Abstract
A cell's reaction to any change in the endogenous or exogenous conditions often involves a complex response that eventually either leads to cell adaptation and survival or to the initiation and execution of (programmed) cell death. The molecular decision whether to live or die, while depending on a cell's genome, is fundamentally influenced by its actual metabolic status. Thus, the collection of all metabolites present in a biological system at a certain time point (the so-called metabolome) defines its physiological, developmental and pathological state and determines its fate during changing and stressful conditions. The budding yeast Saccharomyces cerevisiae is a unicellular organism that allows to easily modify and monitor conditions affecting the cell's metabolome, for instance through a simple change of the nutrition source. Such changes can be used to mimic and study (patho)physiological scenarios, including caloric restriction and longevity, the Warburg effect in cancer cells or changes in mitochondrial mass affecting cell death. In addition, disruption of single genes or generation of respiratory deficiency (via abrogation of mitochondrial DNA) assists in revealing connections between metabolism and apoptosis. In this minireview, we discuss recent studies using the potential of the yeast model to provide new insights into the processes of stress defense, cell death and longevity.
Keywords: Yeast, Apoptosis, Metabolome, Warburg effect, Starvation, Glucose
Introduction
The shamanistic idea that biological fluids reflect the health of an individual is as old as medicine itself. Whereas ancient doctors recognized a difference in smell, taste and colors of patient fluids, modern analyses allow investigating changes in the metabolic composition at a cellular and molecular level. Following up on genomics, transcriptomics and proteomics the newest trendy addition to the “omics”-family is, in fact, metabolomics, which encompasses the examination of the chemical fingerprint represented by the collection of all metabolites in a biological system at a certain time point (metabolome). By systematic screening of the metabolome, data-based biomedical research has gained more input to form hypotheses on complex biomedical problems.
Metabolites are formed in association with ongoing metabolism, a term derived from the Greek word metabole, “change”. Their number in mammalian cells ranges from 3000 to 8000 (Human Metabolome Database) [1], of which only a small fraction is currently quantitatively detectable. Intracellular metabolites can either be generated by a cell itself in response to a certain stimulus or are of exogenous origin like compounds derived from the intestinal microbiome, drugs, pesticides, and environmental pollutants. Being the intermediates of biochemical reactions, metabolites play crucial roles in connecting many different pathways that operate within a living cell. Their presence and quantity provide information on the current cellular metabolic status and define the phenotype in response to genetic or environmental changes. Thus, the levels of a given metabolite depend on and therefore reflect the physiological, developmental and pathological state of a cell, tissue or organism.
The budding yeast Saccharomyces cerevisiae with its manifold advantages as a model system is a promising tool to analyze metabolic changes under diverse conditions. Indeed, yeast has been extensively used to investigate complex scenarios of stress response, longevity and cell death. The usage of the yeast model system has uncovered various genes and pathways that regulate cellular survival in a fashion comparable to mammalian cells during the aging process, upon various apoptotic and necrotic stresses and upon heterologous expression of human disease-related proteins [2–8]. In that context, metabolome analyses are expected to generate valuable insights. Modeled closely to the Human Metabolome Database, the Yeast Metabolome Database (YMDB) contains more than 2000 metabolites with links to 995 different genes/proteins, including enzymes and transporters [9].
Besides investigating the cellular metabolome during different scenarios, which normally leads to the identification of correlative, yet diagnostic changes in metabolite abundance, yeast gives the handy possibility to additionally explore the causative and functional consequences of endogenous (genetic) or exogenous metabolite modulation. This can be achieved by overexpression or deletion of specific yeast genes as well as by heterologous expression of human genes. In addition, the metabolic state of yeast cells can be altered by simply changing the nutritional composition of the culture medium (e.g. via a switch in the carbon source during cell growth) or by deleting mitochondrial DNA, which gives rise to cells with compromised mitochondria and dysfunctional respiration, an almost unique advantage when using yeast. By changing the yeast metabolome it is possible to mimic physiological, tissue-specific, developmental and pathological states of human cells during stress response, aging, cell death or cell survival. Along these lines, several studies in yeast have demonstrated that the external nutritional molecules as well as the intracellular metabolites regulate life, death, longevity and stress defense in a complex and powerful way. In this minireview we focus on selected studies, which have given new insights into this emerging field.
