Abstract
Background
Ethanol is metabolized by a two-step process in which alcohol dehydrogenase (ADH) oxidizes ethanol to acetaldehyde, which is further oxidized to acetate by aldehyde dehydrogenase (ALDH). Although variation in ethanol metabolism in humans strongly influences the propensity to chronically abuse alcohol, few data exist on the behavioral effects of altered ethanol metabolism. Here, we used the nematode C. elegans to directly examine how changes in ethanol metabolism alter behavioral responses to alcohol during an acute exposure. Additionally, we investigated ethanol solution osmolarity as a potential explanation for contrasting published data on C. elegans ethanol sensitivity.
Methods
We developed a gas chromatography assay and validated a spectrophotometric method to measure internal ethanol in ethanol-exposed worms. Further, we tested the effects of mutations in ADH and ALDH genes on ethanol tissue accumulation and behavioral sensitivity to the drug. Finally, we tested the effects of ethanol solution osmolarity on behavioral responses and tissue ethanol accumulation.
Results
Only a small amount of exogenously applied ethanol accumulated in the tissues of C. elegans and consequently their tissue concentrations were similar to those that intoxicate humans. Independent inactivation of an ADH-encoding gene (sodh-1) or an ALDH-encoding gene (alh-6 or alh-13) increased the ethanol concentration in worms and caused hypersensitivity to the acute sedative effects of ethanol on locomotion. We also found that the sensitivity to the depressive effects of ethanol on locomotion is strongly influenced by the osmolarity of the exogenous ethanol solution.
Conclusions
Our results indicate that ethanol metabolism via ADH and ALDH has a statistically discernable but surprisingly minor influence on ethanol sedation and internal ethanol accumulation in worms. In contrast, the osmolarity of the medium in which ethanol is delivered to the animals has a more substantial effect on the observed sensitivity to ethanol.
Introduction
Alcohol abuse is a common disorder influenced by both genetics and environment. Despite strong evidence of a role for genetics in abuse liability, few specific susceptibility candidate genes for alcoholism have emerged. Natural variations in components of the ethanol metabolism machinery are among the very few identified genetic causes of the variation in alcohol abuse liability in humans (recently reviewed by Pautassi et al., 2010). Ethanol is metabolized to acetaldehyde by alcohol dehydrogenase (ADH); subsequently, acetaldehyde is metabolized to acetate by an aldehyde dehydrogenase (ALDH). Certain isoforms of ADH that are more enzymatically active are protective for susceptibility for alcoholism. Similarly, ALDH isoforms with decreased enzymatic activity also decrease disease liability. Both types of alleles are predicted to increase acetaldehyde levels, which appear to be aversive, suggesting a mechanism for the decrease in susceptibility to alcohol abuse disorders in individuals carrying them. While there are significant correlations between inheritance of these ADH or ALDH alleles and rates of alcohol abuse (Chen et al., 2009; Crabb et al., 2004; Edenberg et al., 2006; Kuo et al., 2008), there is little experimental detail on the behavioral consequences during acute ethanol treatment of variation in alcohol metabolism.
The nematode worm, C. elegans, has been increasingly exploited as a behavioral model for understanding the genetic contributions to ethanol responses (Bettinger and McIntire, 2004; Davies et al., 2003, 2004; Davis et al., 2008; Graham et al., 2009; Kapfhamer et al., 2008; Lee et al., 2009; Mitchell et al., 2007, 2010; Morgan and Sedensky, 1995; Speca et al., 2010). Worms are an excellent model for this work because of the extremely well conserved neurobiology between worms and humans (Bargmann, 1998). Worms show a dose-dependent depression of several behaviors when treated with ethanol (Davies et al., 2003, 2004; Morgan and Sedensky, 1995). Several laboratories are now exploring the mechanisms by which ethanol exerts its behavioral or developmental effects, but little effort has been made to determine if metabolism has a significant role in modulating behavioral responses to ethanol in this model. Here, we directly examine the effect of altering ethanol metabolism on the behavioral response to ethanol in the worm.
Methods
Nematode culture and strains
C. elegans were maintained using standard methods (Brenner, 1974). Strains used were: N2 var. Bristol, sodh-1(ok2799), sodh-1(bet20).
RNAi
RNAi induction was performed as described (Kamath et al., 2001). Cultures of bacteria containing RNAi vectors from the RNAi feeding library generated by J. Ahringer at the University of Cambridge (Geneservice, Cambridge, UK) were grown in LB supplemented with 50 μg/mL ampicillin for 12 - 18 hours at 37°C with shaking. Nematode Growth Medium (NGM) plates containing 1mM IPTG and 25 μg/mL ampicillin were seeded with bacteria containing RNAi vectors, and were incubated at room temperature for 24 hours. 3-5 L4 stage wild-type N2 or sodh-1(ok2799) worms were placed on the seeded plates and incubated at 20°C for 36-40 hours. Adult worms were moved to new RNAi plates and allowed to lay eggs for 1-2 hours. Plates were incubated at 20°C to allow worms to develop to adulthood. First day adults were collected and subjected to allyl-alcohol survival assays. Knockdown of target gene expression levels was confirmed using quantitative RT-PCR (Table 2). Primers used in quantitative RT-PCR experiments are in Supplemental Table 1.
Table 2.
Gene Expression Ratios of Wild-Type Worms Treated with RNAi Targeted Against ADH or ALDH Genes.
| Strain | Target gene | Gene expression ratio | SEM |
|---|---|---|---|
| sodh-1(RNAi) | sodh-1 | 0.30 | 0.09 |
| D2063.1(RNAi) | D2063.1 | 0.02 | 0.01 |
| H24K24.3(RNAi) | H24K24.3 | 0.39 | 0.04 |
| alh-1(RNAi) | alh-1 | 0.17 | 0.01 |
| alh-3(RNAi) | alh-3 | 0.08 | 0.03 |
| alh-5(RNAi) | alh-5 | 0.25 | 0.03 |
| alh-6(RNAi) | alh-6 | 0.40 | 0.04 |
| alh-7(RNAi) | alh-7 | 0.47 | 0.08 |
| alh-8(RNAi) | alh-8 | 0.09 | 0.01 |
| alh-9(RNAi) | alh-9 | 0.02 | 0.01 |
| alh-10(RNAi) | alh-10 | 0.50 | 0.07 |
| alh-11(RNAi) | alh-11 | 0.45 | 0.06 |
| alh-13(RNAi) | alh-13 | 0.33 | 0.05 |
Allyl-alcohol survival assays
Allyl-alcohol survival was used to assay the function of ADH as described (Williamson et al., 1991).
Analysis of Locomotion (crawling)
Age-matched 55 hour-old adult animals reared at 20°C were used. Locomotion on plates was assayed as described (Davies et al., 2003). Ten worms for each strain were tested. Two minute movies were recorded, and movies were analyzed using ImagePro Plus Version 6 (Media Cybernetics, Bethesda, MD, U.S.A.). To account for differences in basal speeds, we calculated a relative speed (ethanol-treated average speed / untreated average speed × 100).
