doi: 10.1016/j.bbamem.2007.08.029. [DOI] [PubMed] [Google Scholar]
- 2.Pieczynski J, Margolis B. Protein complexes that control renal epithelial polarity. Am J Physiol Renal Physiol. 2011;300(3):F589–601. doi: 10.1152/ajprenal.00615.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial–mesenchymal transitions. Nat Rev Mol Cell Bio. 2006;7(2):131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 4.Nelson WJ. Remodeling epithelial cell organization: transitions between front-rear and apical-basal polarity. Cold Spring Harb Perspect Biol. 2009;1(1):a000513. doi: 10.1101/cshperspect.a000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wodarz A, Näthke I. Cell polarity in development and cancer. Nat Cell Biol. 2007;9(9):1016–1024. doi: 10.1038/ncb433. [DOI] [PubMed] [Google Scholar]
- 6.Etienne-Manneville S. Polarity proteins in migration and invasion. Oncogene. 2008;27(55):6970–6980. doi: 10.1038/onc.2008.347. [DOI] [PubMed] [Google Scholar]
- 7.Tanos B, Rodriguez-Boulan E. The epithelial polarity program: machineries involved and their hijacking by cancer. Oncogene. 2008;27(55):6939–6957. doi: 10.1038/onc.2008.345. [DOI] [PubMed] [Google Scholar]
- 8.Huang L, Muthuswamy SK. Polarity protein alterations in carcinoma: a focus on emerging roles for polarity regulators. Curr Opin Genet Dev. 2010;20(1):41–50. doi: 10.1016/j.gde.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wodarz A, Hinz U, Engelbert M, Knust E. Expression of crumbs confers apical character on plasma membrane domains of ectodermal epithelia of Drosophila. Cell. 1995;82(1):67–76. doi: 10.1016/0092-8674(95)90053-5. [DOI] [PubMed] [Google Scholar]
- 10.Makarova O, Roh MH, Liu CJ, Laurinec S, Margolis B. Mammalian Crumbs3 is a small transmembrane protein linked to protein associated with Lin-7 (Pals1) Gene. 2003;302(1–2):21–29. doi: 10.1016/s0378111902010843. [DOI] [PubMed] [Google Scholar]
- 11.Lu H, Bilder D. Endocytic control of epithelial polarity and proliferation in Drosophila. Nat Cell Biol. 2005;7(12):1232–1239. doi: 10.1038/ncb1324. [DOI] [PubMed] [Google Scholar]
- 12.Omori Y, Malicki J. oko meduzy and related crumbs genes are determinants of apical cell features in the vertebrate embryo. Curr Biol. 2006;16(10):945–957. doi: 10.1016/j.cub.2006.03.058. [DOI] [PubMed] [Google Scholar]
- 13.Laprise P, Beronja S, Silva-Gagliardi NF, Pellikka M, Jensen AM, McGlade CJ, Tepass U. The FERM protein Yurt is a negative regulatory component of the Crumbs complex that controls epithelial polarity and apical membrane size. Dev Cell. 2006;11(3):363–374. doi: 10.1016/j.devcel.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richardson EC, Pichaud F. Crumbs is required to achieve proper organ size control during Drosophila head development. Development. 2010;137(4):641–650. doi: 10.1242/dev.041913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parsons LM, Grzeschik NA, Allott ML, Richardson HE. Lgl/aPKC and Crb regulate the Salvador/Warts/Hippo pathway. Fly (Austin) 2010;4(4):288–293. doi: 10.4161/fly.4.4.13116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan S, Hurd TW, Liu CJ, Straight SW, Weimbs T, Hurd EA, Domino SE, Margolis B. Polarity proteins control ciliogenesis via kinesin motor interactions. Curr Biol. 2004;14(16):1451–1461. doi: 10.1016/j.cub.2004.08.025. [DOI] [PubMed] [Google Scholar]