Different media guide life and death decisions through enhanced fermentation or respiration
Fermentation and respiration in S. cerevisiae
S. cerevisiae bears a glucose repression system that drastically suppresses respiration independently of oxygen availability (also known as the Crabtree effect). Thus, it preferentially consumes glucose through the process of alcoholic fermentation. During this process, glycolysis-derived pyruvate is converted to ethanol leading to the oxidation of NADH to NAD+. Here, pyruvate is first decarboxylated by pyruvate decarboxylase to acetaldehyde, followed by the reduction to ethanol via alcohol dehydrogenase (ADH). Eventually (after the diauxic shift, see below), ethanol is re-metabolized via respiration, which results in a total of 38 ATP per glucose molecule—before this, however, the fermentative energy output (derived from glycolysis) amounts to only 2 ATP. Yeast can also metabolize a wide variety of other carbon sources, including non-fermentable compounds such as ethanol or glycerol. While the oxidative metabolism of such non-fermentative carbons via the tricarboxylic acid (TCA) cycle and mitochondrial electron transport chain is more efficient in producing ATP, it also generates potentially harmful reactive oxygen species (ROS), such as the superoxide anion [10]. The enzymatic pathways required for the specific utilization of these carbon compounds are well characterized and primarily produce pyruvate [11]. In the presence of oxygen and absence of glucose repression, pyruvate (either generated from glycolysis or through alternative pathways) enters the mitochondria and is further oxidatively decarboxylated to acetyl-CoA by the pyruvate dehydrogenase (PDH) complex. In the TCA cycle, acetyl-CoA is completely oxidized, producing carbon dioxide, NADH and FADH2. The redox carriers (NADH and FADH2) are re-oxidized in the respiratory (electron transport) chain located in the inner mitochondrial membrane. The transfer of electrons is coupled to the process of oxidative phosphorylation through the ATP synthase, an enzyme complex which is also located in the inner mitochondrial membrane and designed to synthesize ATP from ADP and inorganic phosphate.
Enzymes needed for a specific pathway are mostly produced by regulation at the transcriptional level. For instance, galactose-induced expression of genes required for catabolism of this sugar is regulated by the transcriptional activator Gal4p [12], which in turn is strongly repressed in the presence of glucose. A further example is the group of alcohol dehydrogenase (ADH) enzymes, which are regulated through the presence of glucose (Fig. 1). Four (Adh1p and Adh3-5p) out of the five ADH enzymes in yeast reduce acetaldehyde to ethanol in the presence of high glucose concentrations (fermentation). In contrast, Adh2p is active at low levels or absence of glucose and catalyzes the reverse reaction from ethanol to acetaldehyde, thus driving the production of pyruvate for energy supply via respiration [13]. Additionally, posttranscriptional modifications, including but not limited to protein phosphorylation, acetylation and ubiquitination, are also used by cells to rapidly alter the activity of metabolic enzymes. Such modifications – in many cases the first reaction to a sudden environmental change – can function as an on–off switch, regulate protein translocations or vary the speed of an enzyme's degradation.
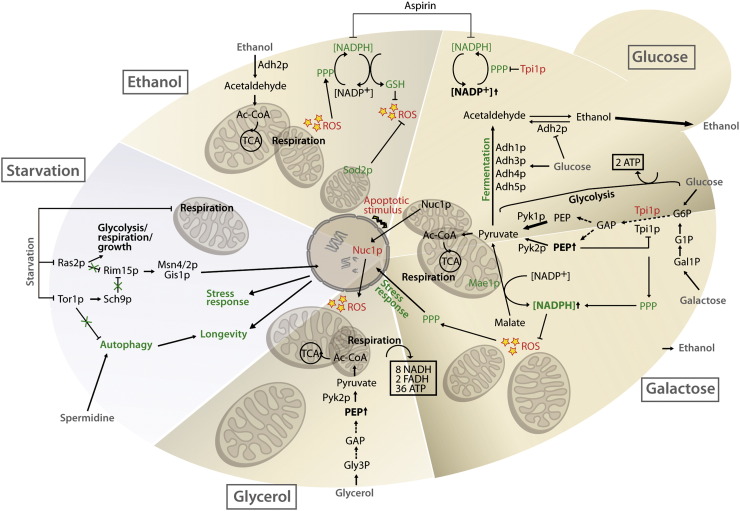
Fig. 1.
Schematic representation of metabolic fluxes upon different carbon sources. Glucose uptake results in fermentation processes via activation of high efficient Pyk1p and thus a rapid turnover of phosphoenolpyruvate (PEP). Inhibition of the pentose phosphate pathway by triose phosphate isomerase causes an increase in NADP+, but not in NADPH. High glucose activates Adh1/3/4/5p which forces the formation of ethanol upon fermentation processes. Additionally, glucose represses Adh2p and thus inhibits the vice-versa production of acetaldehyde from ethanol. Ethanol, galactose and glycerol stimulate expression of the less active Pyk2p, which furthermore enhances intracellular levels of PEP. Enhanced PEP leads to an activation of the PPP, increases respiratory metabolism via pyruvate and acetyl-CoA intermediates. Malate is converted to pyruvate by mitochondrial Mae1p and thereby reductive NADPH is synthesized. Upon strict carbon starvation metabolic fluxes are restricted. At the same time, stress response mechanisms and autophagy are induced, which ultimately results in longevity. Factors and processes involved in the promotion of cell death or exerting pro-survival functions are marked in red and green, respectively. Abbreviations: Ac-CoA, acetyl coenzyme A; GAP, glyceraldehyde 3-phosphate; Gal1P, galactose 1-phosphate; G1P, glucose 1-phosphate; G6P, glucose 6-phosphate; Gly3P, glycerol 3-phosphate; GSH, glutathione; NADP, nicotinamide adenine dinucleotide phosphate; PEP, phosphoenolpyruvate; PPP, pentose phosphate pathway; ROS, reactive oxygen species; TCA, tricarboxylic acid cycle.