Test strains were compared to controls assayed simultaneously on the same plates. A one-way ANOVA was performed with a significance value of P < 0.05 with Dunnett's post-hoc tests comparing each mutant and knockdown strain to N2. t-tests were performed to determine significant differences between timepoints.
Time course analysis was performed as above except that single animals were placed on plates and video images were recorded every 0.5 seconds for 15 minutes. The speed for each interval was averaged for 6 worms. The speeds were binned for each 1-minute period and an average was calculated. A one-way ANOVA was performed with a significance value of P < 0.01 using Prism v5.0 (GraphPad Software), with Bonferroni post-hoc tests comparing all bins.
The effects of osmolarity on crawling were assessed as above, except that assay plates were either made of standard NGM or of Dent's Saline Solution (10 mM D-glucose, 10 mM HEPES, 140 mM NaCl, 6 mM KCl, 3 mM CaCl2, 1 mM MgCl2, pH 7.4) with 2% agar.
Analysis of Locomotion (swimming)
The effect of ethanol on swimming was assessed for both individuals and groups of animals. Individuals were video recorded (30 frames per second) in liquid medium (3 mL) over a 2% agarose surface in 6 cm diameter plates to promote continuous swimming. The midlines of the worms were determined with a custom-written image analysis macro using ImagePro as described (Pierce-Shimomura et al., 2008). Animals were gently transferred to identical conditions that contained 400 mM ethanol. Groups of animals were video recorded in identical conditions. The number of head bends made by each individual in a 30-second time window was quantified at 5, 10, 15 and 20 minutes of exposure to ethanol.
Internal Ethanol Concentration calculations
Exposure to ethanol
55-hour old worms reared at 20°C were used. Several hundred worms were placed on unseeded plates containing the appropriate concentration of ethanol (prepared as for locomotion assays, see above) for 10 or 50 minutes. Worms were photographed for subsequent size analysis. Exactly 200 worms were picked from plates to a tube containing 20 μl ddH2O. Tubes were placed at −80°C until analysis. Worms were thawed on ice and ground in the tube with a pestle (Kontes pellet pestle, Fisher Scientific, USA). Worm homogenate was stored at −20°C.
For 200 worms: Picking took an average of 1.5 minutes per condition. Picking itself caused a loss of between 0 μL and 0.25 μL of the water, evaporation in that time caused an undetectable loss of volume.
Size calculation
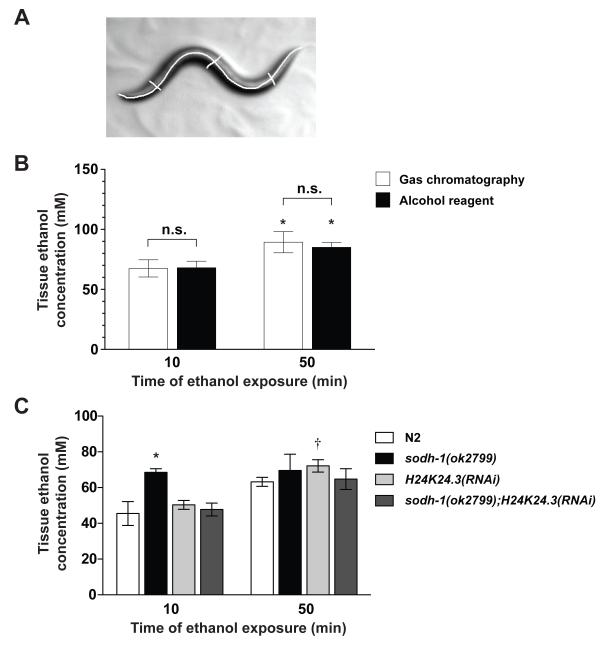
Photographs were analyzed using ImagePro Plus v6. Ten worms were used per strain per condition. A midline was drawn to determine the height, h. Three diameters were drawn, one at the vulva (the approximate center of the animal), one each at the approximate midpoint between the vulva and head or tail (Figure 2a), these lengths were averaged and used as the diameter, ½ of this length was the radius, r. Volume was determined as for a cylinder: volume = πr2h.
Figure 2. Determination of internal ethanol concentration in wild-type and ADH compromised animals.
(a) An example of worm volume measurements. The volume of a worm was determined by taking photographs of animals immediately before they were used for the biochemical analysis. Each animal was traced (n=10 for each strain) to determine the length, h. The diameter of the animal, d, was the average of three widths, one at the vulva, and one each at the midpoint between the vulva and head or tail. The volume was calculated using formula for the volume of a cylinder (volume = πr2h). (b) There is no difference in internal ethanol concentration measurements using gas chromatograph (GC) and spectrophotometric analysis at 500 mM when actual worm volume was used in the calculation. A significant difference is observed between time points (GC: 10 minutes vs. 50 minutes P < 0.05; Spectrophotometric analysis: 10 minutes vs. 50 minutes P < 0.05). (c) Internal ethanol measurements of 200 worms, treated with 400 mM exogenous ethanol, calculated from spectrophotometric analysis. sodh-1(ok2799) was different from N2 at 10 minutes but not at 50 minutes. H24K24.3(RNAi) was not different from N2 at either timepoint. sodh-1(ok2799);H24K24.3(RNAi) was not different from N2 at either timepoint. * Significantly different from N2 at the same timepoint (P < 0.05) † Significantly different from the same strain across timepoints (P < 0.05).
Calculation of internal ethanol concentration
We calculated the internal ethanol concentration using the equation C1V1 = C2V2, where C1 = concentration of ethanol in homogenate, V1 = (20 μL + volume of 200 worms), V2 = volume of 200 worms. We solved for C2.
Gas Chromatography ethanol concentration analysis
Homogenates were tested for ethanol concentration using a Hewlett Packard model 5890A gas chromatograph (GC) equipped with a flame ionization detector and 2 meter 5% Carbowax 20M 80/120 mesh packed column (Restek, Bellefonte, PA). Samples were kept frozen at −20°C until analysis. Injections were accomplished manually with a 10 μl gas-tight glass syringe. The GC parameters were: 5 μl injection volume, 7 minute sample run time, injector temperature 200°C, oven temperature isothermal 90°C, detector temperature 220°C, helium carrier gas flow rate 30 ml/min, hydrogen flame flow rate 25 ml/min and air flow rate 400 ml/min. Data were collected and analyzed by Clarity GC software (Apex Data Systems, Prague, CZ) using a linear regression analysis with no weighting. A 7 point calibration curve preceded the analysis of ethanol concentrations. Ethanol concentrations were calculated by the external standard method. Quality control ethanol standards preceded and followed each pair of samples. Accuracy of results was inferred if the assayed ethanol concentrations from quality control ethanol standards varied by no more than 10% from actual concentrations. All quality control standards met this criterion. If sufficient homogenate was available, each sample was tested in duplicate and a mean of the two values was used in subsequent calculations. Each ethanol concentration data point represents a mean (±SEM) generated from three independent samples.
Internal Ethanol concentration analysis using spectrophotometric analysis
C. elegans homogenates were tested for ethanol concentration according to the manufacturer's directions using an Alcohol Reagent Kit (Pointe Scientific, Canton, MI, USA). 1.5 μL of worm homogenate was added to 300 μL ice-cold alcohol reagent, incubated for five minutes at 30°C, and the reaction was stopped by putting the tube into an ice-cold aluminum block. Alcohol concentration was determined by measuring the absorbance at 340 nm.