- 17.Karp CM, Tan TT, Mathew R, Nelson D, Mukherjee C, Degenhardt K, Karantza-Wadsworth V, White E. Role of the polarity determinant crumbs in suppressing mammalian epithelial tumor progression. Cancer Res. 2008;68(11):4105–4115. doi: 10.1158/0008-5472.CAN-07-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemmers C, Michel D, Lane-Guermonprez L, Delgrossi MH, Medina E, Arsanto JP, Le Bivic A. CRB3 binds directly to Par6 and regulates the morphogenesis of the tight junctions in mammalian epithelial cells. Mol Biol Cell. 2004;15(3):1324–1333. doi: 10.1091/mbc.E03-04-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan S, Fogg V, Wang Q, Chen XW, Liu CJ, Margolis B. A novel Crumbs3 isoform regulates cell division and ciliogenesis via importin beta interactions. J Cell Biol. 2007;178(3):387–398. doi: 10.1083/jcb.200609096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bilder D, Schober M, Perrimon N. Integrated activity of PDZ protein complexes regulates epithelial polarity. Nat Cell Biol. 2003;5(1):53–58. doi: 10.1038/ncb897. [DOI] [PubMed] [Google Scholar]
- 21.Chalmers AD, Pambos M, Mason J, Lang S, Wylie C, Papalopulu N. aPKC, Crumbs3 and Lgl2 control apicobasal polarity in early vertebrate development. Development. 2005;132(5):977–986. doi: 10.1242/dev.01645. [DOI] [PubMed] [Google Scholar]
- 22.Yamanaka T, Ohno S. Role of Lgl/Dlg/Scribble in the regulation of epithelial junction, polarity and growth. Front Biosci. 2008;13:6693–6707. doi: 10.2741/3182. [DOI] [PubMed] [Google Scholar]
- 23.Ozdamar B, Bose R, Barrios-Rodiles M, Wang HR, Zhang Y, Wrana JL. Regulation of the polarity protein Par6 by TGFbeta receptors controls epithelial cell plasticity. Science. 2005;307(5715):1603–1609. doi: 10.1126/science.1105718. [DOI] [PubMed] [Google Scholar]
- 24.Javier RT. Cell polarity proteins: common targets for tumorigenic human viruses. Oncogene. 2008;27(55):7031–7046. doi: 10.1038/onc.2008.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCaffrey LM, Macara IG. Epithelial organization, cell polarity and tumorigenesis. Trends Cell Biol. 2011;21(12):727–735. doi: 10.1016/j.tcb.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 26.Moreno-Bueno G, Portillo F, Cano A. Transcriptional regulation of cell polarity in EMT and cancer. Oncogene. 2008;27(55):6958–6969. doi: 10.1038/onc.2008.346. [DOI] [PubMed] [Google Scholar]
- 27.Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2(2):76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 28.Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, Garcia De Herreros A. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2(2):84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 29.Whiteman EL, Liu CJ, Fearon ER, Margolis B. The transcription factor snail represses Crumbs3 expression and disrupts apico-basal polarity complexes. Oncogene. 2008;27(27):3875–3879. doi: 10.1038/onc.2008.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer. 2005;5(9):675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 31.Leroy P, Mostov KE. Slug is required for cell survival during partial epithelial-mesenchymal transition of HGF-induced tubulogenesis. Mol Biol Cell. 2007;18(5):1943–1952. doi: 10.1091/mbc.E06-09-0823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fogg VC, Liu CJ, Margolis B. Multiple regions of Crumbs3 are required for tight junction formation in MCF10A cells. J Cell Sci. 2005;118(Pt 13):2859–2869. doi: 10.1242/jcs.02412. [DOI] [PubMed] [Google Scholar]