The change from one carbon source to another and the corresponding adaptation process is described as the diauxic shift. Regarding yeast, the term is used for metabolic adaptation during the transition from fermentable glucose to non-fermentable ethanol. This shift results in massive reprogramming of gene expression, including genes involved in gluconeogenesis, the TCA cycle and the glyoxylate cycle [14]. In addition, mitochondrial mass and respiration are increased, both causing substantial changes in metabolites crucial for cellular function and survival. During the diauxic shift, for example, the up-regulation of gluconeogenic gene expression is crucial for the production of glucose-6-phosphate, an essential metabolite for cell growth [11]. At the same time, an increased amount of mitochondrial gene expression is required for efficient oxidative phosphorylation being the only energy supply when glycolysis decays. The crosstalk between these pathways does not only guarantee the most efficient way of energy production, but also regulates the reductive power, which is determinant for proper ROS detoxification under respiratory conditions.
Changes in growth media and their effects on cell death
Having the powerful opportunity to diversely coordinate the nutritional and metabolic status of yeast cells by a simple change of growth media (Fig. 1), the response to various stresses can be used to study distinct cell death pathways under these conditions. For instance, the special ability of S. cerevisiae to switch on and off respiration in response to changes in the carbon source, allows the stringent genetic evaluation of the mitochondrial role during cell death [10]. Additionally, yeast offers the almost unique possibility to generate viable strains lacking mitochondrial DNA (rho0), which display a partial loss of mitochondrial function, including respiration [8,10].
Similar to mammalian apoptosis, yeast apoptosis is under mitochondrial control [15,16]. The possibility to modify respiration efficiency and thus mitochondrial size and number in yeast cells was used by Büttner and colleagues (2007) to distinguish between the life and death functions of Nuc1p, the yeast homolog of mammalian Endonuclease G (EndoG). Specifically, the response of NUC1-deleted cells during chronological aging was compared on glucose and glycerol. While cultivation on glucose does not induce mitochondrial respiration during initial growth phases, energy demands upon growth on glycerol are exclusively met by oxidative phosphorylation during each growth phase, and therefore the number, size and mass of mitochondria are enhanced [6]. Intriguingly, nuc1 mutant cells displayed a survival disadvantage resulting in increased rates of necrotic cell death when aged on glucose media, an effect probably due to the lack of Nuc1p's vital function. In contrast, culturing these cells on glycerol diminished the age-induced apoptotic phenotype possibly due to the lack of the mitochondrial death effector Nuc1p (Fig. 1). Similar to Nuc1p/EndoG another mitochondrial protein displays dual function within the cell regarding life and death. The apoptosis-inducing factor (AIF) is a highly conserved protein from yeast to human, which after apoptosis induction translocates to the nucleus where it participates in apoptotic chromatinolysis [15,17]. Human and mouse cells lacking AIF show a decrease in respiratory function due to reduced content of complex I, pointing to a role of AIF in the biogenesis and/or maintenance of this polyprotein complex [18]. AIF displays a redox activity, which depends on the C-terminally located NADH oxidase domain and which is essential for its vital function, as it is the only NADH oxidase in the mitochondrial intermembrane space [19]. On the one hand, abolishing the oxidoreductase function by removal of the FAD group does not influence the apoptogenic effects. On the other hand, inhibition of the apoptogenic effect by means of para-chloromercuriphenylsulfonic acid does not influence its NADH oxidase activity. These results confirm the dual function of this protein regarding life and death and clarify that these two functions are functionally separable [19]. In yeast, deletion of AIF1 negatively influences growth on the non-fermentable media glycerol suggesting a respiratory function probably located to the oxidoreductase domain of the protein. These data support the above mentioned mammalian cell culture data regarding a highly conserved vital role of AIF also in the mitochondrial respiratory function [18].