Results
Identification and inactivation of alcohol dehydrogenase genes
There are many genes with homology to human ADH in the C. elegans genome, and ADH activity has been directly demonstrated in worms using a spectrophotometric assay (Williamson et al., 1991). Our goal was to examine the behavioral consequences of impairing alcohol metabolism, and therefore we chose a subset of the genes with strong homology to human liver ADHs as good candidates for subsequent analysis. We used two methods to identify potential ADHs that metabolize ethanol in the worm. First, we identified sodh-1, a gene that showed transcriptional regulation in response to a prolonged treatment with very high concentrations (7%) of ethanol (Kwon et al., 2004) and has been annotated as an ADH. We also tested the two genes with highest homology to sodh-1: sodh-2 and D2063.1. Second, we used the primary amino acid sequence of human liver ADH (Human liver ADH proteins, ADH1A (Accession: NP_000658.1), ADH1B (Accession: NP_000659.2), and ADH1C (Accession: NP_000660.1)) in BLAST searches to identify the C. elegans protein with the highest homology, H24K24.3. Although previous work had identified transcripts from both sodh-1 and H24K24.3 as being expressed from ADH-encoding genes, these genes have not previously been functionally characterized (Glasner et al., 1995; Waterston et al., 1992).
Disruption of either of two ADH genes, sodh-1 or H24K24.3, confers resistance to allyl-alcohol toxicity
We tested a subset of the genes identified by homology above for their ability to metabolize ethanol using an allyl-alcohol toxicity assay that has been previously used extensively to identify mutants with defects in ethanol metabolism in worms and in other organisms (Williamson et al. 1991). Allyl-alcohol is metabolized by ADH into the toxin acrolein, and wild-type worms that are grown on allyl-alcohol plates die within 24 hours, whereas worms with defects in ADH function are resistant to allyl-alcohol toxicity ((Williamson et al., 1991) and Table 1). Inactivation of sodh-1 conferred profound resistance to allyl-alcohol-induced lethality (Table 1). Previously, Williamson et al. (1991) performed a genetic selection using allyl-alcohol survival to isolate mutations in putative ADHs in the worm, and identified AL2B, and we replicated the allyl-alcohol resistance of this mutant (Table 1). We identified a G to A point mutation in sodh-1 in AL2B, which would result in a Glycine 158 to Glutamate missense mutation (for consistency of nomenclature, AL2B is hereafter referred to as sodh-1(bet20)). Additionally, RNA inactivation of H24K24.3 conferred resistance to allyl-alcohol, however, knock-down of function with RNAi of neither sodh-2 nor D2063.1 was able to confer strong resistance to allyl-alcohol (Table 1). It is important to note that these RNAi experiments do not unambiguously rule out a role for sodh-2 or D2063.1 in the metabolism of ethanol. This RNAi treatment does not completely eliminate the mRNA for either candidate ADH (Table 2), and the residual mRNA may be sufficient to confer function of the candidate. In addition, C. elegans tissues are differentially susceptible to RNAi. Worm neurons are quite refractive to RNAi using this method, and it is possible that expression of the candidate gene is preserved in neurons. In this case, expression of the gene in neurons may be sufficient to provide function. Because inactivation of both sodh-1 and H24K24.3 altered allyl-alcohol sensitivity, we therefore used strains inactivated for sodh-1 and H24K24.3 in our subsequent studies.
Table 1. Sensitivity of Different Genotypes to 0.35% Allyl-alcohol Induced Lethality.
| Genotype | Lethality (%) ± SEM | Number of trials |
|---|---|---|
| N2 (wild type) | 96.6 ± 0.03 | 17 |
| sodh-1(ok2799) | 1.4 ± 0.01 | 14 |
| sodh-1(RNAi) | 0 ± 0.00 | 6 |
| sodh-1(bet20)/AL2B | 0 ± 0.00 | 2 |
| H24K24.3(RNAi) | 17.7 ± 0.05 | 11 |
| sodh-2(RNAi) | 80.0 ± 0.02 | 4 |
| D2063.1(RNAi) | 85.0 ± 0.09 | 5 |
Young-adult worms were exposed to 0.35% allyl-alcohol for 24 hours. Lethality was assessed at 24 hours of exposure.
Determination of the timecourse of intoxication in wild-type animals
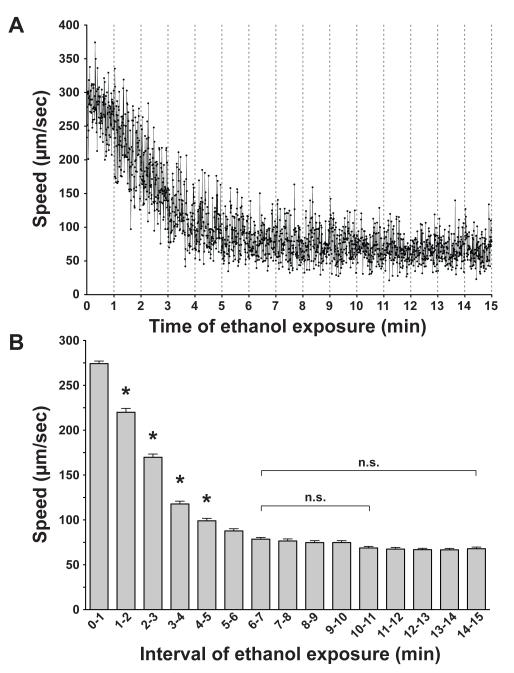
To determine the appropriate time points during ethanol treatment at which to assess the biochemical and behavioral effects of compromising ADH function, we observed the kinetics of the onset of intoxication in wild-type animals. We placed individual worms (n=6) in a copper ring on a plate with 500 mM exogenous ethanol, and recorded their locomotion continuously for 15 minutes, starting immediately after the animals had been placed on the plate. Animals rapidly decrease speed in the first few minutes of ethanol exposure, and reach a plateau at 6 minutes, suggesting that the animals accumulate enough ethanol in this time to become maximally impaired (Figure 1a). When we examined the data in 1-minute bins, we found that the neighboring bins were not different after 6 minutes (P > 0.05, Figure 1b). We chose 10 minutes exposure as an initial time point for biochemical and behavioral testing for convenience and consistency with previous experimental designs (Davies et al., 2003, 2004; Kapfhamer et al., 2008).
Figure 1. Time course for ethanol effects on speed of locomotion.
(a) The mean speed (μm/sec ± SEM) of young adult wild-type animals (n = 6) exposed to 500 mM exogenous ethanol in the absence of food is shown for every 0.5 seconds over a 15-minute period beginning immediately after ethanol exposure. (b) Speeds within each minute of ethanol exposure were binned and a mean (n = 120) calculated for each bin. Significant decreases in speed are only seen in the first 5 minutes when each bin is compared with the previous bin (*, P < 0.01). Bin 6-7 minutes is not significantly different from any bin that follows. n.s., not significantly different.