- 33.Dhasarathy A, Kajita M, Wade PA. The transcription factor snail mediates epithelial to mesenchymal transitions by repression of estrogen receptor-alpha. Mol Endocrinol. 2007;21(12):2907–2918. doi: 10.1210/me.2007-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng L, Zha Z, Lang B, Liu J, Yao X. Heregulin-beta1 promotes metastasis of breast cancer cell line SKBR3 through upregulation of Snail and induction of epithelial-mesenchymal transition. Cancer Lett. 2009;280(1):50–60. doi: 10.1016/j.canlet.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 35.Ohkubo T, Ozawa M. The transcription factor Snail downregulates the tight junction components independently of E-cadherin downregulation. J Cell Sci. 2004;117(Pt 9):1675–1685. doi: 10.1242/jcs.01004. [DOI] [PubMed] [Google Scholar]
- 36.Barrallo-Gimeno A, Nieto MA. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 2005;132(14):3151–3161. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- 37.Wu Y, Zhou BP. Snail: More than EMT. Cell Adh Migr. 2010;4(2):199–203. doi: 10.4161/cam.4.2.10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schlüter MA, Pfarr CS, Pieczynski J, Whiteman EL, Hurd TW, Fan S, Liu CJ, Margolis B. Trafficking of Crumbs3 during cytokinesis is crucial for lumen formation. Mol Biology Cell. 2009;20(22):4652–4663. doi: 10.1091/mbc.E09-02-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weisz OA. Acidification and protein traffic. Int Rev Cytol. 2003;226:259–319. doi: 10.1016/s0074-7696(03)01005-2. [DOI] [PubMed] [Google Scholar]
- 40.Vastag M, Neuhofer W, Nagel W, Beck FX. Ammonium affects tight junctions and the cytoskeleton in MDCK cells. Pflugers Arch. 2005;449(4):384–391. doi: 10.1007/s00424-004-1341-y. [DOI] [PubMed] [Google Scholar]
- 41.Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA. Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. J Clin Invest. 2009;119(6):1438–1449. doi: 10.1172/JCI38019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300(5618):445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 43.Dall’Olio F, Chiricolo M. Sialyltransferases in cancer. Glycoconj J. 2001;18(11–12):841–850. doi: 10.1023/a:1022288022969. [DOI] [PubMed] [Google Scholar]
- 44.Wang PH. Altered Glycosylation in Cancer: Sialic Acids and Sialyltransferases. J Cancer Mol. 2005;1(2):73–81. [Google Scholar]
- 45.Geisler C, Jarvis DL. Letter to the Glyco-Forum: Effective glycoanalysis with Maackia amurensis lectins requires a clear understanding of their binding specificities. Glycobiology. 2011;21(8):988–993. doi: 10.1093/glycob/cwr080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verges M. Retromer: multipurpose sorting and specialization in polarized transport. Int Rev Cell Mol Biol. 2008;271:153–198. doi: 10.1016/S1937-6448(08)01204-5. [DOI] [PubMed] [Google Scholar]