Oxidant defense is crucial for cell survival and depends on the reductive power of the cell, which is determined by ROS-metabolizing enzymes, like superoxide dismutase (SOD) and catalase, and/or by enzymes generating reductive metabolites, like those in the pentose phosphate pathway (PPP) (Fig. 1). These metabolites include, but are likely not limited to NADH, NADPH and reduced glutathione (GSH). The ratio between their reductive and oxidized forms is generally taken to express the reductive potential of a cell. While NADH, derived from glycolysis and the TCA cycle, is mostly used for ATP production during oxidative phosphorylation, NADPH provides the reducing equivalents for the biosynthesis of lipids and the reductive capability involved in the protection against ROS, which is mainly determined by GSH. NADPH is generated during the PPP, which branches off from glycolysis at the level of glucose 6-phosphate (Fig. 1) due to Zwf1p. Compared to other yeast species, S. cerevisiae has very low catabolic fluxes through the PPP [20], indicating that its primary role in budding yeast is the production of NADPH for refilling reductive power rather than its use for anabolic and catabolic reactions. When cells are exposed to oxidative stress or an environment that requests massive respiration (therefore enhancing ROS production), a high reductive power and thus more NADPH is needed. Thereby, cells undergo the so-called metabolic switch, which describes a drift from glycolysis to the PPP and thus increased NADPH production. This shift in primary carbon metabolism has been suggested to be the fastest response to oxidative stress, occurring just within seconds. Besides Zwf1p, also the aldehyde dehydrogenase Ald6p as well as the isocitrate dehydrogenase Idp2p is responsible for NADPH production to generate reductive power within the cytosol [21]. These factors' activity depends on the carbon source: Zwf1p and Ald6p have been postulated to be responsible for NADPH production upon growth on glucose, whereas Idp2p is only expressed during the diauxic shift or under non-fermentable conditions [21]. NADPH availability seems to be determinant during aspirin-induced cell death on the non-fermentable carbon source ethanol. Ethanol is produced routinely in the cell as a consequence of alcoholic fermentation and is also thought to enter the cell by passive diffusion. Aspirin-induced cell death in yeast cells lacking Sod2p, the mitochondrial MnSOD, occurred only upon growth on ethanol (Fig. 1), but neither on other non-fermentable carbons, like glycerol and acetate, nor on glucose [22]. On ethanol, aspirin led to the inactivation of catalase activity [22]. Upon treatment with aspirin, total NADP(H) content decreased in all cells and conditions; MnSOD-deficient cells, however, additionally displayed an initial decrease in the NADPH/NADP+ ratio when grown on non-fermentable carbon sources. Cultured in glycerol and acetate medium, but not in ethanol, this loss of NADPH/NADP+ ratio was balanced with an increase of NADP(H) levels. Thus, according to the principle “the straw that breaks the camel's back”, MnSOD-deficient cells grown on ethanol, showed a decreased NADPH/NADP+ ratio as well as low total NADP(H) levels, and accumulated a lethal amount of ROS, when an additional reduction of the NADP(H) content occurred through treatment with aspirin. Although acetate, glycerol and ethanol are all strictly non-fermentable carbon sources the difference in total NADP(H) levels reflects that the metabolic pathways resulting in the generation of acetyl coenzyme A are different and thus produce different types and amounts of metabolites. These metabolites as well as the involved enzymes may have an impact on other pathways and consequently modulate metabolite pools differently, such as the levels of NADP(H). Interestingly, the first step of ethanol utilization involves the reductive conversion of NAD to NADH, which could directly influence the balance of NAD/NADP pools.
Applying state of the art 13C-labeling methods and using MALDI-TOF-mass spectrometry, Velagapudi and colleagues (2007) determined metabolic flux distributions – particularly concerning such branching into the PPP – of 27 deletion strains cultivated on glucose and galactose [23]. On glucose, the growth was predominantly fermentative, whereas on galactose respiration was more active with a corresponding lower ethanol production. Cells lacking malic enzyme gene (MAE1) showed an enormously increased flux from glucose-6-phophate into the PPP when grown on galactose. The authors concluded a role of Mae1p in NADPH-supply during respiratory growth (Fig. 1).
Besides NADPH production, the PPP seems to have a second, largely unexplored role in antioxidant response. Krüger and colleagues (2011) established that hyperactivation or deletion of the PPP enhances or lowers, respectively, the expression of genes involved in the oxidative response (Fig. 1). This suggests PPP as a redox sensor that functions as a metabolic signal required for balancing and timing of the oxidative response [24]. In line with these results, Grüning et al. (2011) could show that a metabolic feedback loop initiated by pyruvate kinase (PYK), a regulator of aerobic to anaerobic metabolism, stimulates redox metabolism in respiring cells [25]. Yeast possesses two PYK paralogues (PYK1, PYK2), which are differentially expressed in fermentative and oxidative metabolism (Fig. 1) [16]. If cells are grown on glucose, PYK1 is active and converts phosphoenolpyruvate (PEP) to pyruvate, which is further metabolized to ethanol (fermentation). Accordingly, PYK1 activity is positively enhanced in the presence of fructose-1,6-bisphosphate (FBP), a product of glucose catabolism. On the contrary, PYK2 expression is blocked in the presence of glucose. In fact, a functional copy of PYK1 is required for growth on glucose as the sole carbon source, whereas deletion of PYK2 has no effect on the specific growth rate on glucose [26]. A switch from PYK1 to PYK2 causes a shift from fermentative to oxidative metabolism: The lower metabolic rate of Pyk2p, which is only active upon growth on a non-fermentable carbon source, provokes respiration. Nevertheless, no increase in ROS levels can be detected when respiration is activated [25]. The low metabolic rate of Pyk2p provides higher levels of PEP. PEP accumulation inactivates the important glycolytic enzyme triosephosphateisomerase (Tpi1p), whose inhibition stimulates the PPP, increases antioxidative metabolism and prevents ROS accumulation (Fig. 1) [25]. Thus, if respiration is activated the cell simultaneously increases its reductive power supply by a metabolic feedback activation of the PPP via PYK in order to guarantee survival.
The energy-supplying pathways encompassing glycolysis and respiration (TCA cycle and oxidative phosphorylation) generate mitochondrial ROS, which can harm the cell and lead to a decreased life span during aging. To counteract these hazardous molecules, metabolic pathway intermediates simultaneously enhance the redox response of the cell, e.g. by increasing NADPH levels through the PPP or by activating stress response transcription factors that induce ROS-detoxifying enzymes (i.e. SODs, catalase). Furthermore, stress is reduced and survival enhanced by avoiding mitochondrial oxidative phosphorylation and by exclusively using glycolysis combined with the subsequent production of lactate (mammals) or ethanol (yeast), as well as by switching to alternative metabolic pathways to provide biosynthetic molecules. This strategy is particularly used by fast-proliferating cells like cancer cells, which can be modeled in yeast via a switch from respiratory (non-fermentable) to fermentable media.