Determination of the internal ethanol concentration in wild-type and ADH-compromised animals
Worms generate a cuticle that is impermeable to many pharmacological agents (Burns et al., 2010; Cox et al., 1981), and we have previously measured internal concentrations approximately 20x lower than the exogenous dose (Davies et al., 2003, 2004; Kapfhamer et al., 2008). However, the determination of internal ethanol concentration has been the subject of some conflicting reports; using different methodology, others have reported that more than half of the exogenous ethanol accumulates in internal tissues within 20 minutes (Mitchell et al., 2007). To unambiguously determine the internal concentration of ethanol that animals achieve in our behavioral assays, we exposed animals to ethanol on plates as we would for behavioral assays, picked exactly 200 worms from the plate into a known volume of liquid, homogenized them, and performed gas chromatography analysis on the homogenate. All ethanol in the solution would come from the worms, and all ethanol that was brought with the worms should be captured in the solution, so by determining the volume of the worms we could calculate the ethanol concentration of the worms. This calculation of internal ethanol concentration is dependent on the size of the worms tested and animals of different genotypes differed considerably in size. We estimated the volume of the worms by calculating the volume of a cylinder of the length and average diameter of the worms we tested (Figure 2a). Consistent with our previously reported results, we found that internal ethanol concentration remains much lower than the exogenous dose. However, using this very precise method, we determined that the internal concentration of ethanol in these experiments is approximately 2x what we have observed in the past using different tissue preparation methods. For wild-type N2 animals exposed to 500 mM exogenous ethanol, at 10 minutes of exposure the tissue concentration was 67.5 ± 7.1 mM. The tissue concentration of ethanol increased over time; at 50 minutes of exposure, the tissue concentration was 89.3 ± 8.8 mM (P = 0.02 vs. 10 minutes; Figure 2b). This indicates that the development of acute tolerance that we observe over 50 minutes of exposure (Davies et al., 2004) does not reflect a decrease in internal tissue ethanol.
We sought an explanation for the differences in our previously published concentrations and what we have found here using this very sensitive protocol (Davies et al., 2003, 2004; Kapfhamer et al., 2008). Examining our previous protocol, we identified a major confound in the estimation of worm volume in the assay; we made an assumption that, in a pellet of worms that remains following centrifugation and removal of the supernatant, worms made up the vast majority of the volume of the pellet. However, when we calculated the volume of worms in a pellet by counting the worms and determining their volume, we found that this volume could vary by more than 2 fold depending on the size of the pellet and adult age of the worms, and at the most accounted for less than half of the volume (data not shown). When we used a known number of worms of known volume, our original spectrophotometric assay protocol closely recapitulated the internal ethanol concentration determined by gas chromatography (Figure 2b), demonstrating that use of this simple protocol is acceptable for future internal ethanol concentration measurements in C elegans.
We predicted that the internal concentration of ethanol would be higher in animals in which ethanol metabolism is compromised. We determined the effect of inactivation of the two ADHs both singly and in combination on the accumulation of ethanol at the dose of exogenous ethanol at which we tested behavior, 400 mM. We determined the internal ethanol concentration after 10 and 50 minutes of ethanol exposure, to observe an acute accumulation of ethanol (10 minutes) as well as accumulation that could potentially be altered by the development of acute tolerance (50 minutes). Loss of sodh-1 significantly increased the internal ethanol concentration in animals relative to wild type (Figure 2c). However, inactivation of H24K24.3 did not significantly increase the internal ethanol concentration (Figure 2c). Interestingly, when we examined animals in which both ADHs were inactivated, sodh-1(ok2799);H24K24.3(RNAi), the concentration of ethanol that the animals accumulated was similar to that of wild type at 10 minutes (Figure 2c), suggesting the possibility that loss of H24K24.3 may trigger a compensatory response that minimizes the effect seen for loss of sodh-1 alone.
Assessment of the ethanol-responsive behavioral consequences of compromising ADH function
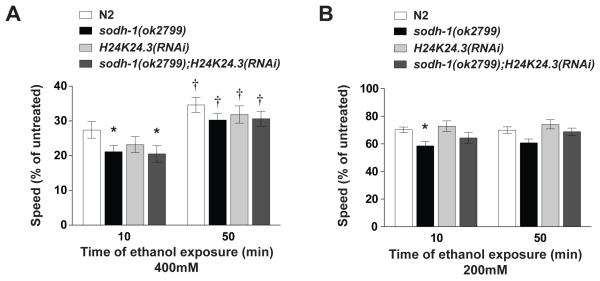
We assessed the behavioral effects of altering ethanol metabolism on the locomotion of worms. We exposed wild-type and ADH-defective animals to exogenous ethanol and recorded their locomotion on plates after 10 minutes of exposure. 400 mM was chosen as the upper dose for locomotion analyses using mutant or knockdown strains rather than 500 mM because we hypothesized that the metabolism-defective animals might show hypersensitivity to ethanol and we wanted to eliminate possible floor effects at 500 mM. Loss of sodh-1 conferred mild but significant hypersensitivity to ethanol when the animals were tested on low (200 mM) and high concentrations (400 mM) of the drug (Figure 3a and 3b). Reduction of function of H24K24.3 did not change the effects of ethanol on locomotion significantly, although there may be a trend towards increased sensitivity. Inactivating both sodh-1 and H24K24.3 did not increase the effect of ethanol on locomotion compared with the sodh-1(ok2799) mutation alone (Figure 3a). These results indicate that the difference in ethanol accumulation seen with sodh-1(ok2799) mutant animals is sufficient to alter the behavior of the animals when they are exposed to ethanol. Surprisingly, the measured internal ethanol concentration for animals with a loss of both sodh-1 and H24K24.3 was not different from wild-type animals (Figure 2c) and yet these animals show a significant increase in ethanol sensitivity compared with wild-type animals (Figure 3a).
Figure 3. ADH compromised animals demonstrate behavioral sensitivity to ethanol, and develop acute functional tolerance to ethanol.
Animals were treated continuously with exogenous ethanol, beginning at 10 minutes and at 50 minutes of exposure, two-minute digital movies were recorded, and speed was determined by ImagePro image analysis software. A % relative speed was calculated by dividing treated speed by untreated speed, to account for any baseline speed differences. (a) Animals treated with 400 mM exogenous ethanol. Locomotion of wild-type N2 worms is strongly suppressed by this dose of ethanol. sodh-1(ok2799) and sodh-1(ok2799);H24K24.3(RNAi) strains are more strongly affected than N2 by ethanol at this dose, but H24K24.3(RNAi) animals are not significantly different from N2. Additionally, at this dose of ethanol, ADH mutant animals develop acute functional tolerance; each strain moved significantly faster at 50 minutes than at 10 minutes. (b) Animals treated with 200 mM exogenous ethanol. At this dose, sodh-1(ok2799) and sodh-1(ok2799);H24K24.3(RNAi) strains showed enhanced behavioral sensitivity to ethanol relative to N2, but H24K24.3(RNAi) was not different from N2. At this dose, we do not observe development of acute functional tolerance.