- 47.Pocha SM, Wassmer T, Niehage C, Hoflack B, Knust E. Retromer controls epithelial cell polarity by trafficking the apical determinant Crumbs. Curr Biol. 2011;21(13):1111–1117. doi: 10.1016/j.cub.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 48.Zhou B, Wu Y, Lin X. Retromer regulates apical-basal polarity through recycling Crumbs. Dev Biol. 2011;360(1):87–95. doi: 10.1016/j.ydbio.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 49.Yu CY, Chen JY, Lin YY, Shen KF, Lin WL, Chien CL, ter Beest MB, Jou TS. A bipartite signal regulates the faithful delivery of apical domain marker podocalyxin/Gp135. Mol Biol Cell. 2007;18(5):1710–1722. doi: 10.1091/mbc.E06-07-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shore EM, Nelson WJ. Biosynthesis of the cell adhesion molecule uvomorulin (E-cadherin) in Madin-Darby canine kidney epithelial cells. J Biol Chem. 1991;266(29):19672–19680. [PubMed] [Google Scholar]
- 51.Le TL, Yap AS, Stow JL. Recycling of E-cadherin: a potential mechanism for regulating cadherin dynamics. J Cell Biol. 1999;146(1):219–232. [PMC free article] [PubMed] [Google Scholar]
- 52.Desclozeaux M, Venturato J, Wylie FG, Kay JG, Joseph SR, Le HT, Stow JL. Active Rab11 and functional recycling endosome are required for E-cadherin trafficking and lumen formation during epithelial morphogenesis. Am J Physiol Cell Physiol. 2008;295(2):C545–556. doi: 10.1152/ajpcell.00097.2008. [DOI] [PubMed] [Google Scholar]
- 53.Morimoto S, Nishimura N, Terai T, Manabe S, Yamamoto Y, Shinahara W, Miyake H, Tashiro S, Shimada M, Sasaki T. Rab13 mediates the continuous endocytic recycling of occludin to the cell surface. J Biol Chem. 2005;280(3):2220–2228. doi: 10.1074/jbc.M406906200. [DOI] [PubMed] [Google Scholar]
- 54.Fujita Y, Krause G, Scheffner M, Zechner D, Leddy HE, Behrens J, Sommer T, Birchmeier W. Hakai, a c-Cbl-like protein, ubiquitinates and induces endocytosis of the E-cadherin complex. Nat Cell Biol. 2002;4(3):222–231. doi: 10.1038/ncb758. [DOI] [PubMed] [Google Scholar]
- 55.Traweger A, Fang D, Liu YC, Stelzhammer W, Krizbai IA, Fresser F, Bauer HC, Bauer H. The tight junction-specific protein occludin is a functional target of the E3 ubiquitin-protein ligase itch. J Biol Chem. 2002;277(12):10201–10208. doi: 10.1074/jbc.M111384200. [DOI] [PubMed] [Google Scholar]
- 56.Lin Y, Wu Y, Li J, Dong C, Ye X, Chi YI, Evers BM, Zhou BP. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J. 2010;29(11):1803–1816. doi: 10.1038/emboj.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Amith SR, Jayanth P, Franchuk S, Finlay T, Seyrantepe V, Beyaert R, Pshezhetsky AV, Szewczuk MR. Neu1 desialylation of sialyl alpha-2,3-linked beta-galactosyl residues of TOLL-like receptor 4 is essential for receptor activation and cellular signaling. Cell Signal. 2010;22(2):314–324. doi: 10.1016/j.cellsig.2009.09.038. [DOI] [PubMed] [Google Scholar]
- 58.Morell AG, Gregoriadis G, Scheinberg IH, Hickman J, Ashwell G. The role of sialic acid in determining the survival of glycoproteins in the circulation. J Biol Chem. 1971;246(5):1461–1467. [PubMed] [Google Scholar]
- 59.Richards AA, Colgrave ML, Zhang J, Webster J, Simpson F, Preston E, Wilks D, Hoehn KL, Stephenson M, Macdonald GA, Prins JB, Cooney GJ, Xu A, Whitehead JP. Sialic acid modification of adiponectin is not required for multimerization or secretion but determines half-life in circulation. Mol Endocrinol. 2010;24(1):229–239. doi: 10.1210/me.2009-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brockhausen I. Mucin-type O-glycans in human colon and breast cancer: glycodynamics and functions. EMBO Rep. 2006;7(6):599–604. doi: 10.1038/sj.embor.7400705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Harduin-Lepers A, Krzewinski-Recchi MA, Colomb F, Foulquier F, Groux-Degroote S, Delannoy P. Sialyltransferases functions in cancers. Front Biosci (Elite Ed) 2012;4:499–515. doi: 10.2741/e396. [DOI] [PubMed] [Google Scholar]