Besides modifications in the carbon source, also other nutrients can be either replaced, displaced or additionally added to yeast cultures in order to mimic certain (patho)physiological aspects and study cell death pathways as well as longevity. Apart from partial (either nitrogen or carbons) or total removal (culture in water) of nutrients, which will be discussed in the next section, addition of alternative energy supplies is possible. For instance, using various lipase-treated cooking oils, externally added free fatty acids (FFA), or genetic mutations setting yeast on a fatty diet, we previously established a yeast aging model for obesity-induced tissue and/or cellular damage [27]. The presence of FFA results in the up-regulation of genes encoding enzymes for fatty acid beta-oxidation and proteins involved in the enlargement of peroxisomes [28,29]. Using a quadruple knockout (lacking the acyltransferases LRO1, DGA1, ARE1 and ARE2) to avoid metabolism and thus detoxification of FFA, we demonstrated that the severity of FFA-induced cell death correlates with the degree of unsaturation. This cell death shows a necrotic phenotype and depends on mitochondrial function [27,30]. Altogether, these results outline the detrimental metabolic consequences of increased energy supply by diverse nutrients.
The Warburg effect: from yeast to man
A model for tumor growth and metabolism was recently presented by our group. Metabolic states were forced by growth on distinct carbon sources either favoring high glycolysis (on glucose), glycolysis in combination with respiration (on galactose), or respiration only (on glycerol) [31]. The most common biochemical phenotype of tumor cells is dominated by high glycolytic flux accompanied by lactate production as well as drastically reduced mitochondrial respiration, even under normoxic conditions. Otto Warburg described this phenotype as the Warburg effect in the 1920's. This shift to aerobic glycolysis accompanied by lactate production and coupled with increased glucose uptake, is likely used by proliferating cells to promote the efficient conversion of glucose into the macromolecules needed to construct a new cell [32]. As mentioned above, S. cerevisiae possesses a unique glucose repression system that drastically suppresses respiration independently of oxygen availability upon growth on glucose (Crabtree effect). Rapidly proliferating yeasts show comparable fermentation values/rates as tumor cells [33], thus serving as a model to study the relationship between energy metabolism and maximal proliferation. We could demonstrate that a transfer to the same highly respiratory media (from liquid to solid glycerol) leads to a diminished colony formation of about 70%, whereas transfer to a fermentable carbon source, and consequently shutting off respiration (from liquid glycerol to solid glucose), results in highly efficient initiation of colony growth. This was further confirmed by using respiration-deficient strains disrupted in the mitochondrial DNA (rho0), the mitochondrial inner membrane insertase Oxa1p, and the mitochondrial GTPase Mgm1p, respectively, all three showed an advantage in survival during early development of these fast-proliferating solid yeast cell populations accompanied by decreased ROS production. When highly proliferative, fermentatively growing cells were transferred to respiratory media (from liquid glucose to solid galactose or glycerol) and thus challenged with abrupt induction of respiration, cell survival (as monitored by colony formation) was severely reduced. Accordingly, addition of GSH, a major cellular antioxidant, rescued cell survival and, in addition, reduced ROS production during early colony development. Thus, the Warburg effect might directly contribute to the initiation of cancer formation, not only by enhanced glycolysis but also via decreased respiration in the presence of oxygen, which suppresses apoptosis [31].
Starvation, caloric restriction and longevity—at the crossroads of autophagy and stress response
There exist several starvation scenarios: nitrogen, amino acid, carbon and complete starvation, the latter one being a shift of the cells into water. Initially, starvation studies in yeast led to the paramount discovery and characterization of the autophagy-related genes (ATGs). Starvation is also used as a model for caloric restriction and aging studies [34,35]. Shifting yeast cells into starvation conditions results in longevity in many cases [35,36]. This long-lived cells show changes in their metabolism, autophagy rates, stress response, cell division, as well as in the quality and quantity of organelles (e.g. mitochondria) (Fig. 1). Longevity is also associated with changes in protein levels, caused by altered transcription, posttranscriptional modification, turnover or translocation. Of note, exogenous media change can impact longevity by changing the intracellular metabolite pattern. The identification of aging-related metabolites and the mechanisms of how energy metabolism connects to various starvation-induced pathways are pivotal to understand how caloric restriction exerts its beneficial effects on stress response and longevity.