Assessment of the ethanol-responsive behavioral consequences of compromising ALDH function
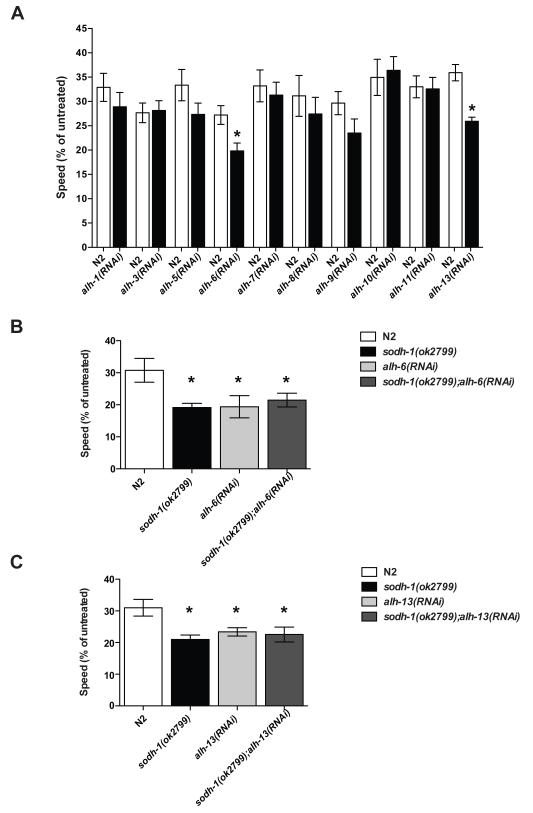
The second step of ethanol metabolism is the oxidation of acetaldehyde to acetate, catalyzed by aldehyde dehydrogenase (ALDH). We tested the effects of knocking down function of each of 10 C. elegans ALDH-like enzymes using RNAi. Inactivation of either alh-6 or alh-13 caused ethanol hypersensitivity (Figure 4a).
Figure 4. Behavioral effects of intoxication in wild-type and ALDH compromised animals.
Animals were treated with exogenous ethanol for 10 minutes, two-minute digital movies were recorded, and speed was determined by ImagePro image analysis software. A % relative speed was calculated by dividing treated speed by untreated speed, to account for any baseline speed differences. In each case, the alh knockdown strain was compared to N2 animals tested on the same plates. (a) Knockdown of either alh-6 or alh-13 conferred hypersensitivity to the effects of ethanol on locomotion (*, P < 0.05). (b, c) Knockdown of neither alh-6 nor alh-13 in the background of sodh-1(ok2799) was able to enhance the ethanol sensitivity beyond that of sodh-1(ok2799) alone.
If ADH and ALDH act in a linear pathway, the phenotype of loss of the upstream gene should not be enhanced by loss of the downstream gene. We tested this by knocking down function of either alh-6 or alh-13 in a sodh-1(ok2799) background, and found that the phenotype of the combinations was not different from that of sodh-1 alone (Figure 4b, 4c). This result provides further support for the linear nature of the metabolism pathway, and further strongly suggests that the ethanol hypersensitivity phenotypes of sodh-1(ok2799), alh-6(RNAi) and alh-13(RNAi) are due to changing ethanol metabolism.
The action of ADH is reversible; one possibility for the increase in ethanol sensitivity in alh-6 and alh-13 knock down animals is that the excess acetaldehyde in these animals is converted back to ethanol, thereby increasing the internal ethanol concentration. We observed an increase in internal ethanol concentration alh-6(RNAi) and alh-13(RNAi) but not alh-1(RNAi) (which did not show a change in behavior on ethanol) (Supplemental Figure 1), suggesting that one effect of loss of ALDH function may be through increasing the effective ethanol dose.
Ethanol responses in C. elegans are sensitive to osmolarity
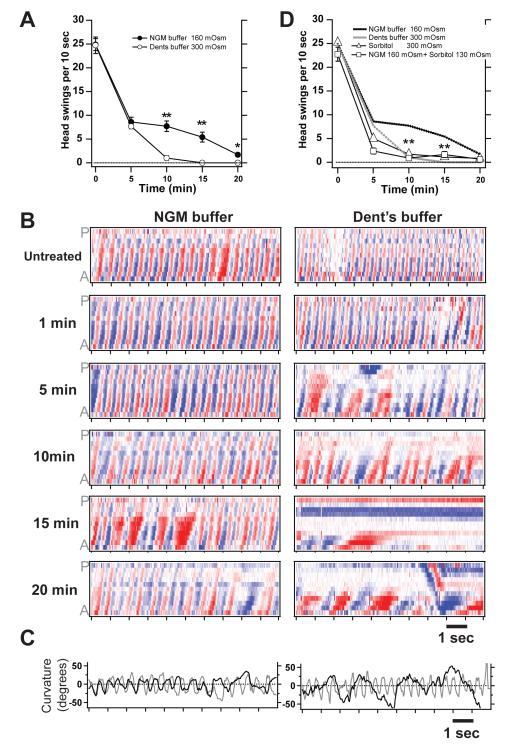
We next determined how ethanol affects C. elegans swimming in liquid. We found that ethanol inhibits swimming, although at a substantially higher dose than is required to inhibit crawling (Figure 5). While our results are in agreement with the first study of ethanol's effects on swimming in C. elegans (Morgan and Sedensky, 1995), they differ significantly from those reported by Mitchell et al. (2007), who used slightly different conditions. We found that the differences in our results from those of Mitchell et al. (2007) could be explained by the buffers used in our swimming analyses. Our study used Nematode Growth Media (NGM), which contains the same salts as the medium that animals are cultured on (Brenner, 1974), whereas the Mitchell study used Dent's buffer, a physiological saline normally used to record the electrical activity of dissected muscle (Avery et al., 1995). While the basal swimming behavior of worms in the two buffers was indistinguishable (data not shown), we observed a striking difference in behavior in the two buffers when ethanol was added. Figure 5a shows the time course of intoxication in 500 mM ethanol in the two buffers. Swimming movement was quantified as the number of head swings during a 10 second time window. The motion of the animals exposed to ethanol decreased in both conditions by 5 minutes of exposure. However, animals assayed in Dent's buffer became essentially immotile by 10 minutes, whereas animals assayed in NGM buffer decreased motion but remained motile during the entire 20 minutes of treatment.
Figure 5. Sensitivity to intoxication while swimming depends on exogenous osmolarity.
(a) Time course of intoxication while animals were swimming in 500 mM ethanol. Animals become significantly more immobilized by the same concentration of ethanol in Dent's buffer than in NGM buffer (**, P <0.001). (b) Body curvature matrices during intoxication for one representative animal in NGM buffer and one representative animal in Dent's buffer. Color intensity along the anterior-posterior (A-P) axis versus time represents the amount of bending at given points along the body (red = ventral, white = no bend, blue = dorsal). (c) Plots of neck curvature versus time. Untreated (grey) and 20 minute treatment in ethanol (black). (d) Time course of intoxication while animals were swimming in 500 mM ethanol. NGM and Dent's buffer data are replotted from panel (a). Animals treated with NGM + sorbitol or sorbitol alone were as sensitive to ethanol as those animals treated in Dent's buffer.