- 62.Burchell J, Poulsom R, Hanby A, Whitehouse C, Cooper L, Clausen H, Miles D, Taylor-Papadimitriou J. An alpha2,3 sialyltransferase (ST3Gal I) is elevated in primary breast carcinomas. Glycobiology. 1999;9(12):1307–1311. doi: 10.1093/glycob/9.12.1307. [DOI] [PubMed] [Google Scholar]
- 63.Perez-Garay M, Arteta B, Pages L, de Llorens R, de Bolos C, Vidal-Vanaclocha F, Peracaula R. alpha2,3-sialyltransferase ST3Gal III modulates pancreatic cancer cell motility and adhesion in vitro and enhances its metastatic potential in vivo. PLoS One. 2010;5(9) doi: 10.1371/journal.pone.0012524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miyagi T, Wada T, Yamaguchi K, Shiozaki K, Sato I, Kakugawa Y, Yamanami H, Fujiya T. Human sialidase as a cancer marker. Proteomics. 2008;8(16):3303–3311. doi: 10.1002/pmic.200800248. [DOI] [PubMed] [Google Scholar]
- 65.Hedlund M, Ng E, Varki A, Varki NM. alpha 2–6-Linked sialic acids on N-glycans modulate carcinoma differentiation in vivo. Cancer Res. 2008;68(2):388–394. doi: 10.1158/0008-5472.CAN-07-1340. [DOI] [PubMed] [Google Scholar]
- 66.Sata T, Roth J, Zuber C, Stamm B, Heitz PU. Expression of alpha 2,6-linked sialic acid residues in neoplastic but not in normal human colonic mucosa. A lectin-gold cytochemical study with Sambucus nigra and Maackia amurensis lectins. Am J Pathol. 1991;139(6):1435–1448. [PMC free article] [PubMed] [Google Scholar]
- 67.Natsugoe S, Uchikado Y, Okumura H, Matsumoto M, Setoyama T, Tamotsu K, Kita Y, Sakamoto A, Owaki T, Ishigami S, Aikou T. Snail plays a key role in E-cadherin-preserved esophageal squamous cell carcinoma. Oncol Rep. 2007;17(3):517–523. [PubMed] [Google Scholar]
- 68.Hakomori S. Glycosylation defining cancer malignancy: new wine in an old bottle. Proc Natl Acad Sci U S A. 2002;99(16):10231–10233. doi: 10.1073/pnas.172380699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reis CA, Osorio H, Silva L, Gomes C, David L. Alterations in glycosylation as biomarkers for cancer detection. J Clin Pathol. 2010;63(4):322–329. doi: 10.1136/jcp.2009.071035. [DOI] [PubMed] [Google Scholar]
- 70.Maupin KA, Sinha A, Eugster E, Miller J, Ross J, Paulino V, Keshamouni VG, Tran N, Berens M, Webb C, Haab BB. Glycogene expression alterations associated with pancreatic cancer epithelial-mesenchymal transition in complementary model systems. PLoS One. 2010;5(9):e13002. doi: 10.1371/journal.pone.0013002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Whiteman EL, Chen JJ, Birnbaum MJ. Platelet-derived growth factor (PDGF) stimulates glucose transport in 3T3-L1 adipocytes overexpressing PDGF receptor by a pathway independent of insulin receptor substrates. Endocrinology. 2003;144(9):3811–3820. doi: 10.1210/en.2003-0480. [DOI] [PubMed] [Google Scholar]
- 72.Straight SW, Pieczynski JN, Whiteman EL, Liu CJ, Margolis B. Mammalian lin-7 stabilizes polarity protein complexes. J Biol Chem. 2006;281(49):37738–37747. doi: 10.1074/jbc.M607059200. [DOI] [PubMed] [Google Scholar]
- 73.Marshall OJ. PerlPrimer: cross-platform, graphical primer design for standard, bisulphite and real-time PCR. Bioinformatics. 2004;20(15):2471–2472. doi: 10.1093/bioinformatics/bth254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.