Autophagy (a cellular self-digestion process) is a suggested key component of longevity that is crucial for metabolic homeostasis. Recent studies with life span-extending deletion strains (e.g. such deleted in the target of rapamycin TOR1 or the protein kinase SCH9) have revealed overlapping activities of autophagy and nutrient-responsive stress response pathways in the context of longevity [35,37]. Moreover, autophagy-stimulating substances (resveratrol, rapamycin and spermidine) have been shown to elongate life span or health span from yeast to mammals in an autophagy-dependent fashion [38–41]. ROS is known to shorten chronological life span, and superoxide in particular was suggested to play important signaling roles during aging [36,42]. However, early sublethal concentrations of ROS can strengthen the cell and lead to life span extension, a process often referred to as hormesis [43,44]. Under nitrogen starvation conditions, accumulation of mitochondrial H2O2 is essential for autophagy induction. Here, the oxidative environment of mitochondria inhibits the activity of Atg4p by oxidizing a regulatory cysteine residue, which inhibits delipidation of a further protein essential for macroautophagy, Atg8p, thus allowing autophagosome formation. Away from mitochondria, a reducing environment around the vacuole/lysosomes is maintained, where Atg4p can cleave Atg8p from autophagosomes, a process required for the fusion with the vacuole/lysosomes [45]. Since starvation and aging share overlapping pathways for survival maintenance, it is a pivotal yet pending matter to understand the mechanistic relationships between sublethal hormetic and autophagy-inducing oxidative stress, versus the detrimental consequences of high ROS levels.
As mentioned above, loss of viability was initially used to screen for autophagy-defective mutants in S. cerevisiae; however, the mechanism of cell death in these mutants has remained unclear. Suzuki and colleagues (2011) were able to show that, upon starvation, wild type cells upregulate proteins for respiration and ROS-detoxifying enzymes. In contrast, ATG-depleted cells lack this adaptive detoxifying process and consequently accumulate high levels of ROS. This leads to a deficiency in the respiratory function, resulting in the loss of mitochondrial DNA and further cell death [46]. Mitochondria are believed to be one of the main sources of ROS and therefore it seems logical that by removing mitochondria, cell death can be prevented under conditions where respiration is not essential to meet the cell's energy demand. The role of autophagy as a clearing mechanism, which also eliminates damaged mitochondria, has been demonstrated, but a special role for selective mitophagy has not been confirmed [46]. Cells deleted in ATG32, which is essential for selective mitophagy, showed nearly normal respiratory activity, indicating that selective mitochondria-elimination is not responsible for the observed respiratory defects caused in ATG deletion strains under starvation conditions. On the other hand, cells lacking ATG15, which can form autophagosomes but not degrade their contents, led to respiratory-deficient cells.
Several studies in recent years have shown that the nutritional signaling pathways involving the kinases Akt/PKB, Tor and Ras regulate life span of several model organisms, suggesting that the underlying mechanisms of longevity are conserved among eukaryotes [47]. These kinases are part of nutrition and growth factor sensing pathways and control many aspects of cell physiology, including ribosome biogenesis, translation, autophagy, cell growth, and proliferation [48], and suppress the activity of cellular stress responses (e.g. mediated by transcription factors). It was shown in yeast, that both Ras2p and Tor1p influence key anti-stress regulators, such as the protein kinase Rim15p, the transcriptional activator Msn2/4p, and the histone demethylase/transcription factor Gis1p (Fig. 1). These factors directly or indirectly increase the levels of detoxification enzymes (e.g. Sod2p) and protein chaperones (e.g. heat shock proteins) [35]. Moreover, besides regulating stress-resistance genes, Ras also directly regulates mitochondrial respiration and ROS production [43]. Upon starvation, pharmacological treatment (e.g. rapamycin) or gene deletion, the inhibition of these pathways leads to activation of the key transcription factors enhancing stress defense and longevity. Although many reports have linked these pathways to aging [49,50], the mechanisms of caloric restriction-dependent life span extension remain poorly understood. Recently, Wei and colleagues (2009) identified expression changes in a set of genes controlled by Sch9p as well as Tor1p and Ras2p that caused a metabolic switch, which together with the direct regulation of stress resistance was responsible for enhanced cellular protection and life span extension [35]. This metabolic switch resulted in a shift from respiration to glycolysis, causing the removal of pro-aging carbon sources (e.g. glucose) as well as glycerol biosynthesis. Consistently, strains deleted in either Sch9p, Tor1p or Ras2p (mimicking starvation) displayed an up-regulation of genes involved in glycolysis/fermentation, while mitochondrial related genes were down-regulated. As a consequence, mitochondrial function including the TCA cycle and oxidative phosphorylation was diminished and further inhibited generation of ROS [35]. Accordingly, it was hypothesized by Ruckenstuhl et al. (2010) that the glycolytically generated NADH, not used for respiration, is redirected and used for production of glycerol, a scenario normally taking place under anaerobic conditions [51]. New insights into Tor1p-regulated mitochondrial function during growth and in the postdiauxic stationary phase have been provided by Pan et al. (2011). Their study shows that elevated ROS (e.g. superoxide) in TOR1 deletion strains during growth is an adaptive mitochondrial signal that programs the down-regulation of mitochondrial potential and ROS during stationary phase to promote longevity. They also confirmed a role of media metabolites (e.g. acidification), but proposed a mostly intrinsic control of longevity-related pathways [43].
In order to understand the process of aging, age-related disorders like neurodegeneration and lifestyle-associated diseases like diabetes/stroke, it is important to consider the actual metabolome and the respective anabolic and catabolic activities of a cell in addition to simply analyzing pathway activities.