To examine the time course of intoxication during swimming more closely, we plotted matrices that represent body curvature along the anterior-posterior axis versus time (Figure 5b). In this scheme, upward slanting red and blue "waves" in the matrix represent ventral and dorsal bends that pass from head to tail to generate forward motion. The representative plot for an animal swimming in NGM buffer (Figure 5b, left column) shows little difference from its untreated condition even after 10 minutes in ethanol aside from a slight decrease in frequency and dampening of bends, which is evident in the muted red and blue color code. By contrast, the representative plot for an animal swimming in Dent's buffer (Figure 5b, right column) shows severe impairment of coordination, beginning at 5 minutes of exposure, reflected in the fact that many of the bends fail to propagate fully from head to tail. Portions of the animal also become immobile by 15 minutes, which is reflected in the fixed color pattern versus time. We also compared the swimming of untreated (grey line) versus 20-minute exposure to ethanol (black line) by plotting the "neck" curvature versus time (Figure 5c). The animal treated with ethanol in NGM buffer showed a slowing in frequency, while the animal in Dent's buffer showed a much slower bend frequency and a rise in maximal bend amplitudes.
While searching for factors that could explain the vastly different results obtained in the different buffers, we noticed that the two buffers differed greatly in osmolarity: NGM is 160 mOsm, while Dent's buffer is 300 mOsm. We tested if C. elegans' sensitivity to intoxication while swimming depends on osmolarity, and found that worms became rapidly intoxicated when assayed in NGM buffer in which we had adjusted the osmolarity to match Dent's buffer by adding 130 mOsm sorbitol (Figure 5d). Assaying worms in only 300 mOsm sorbitol without any salts reproduced a dose response for intoxication that was characteristic of Dent's buffer (Figure 5d). Moreover, we found that pre-incubation for 20 minutes in Dent's buffer or 300 mOsm sorbitol conferred enhanced sensitivity to intoxication when the animals were assayed in 150 mOsm NGM buffer (data not shown). Together, these results suggest that acute sensitivity to exogenous ethanol in C. elegans depends on osmolarity, that this sensitivity can be dynamically adjusted, and that this is an explanation that can resolve the conflicting reports of dose response sensitivity to ethanol while swimming.
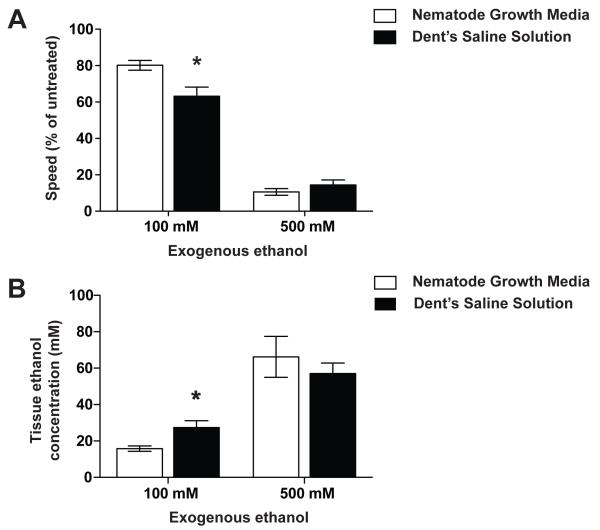
We tested the effect of osmolarity on the dose effect for ethanol on crawling, and found that the effect was significant, but more subtle than for swimming. At 10 minutes of exposure to 100 mM ethanol, the behavior of worms tested on NGM was less affected compared with worms tested on plates made with Dent's Saline (Figure 6a). We asked if the effect of osmolarity on the dose response for ethanol was due to altering the acute accumulation of ethanol. At 10 minutes of 100 mM ethanol exposure, worms on NGM plates accumulated significantly less ethanol than did animals on DS plates (Figure 6b).
Figure 6. Sensitivity to intoxication and tissue accumulation of ethanol while crawling depends on exogenous osmolarity.
(a) Animals were treated with exogenous ethanol for 10 minutes, two-minute digital movies were recorded, and speed was determined by ImagePro image analysis software. A % relative speed was calculated by dividing treated speed by untreated speed, to account for any baseline speed differences. Animals exposed to 100 mM exogenous ethanol for 10 minutes on NGM plates were less affected than animals exposed on Dent's Saline plates. (b) Animals exposed to ethanol on NGM plates accumulated significantly more tissue ethanol than animals exposed on Dent's Saline plates.
Discussion
The ADH enzymatic pathway is shared throughout evolutionary history. Modulation of ethanol metabolism has been shown to have a variety of biological effects across phyla when animals ingest large quantities of exogenous ethanol. In Drosophila melanogaster, functional variations in both ADH (Geer et al., 1989; Ogueta et al., 2010) and ALDH (Wolf et al, 2002; Fry and Saweikis, 2006; Fry et al., 2008) contribute to variation in ethanol sensitivity. In humans, functional variants of both ADH and ALDH have been correlated with altered susceptibility to becoming alcoholic. We have assessed the behavioral effects of altering alcohol metabolism in C. elegans.
We identified two ADHs in worms using a method described by Williamson et al. (1991), in which we exploited the fact that ADH metabolizes allyl-alcohol to the toxin acrolein. Inactivation of either sodh-1 or H24K24.3 them conferred resistance to allyl-alcohol – induced lethality. We inactivated these two genes, both singly and in combination, in our analysis of ADH pathway function in the behavioral responses to ethanol.
To determine if altering ADH function results in differences in tissue levels of ethanol, we developed a method to accurately measure the internal ethanol concentration in ethanol-exposed C. elegans, in which we used a known volume of worm tissue and measured ethanol concentration using gas chromatography. The concentrations that we measured reflect the relative lack of permeability of these nematodes to chemicals in their environment; for 500 mM exogenous ethanol, the wild-type internal concentration is in the range of 70-90 mM (Figure 2b). This translates to blood alcohol concentration (BAC) values of 0.32-0.41%, which would cause profound intoxication in a naïve human drinker. The degree of intoxication associated with these concentrations in C. elegans is also profound, on an agar medium worms exposed to 500 mM ethanol move at approximately 20% of their untreated speeds (Davies et al., 2003, 2004 and see Figure 3a) the amplitude of their body bends is significantly reduced (Davies et al., 2003), and their movement becomes severely uncoordinated.
Previously, we reported internal ethanol concentrations that were approximately half of the concentrations that we observe here (Davies et al., 2003, 2004; Kapfhamer et al., 2008). We found that we had overestimated the volume of worms in a pellet derived from spinning worms out of a solution because we assumed that the vast majority of the pellet was made of worm tissue, which we have found here to be incorrect. This observation can also explain why Mitchell et al. (2007) vastly overestimated the internal concentration in their animals; in their paradigm, they incubated worms in a high concentration of ethanol and then tested a pellet consisting of the resulting worm + ethanol solution. Our results suggest that the mixture probably contained relatively less worm tissue and more ethanol solution than they estimated, which would have contributed a significant amount of ethanol to the final concentration.
There is substantial evidence from human studies that an individual's naïve level of ethanol response is a predisposing factor in the development of alcoholism (Heath et al., 1999; Rodriguez et al., 1993; Schuckit et al., 2001; Schuckit, 1994; Volavka et al., 1996). Factors influencing an individual's level of response to ethanol include both acute sensitivity as well as the magnitude and rate of development of acute tolerance during the ethanol exposure (Hu et al., 2008; Ponomarev and Crabbe, 2002). Here we show that altered ethanol metabolism in C. elegans affects the acute sensitivity of the worms but not the development of acute tolerance. First, ethanol tissue concentrations do not decrease during a single constant exposure (Figure 2b), so metabolism is not sufficient to reduce the concentration of ethanol below that seen at an early time point. Therefore, any reduction in the behavioral effects over this time course is likely to be due to compensatory mechanisms that act in the opposite direction to ethanol or directly limit the action of ethanol. Second, ADH mutant animals develop acute tolerance similar to that displayed by wild-type animals showing that a reduction in metabolism does not impact the mechanisms of acute tolerance development (Figure 3a). This provides further support for the idea that acute tolerance is occurring at the level of the affected tissues, which in C. elegans includes the nervous system (Davies et al., 2003) rather than through a systemic metabolic process.
Our investigation into the reported differences in ethanol sensitivity for swimming behavior led to the unexpected finding that the worms' sensitivity to ethanol depends on the osmolarity of the external medium, and that the worms are able to change their sensitivity to the effects of osmolarity based on experience. Therefore, the constituents of the exogenous medium are critical to note in future alcohol studies using C. elegans. We speculate that the permeability of the worm to exogenous ethanol might change depending on the history of exposure. Dynamic permeability may be adaptive for an animal that must occasionally encounter dangerous chemicals, such as ethanol, while roaming through its natural soil environment. Future study of this phenomenon in C. elegans may give rise to novel strategies to alter excess permeation of ethanol into specific tissues to prevent toxicity.
In both vertebrates and C. elegans, ethanol metabolism makes important contributions to ethanol responsive behaviors. Our results suggest that both metabolism and environmental conditions should be considered in the analysis of mechanisms that contribute to ethanol responsive behaviors.
Supplementary Material
Acknowledgements
We thank V. Williamson for the gift of AL2B. Some strains were provided by the Caenorhabditis Genetics Center (funded by the NIH National Center for Research Support). Support was provided by ABMRF/The Foundation for Alcohol Research, NIH R03AA020195 (JTP), NIH Institutional Training Grant 5T32AA007471 (SJD), Bruce Jones Graduate Fellowships in Addiction Research (SJD), NIH RO1AA016837 (JCB), RO1AA016842 (AGD), P20AA017070 (MG, AGD and JCB) and the Thomas F. & Kate Miller Jeffress Memorial Trust (JCB).
References
- Avery L, Raizen D, Lockery S. Electrophysiological methods. Methods Cell Biol. 1995;48:251–269. [PMC free article] [PubMed] [Google Scholar]
- Bargmann CI. Neurobiology of the Caenorhabditis elegans genome. Science. 1998;282:2028–2033. doi: 10.1126/science.282.5396.2028. [DOI] [PubMed] [Google Scholar]
- Bettinger JC, McIntire SL. State-dependency in C. elegans. Genes Brain Behav. 2004;3:266–272. doi: 10.1111/j.1601-183X.2004.00080.x. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns AR, Wallace IM, Wildenhain J, Tyers M, Giaever G, Bader GD, Nislow C, Cutler SR, Roy PJ. A predictive model for drug bioaccumulation and bioactivity in Caenorhabditis elegans. Nat Chem Biol. 2010;6:549–557. doi: 10.1038/nchembio.380. [DOI] [PubMed] [Google Scholar]
- Chen YC, Peng GS, Wang MF, Tsao TP, Yin SJ. Polymorphism of ethanol-metabolism genes and alcoholism: correlation of allelic variations with the pharmacokinetic and pharmacodynamic consequences. Chem Biol Interact. 2009;178:2–7. doi: 10.1016/j.cbi.2008.10.029. [DOI] [PubMed] [Google Scholar]
- Cox GN, Kusch M, Edgar RS. Cuticle of Caenorhabditis elegans: its isolation and partial characterization. J Cell Biol. 1981;90:7–17. doi: 10.1083/jcb.90.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabb DW, Matsumoto M, Chang D, You M. Overview of the role of alcohol dehydrogenase and aldehyde dehydrogenase and their variants in the genesis of alcohol-related pathology. Proc Nutr Soc. 2004;63:49–63. doi: 10.1079/pns2003327. [DOI] [PubMed] [Google Scholar]
- Davies AG, Bettinger JC, Thiele TR, Judy ME, McIntire SL. Natural variation in the npr-1 gene modifies ethanol responses of wild strains of C. elegans. Neuron. 2004;42:731–743. doi: 10.1016/j.neuron.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Davies AG, Pierce-Shimomura JT, Kim H, VanHoven MK, Thiele TR, Bonci A, Bargmann CI, McIntire SL. A central role of the BK potassium channel in behavioral responses to ethanol in C. elegans. Cell. 2003;115:655–666. doi: 10.1016/s0092-8674(03)00979-6. [DOI] [PubMed] [Google Scholar]
- Davis JR, Li Y, Rankin CH. Effects of developmental exposure to ethanol on Caenorhabditis elegans. Alcohol Clin Exp Res. 2008;32:853–867. doi: 10.1111/j.1530-0277.2008.00639.x. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Xuei X, Chen HJ, Tian H, Wetherill LF, Dick DM, Almasy L, Bierut L, Bucholz KK, Goate A, Hesselbrock V, Kuperman S, Nurnberger J, Porjesz B, Rice J, Schuckit M, Tischfield J, Begleiter H, Foroud T. Association of alcohol dehydrogenase genes with alcohol dependence: a comprehensive analysis. Hum Mol Genet. 2006;15:1539–1549. doi: 10.1093/hmg/ddl073. [DOI] [PubMed] [Google Scholar]
- Fry JD, Donlon K, Saweikis M. A worldwide polymorphism in aldehyde dehydrogenase in Drosophila melanogaster: evidence for selection mediated by dietary ethanol. Evolution. 2008;62:66–75. doi: 10.1111/j.1558-5646.2007.00288.x. [DOI] [PubMed] [Google Scholar]
- Fry JD, Saweikis M. Aldehyde dehydrogenase is essential for both adult and larval ethanol resistance in Drosophila melanogaster. Genet Res. 2006;87:87–92. doi: 10.1017/S0016672306008032. [DOI] [PubMed] [Google Scholar]
- Geer BW, Dybas LK, Shanner LJ. Alcohol dehydrogenase and ethanol tolerance at the cellular level in Drosophila melanogaster. The Journal of experimental zoology. 1989;250:22–39. doi: 10.1002/jez.1402500105. [DOI] [PubMed] [Google Scholar]
- Glasner JD, Kocher TD, Collins JJ. Caenorhabditis elegans contains genes encoding two new members of the Zn-containing alcohol dehydrogenase family. J Mol Evol. 1995;41:46–53. doi: 10.1007/BF00174040. [DOI] [PubMed] [Google Scholar]
- Graham ME, Edwards MR, Holden-Dye L, Morgan A, Burgoyne RD, Barclay JW. UNC-18 modulates ethanol sensitivity in Caenorhabditis elegans. Mol Biol Cell. 2009;20:43–55. doi: 10.1091/mbc.E08-07-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath AC, Madden PA, Bucholz KK, Dinwiddie SH, Slutske WS, Bierut LJ, Rohrbaugh JW, Statham DJ, Dunne MP, Whitfield JB, Martin NG. Genetic differences in alcohol sensitivity and the inheritance of alcoholism risk. Psychol Med. 1999;29:1069–1081. doi: 10.1017/s0033291799008909. [DOI] [PubMed] [Google Scholar]
- Hu W, Saba L, Kechris K, Bhave SV, Hoffman PL, Tabakoff B. Genomic insights into acute alcohol tolerance. J Pharmacol Exp Ther. 2008;326:792–800. doi: 10.1124/jpet.108.137521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biology. 2001;2 doi: 10.1186/gb-2000-2-1-research0002. research0002.1-0002.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapfhamer D, Bettinger JC, Davies AG, Eastman CL, Smail EA, Heberlein U, McIntire SL. Loss of RAB-3/A in Caenorhabditis elegans and the mouse affects behavioral response to ethanol. Genes Brain Behav. 2008;7:669–676. doi: 10.1111/j.1601-183X.2008.00404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo PH, Kalsi G, Prescott CA, Hodgkinson CA, Goldman D, van den Oord EJ, Alexander J, Jiang C, Sullivan PF, Patterson DG, Walsh D, Kendler KS, Riley BP. Association of ADH and ALDH genes with alcohol dependence in the Irish Affected Sib Pair Study of alcohol dependence (IASPSAD) sample. Alcohol Clin Exp Res. 2008;32:785–795. doi: 10.1111/j.1530-0277.2008.00642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon JY, Hong M, Choi MS, Kang S, Duke K, Kim SK, Lee S, Lee J. Ethanol-response genes and their regulation analyzed by a microarray and comparative genomic approach in the nematode Caenorhabditis elegans. Genomics. 2004;83:600–614. doi: 10.1016/j.ygeno.2003.10.008. [DOI] [PubMed] [Google Scholar]
- Lee J, Jee C, McIntire SL. Ethanol preference in C. elegans. Genes Brain Behav. 2009;8:578–585. doi: 10.1111/j.1601-183X.2009.00513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell PH, Bull K, Glautier S, Hopper NA, Holden-Dye L, O’Connor V. The concentration-dependent effects of ethanol on Caenorhabditis elegans behaviour. Pharmacogenomics J. 2007;7:411–417. doi: 10.1038/sj.tpj.6500440. [DOI] [PubMed] [Google Scholar]
- Mitchell P, Mould R, Dillon J, Glautier S, Andrianakis I, James C, Pugh A, Holden-Dye L, O’Connor V. A differential role for neuropeptides in acute and chronic adaptive responses to alcohol: behavioural and genetic analysis in Caenorhabditis elegans. PLoS ONE. 2010;5:e10422. doi: 10.1371/journal.pone.0010422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan PG, Sedensky MM. Mutations affecting sensitivity to ethanol in the nematode, Caenorhabditis elegans. Alcoholism Clinical and Experimental Research. 1995;19:1423–1429. doi: 10.1111/j.1530-0277.1995.tb01002.x. [DOI] [PubMed] [Google Scholar]
- Ogueta M, Cibik O, Eltrop R, Schneider A, Scholz H. The influence of Adh function on ethanol preference and tolerance in adult Drosophila melanogaster. Chemical Senses. 2010;35:813–822. doi: 10.1093/chemse/bjq084. [DOI] [PubMed] [Google Scholar]
- Pautassi RM, Camarini R, Quadros IM, Miczek KA, Israel Y. Genetic and environmental influences on ethanol consumption: perspectives from preclinical research. Alcohol Clin Exp Res. 2010;34:976–987. doi: 10.1111/j.1530-0277.2010.01172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce-Shimomura JT, Chen BL, Mun JJ, Ho R, Sarkis R, McIntire SL. Genetic analysis of crawling and swimming locomotory patterns in C. elegans. PNAS. 2008;105:20982–20987. doi: 10.1073/pnas.0810359105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev I, Crabbe JC. A novel method to assess initial sensitivity and acute functional tolerance to hypnotic effects of ethanol. J Pharmacol Exp Ther. 2002;302:257–263. doi: 10.1124/jpet.302.1.257. [DOI] [PubMed] [Google Scholar]
- Rodriguez LA, Wilson JR, Nagoshi CT. Does psychomotor sensitivity to alcohol predict subsequent alcohol use? Alcohol Clin Exp Res. 1993;17:155–161. doi: 10.1111/j.1530-0277.1993.tb00741.x. [DOI] [PubMed] [Google Scholar]
- Schuckit MA. Low level of response to alcohol as a predictor of future alcoholism. Am J Psychiatry. 1994;151:184–189. doi: 10.1176/ajp.151.2.184. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Edenberg H, Kalmijn J, Flury L, Smith TL, Reich T, Bierut L, Goate A, Foroud T. A genome-wide search for genes that relate to a low level of response to alcohol. Alcoholism Clinical and Experimental Research Article. 2001;25:323–329. [PubMed] [Google Scholar]
- Speca DJ, Chihara D, Ashique AM, Bowers MS, Pierce-Shimomura JT, Lee J, Rabbee N, Speed TP, Gularte RJ, Chitwood J, Medrano JF, Liao M, Sonner JM, Eger EI, Peterson AS, Mcintire SL. Conserved role of unc-79 in ethanol responses in lightweight mutant mice. PLoS Genet. 2010;6:e1001057. doi: 10.1371/journal.pgen.1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volavka J, Czobor P, Goodwin D, Gabrielli W, Penick E, Mednick S, Jensen P, Knop J. The electroencephalogram after alcohol administration in high-risk men and the development of alcohol use disorders 10 years later. Arch Gen Psychiatry. 1996;53:258–263. doi: 10.1001/archpsyc.1996.01830030080012. [DOI] [PubMed] [Google Scholar]
- Waterston R, Martin C, Craxton M, Huynh C, Coulson A, Hillier L, Durbin R, Green P, Shownkeen R, Halloran N, Metzstein M, Hawkins T, Wilson R, Berks M, Du Z, Thomas K, Thierry-Mieg J, Sulston J. A survey of expressed genes in Caenorhabditis elegans. Nat Genet. 1992;1:114–123. doi: 10.1038/ng0592-114. [DOI] [PubMed] [Google Scholar]
- Williamson VM, Long M, Theodoris G. Isolation of Caenorhabditis elegans mutants lacking alcohol dehydrogenase activity. Biochem Genet. 1991;29:313–323. doi: 10.1007/BF00554139. [DOI] [PubMed] [Google Scholar]
- Wolf FW, Rodan AR, Tsai LT, Heberlein U. High-resolution analysis of ethanol-induced locomotor stimulation in Drosophila. J Neurosci. 2002;22:11035–11044. doi: 10.1523/JNEUROSCI.22-24-11035.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.