Acknowledgments
We are grateful to the Austrian Science Fund FWF (S9304-B05 to FM and DC-G, S9302-B05 to FM, P23490 to FM, SFB-LIPOTOX to FM, S9302-B05 to FM, P24381-B20 to FM and TE, GM087346 to JR and W1226-B18 to FM and CS), and the European Commission (Apo-Sys FP7 to FM and TE).
References
- 1.Kind T., Scholz M., Fiehn O. How large is the metabolome? A critical analysis of data exchange practices in chemistry. PLoS One. 2009;4:e5440. doi: 10.1371/journal.pone.0005440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carmona-Gutierrez D., Eisenberg T., Buttner S., Meisinger C., Kroemer G., Madeo F. Apoptosis in yeast: triggers, pathways, subroutines. Cell Death Differ. 2010;17:763–773. doi: 10.1038/cdd.2009.219. [DOI] [PubMed] [Google Scholar]
- 3.Buttner S., Ruli D., Vogtle F.N., Galluzzi L., Moitzi B., Eisenberg T., Kepp O., Habernig L., Carmona-Gutierrez D., Rockenfeller P., Laun P., Breitenbach M., Khoury C., Frohlich K.U., Rechberger G., Meisinger C., Kroemer G., Madeo F. A yeast BH3-only protein mediates the mitochondrial pathway of apoptosis. EMBO J. 2011;30:2779–2792. doi: 10.1038/emboj.2011.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Madeo F., Frohlich E., Ligr M., Grey M., Sigrist S.J., Wolf D.H., Frohlich K.U. Oxygen stress: a regulator of apoptosis in yeast. J. Cell Biol. 1999;145:757–767. doi: 10.1083/jcb.145.4.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Madeo F., Frohlich E., Frohlich K.U. A yeast mutant showing diagnostic markers of early and late apoptosis. J. Cell Biol. 1997;139:729–734. doi: 10.1083/jcb.139.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buttner S., Eisenberg T., Carmona-Gutierrez D., Ruli D., Knauer H., Ruckenstuhl C., Sigrist C., Wissing S., Kollroser M., Frohlich K.U., Sigrist S., Madeo F. Endonuclease G regulates budding yeast life and death. Mol. Cell. 2007;25:233–246. doi: 10.1016/j.molcel.2006.12.021. [DOI] [PubMed] [Google Scholar]
- 7.Eisenberg T., Carmona-Gutierrez D., Buttner S., Tavernarakis N., Madeo F. Necrosis in yeast. Apoptosis. 2010;15:257–268. doi: 10.1007/s10495-009-0453-4. [DOI] [PubMed] [Google Scholar]
- 8.Braun R.J., Buttner S., Ring J., Kroemer G., Madeo F. Nervous yeast: modeling neurotoxic cell death. Trends Biochem. Sci. 2010;35:135–144. doi: 10.1016/j.tibs.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 9.Jewison T., Knox C., Neveu V., Djoumbou Y., Guo A.C., Lee J., Liu P., Mandal R., Krishnamurthy R., Sinelnikov I., Wilson M., Wishart D.S. YMDB: the Yeast Metabolome Database. Nucleic Acids Res. 2012;40:D815–D820. doi: 10.1093/nar/gkr916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eisenberg T., Buttner S., Kroemer G., Madeo F. The mitochondrial pathway in yeast apoptosis. Apoptosis. 2007;12:1011–1023. doi: 10.1007/s10495-007-0758-0. [DOI] [PubMed] [Google Scholar]
- 11.Barnett J.A., Entian K.D. A history of research on yeasts 9: regulation of sugar metabolism. Yeast. 2005;22:835–894. doi: 10.1002/yea.1249. [DOI] [PubMed] [Google Scholar]
- 12.Lohr D., Venkov P., Zlatanova J. Transcriptional regulation in the yeast GAL gene family: a complex genetic network. FASEB J. 1995;9:777–787. doi: 10.1096/fasebj.9.9.7601342. [DOI] [PubMed] [Google Scholar]
- 13.Young E.T., Tachibana C., Chang H.W., Dombek K.M., Arms E.M., Biddick R. Artificial recruitment of mediator by the DNA-binding domain of Adr1 overcomes glucose repression of ADH2 expression. Mol. Cell. Biol. 2008;28:2509–2516. doi: 10.1128/MCB.00658-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turcotte B., Liang X.B., Robert F., Soontorngun N. Transcriptional regulation of nonfermentable carbon utilization in budding yeast. FEMS Yeast Res. 2010;10:2–13. doi: 10.1111/j.1567-1364.2009.00555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wissing S., Ludovico P., Herker E., Buttner S., Engelhardt S.M., Decker T., Link A., Proksch A., Rodrigues F., Corte-Real M., Frohlich K.U., Manns J., Cande C., Sigrist S.J., Kroemer G., Madeo F. An AIF orthologue regulates apoptosis in yeast. J. Cell Biol. 2004;166:969–974. doi: 10.1083/jcb.200404138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ludovico P., Rodrigues F., Almeida A., Silva M.T., Barrientos A., Corte-Real M. Cytochrome c release and mitochondria involvement in programmed cell death induced by acetic acid in Saccharomyces cerevisiae. Mol. Biol. Cell. 2002;13:2598–2606. doi: 10.1091/mbc.E01-12-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Susin S.A., Lorenzo H.K., Zamzami N., Marzo I., Snow B.E., Brothers G.M., Mangion J., Jacotot E., Costantini P., Loeffler M., Larochette N., Goodlett D.R., Aebersold R., Siderovski D.P., Penninger J.M., Kroemer G. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397:441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- 18.Vahsen N., Cande C., Briere J.J., Benit P., Joza N., Larochette N., Mastroberardino P.G., Pequignot M.O., Casares N., Lazar V., Feraud O., Debili N., Wissing S., Engelhardt S., Madeo F., Piacentini M., Penninger J.M., Schagger H., Rustin P., Kroemer G. AIF deficiency compromises oxidative phosphorylation. EMBO J. 2004;23:4679–4689. doi: 10.1038/sj.emboj.7600461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miramar M.D., Costantini P., Ravagnan L., Saraiva L.M., Haouzi D., Brothers G., Penninger J.M., Peleato M.L., Kroemer G., Susin S.A. NADH oxidase activity of mitochondrial apoptosis-inducing factor. J. Biol. Chem. 2001;276:16391–16398. doi: 10.1074/jbc.M010498200. [DOI] [PubMed] [Google Scholar]
- 20.Blank L.M., Sauer U. TCA cycle activity in Saccharomyces cerevisiae is a function of the environmentally determined specific growth and glucose uptake rates. Microbiology. 2004;150:1085–1093. doi: 10.1099/mic.0.26845-0. [DOI] [PubMed] [Google Scholar]
- 21.Minard K.I., McAlister-Henn L. Sources of NADPH in yeast vary with carbon source. J. Biol. Chem. 2005;280:39890–39896. doi: 10.1074/jbc.M509461200. [DOI] [PubMed] [Google Scholar]
- 22.Sapienza K., Balzan R. Metabolic aspects of aspirin-induced apoptosis in yeast. FEMS Yeast Res. 2005;5:1207–1213. doi: 10.1016/j.femsyr.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 23.Velagapudi V.R., Wittmann C., Schneider K., Heinzle E. Metabolic flux screening of Saccharomyces cerevisiae single knockout strains on glucose and galactose supports elucidation of gene function. J. Biotechnol. 2007;132:395–404. doi: 10.1016/j.jbiotec.2007.08.043. [DOI] [PubMed] [Google Scholar]
- 24.Kruger A., Gruning N.M., Wamelink M.M., Kerick M., Kirpy A., Parkhomchuk D., Bluemlein K., Schweiger M.R., Soldatov A., Lehrach H., Jakobs C., Ralser M. The pentose phosphate pathway is a metabolic redox sensor and regulates transcription during the antioxidant response. Antioxid. Redox Signal. 2011;15:311–324. doi: 10.1089/ars.2010.3797. [DOI] [PubMed] [Google Scholar]
- 25.Gruning N.M., Rinnerthaler M., Bluemlein K., Mulleder M., Wamelink M.M., Lehrach H., Jakobs C., Breitenbach M., Ralser M. Pyruvate kinase triggers a metabolic feedback loop that controls redox metabolism in respiring cells. Cell Metab. 2011;14:415–427. doi: 10.1016/j.cmet.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boles E., Schulte F., Miosga T., Freidel K., Schluter E., Zimmermann F.K., Hollenberg C.P., Heinisch J.J. Characterization of a glucose-repressed pyruvate kinase (Pyk2p) in Saccharomyces cerevisiae that is catalytically insensitive to fructose-1,6-bisphosphate. J. Bacteriol. 1997;179:2987–2993. doi: 10.1128/jb.179.9.2987-2993.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rockenfeller P., Ring J., Muschett V., Beranek A., Buettner S., Carmona-Gutierrez D., Eisenberg T., Khoury C., Rechberger G., Kohlwein S.D., Kroemer G., Madeo F. Fatty acids trigger mitochondrion-dependent necrosis. Cell Cycle. 2010;9:2836–2842. doi: 10.4161/cc.9.14.12267. [DOI] [PubMed] [Google Scholar]
- 28.Hiltunen J.K., Mursula A.M., Rottensteiner H., Wierenga R.K., Kastaniotis A.J., Gurvitz A. The biochemistry of peroxisomal beta-oxidation in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2003;27:35–64. doi: 10.1016/S0168-6445(03)00017-2. [DOI] [PubMed] [Google Scholar]
- 29.Gurvitz A., Rottensteiner H. The biochemistry of oleate induction: transcriptional upregulation and peroxisome proliferation. Biochim. Biophys. Acta. 2006;1763:1392–1402. doi: 10.1016/j.bbamcr.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 30.Jungwirth H., Ring J., Mayer T., Schauer A., Buttner S., Eisenberg T., Carmona-Gutierrez D., Kuchler K., Madeo F. Loss of peroxisome function triggers necrosis. FEBS Lett. 2008;582:2882–2886. doi: 10.1016/j.febslet.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 31.Ruckenstuhl C., Buttner S., Carmona-Gutierrez D., Eisenberg T., Kroemer G., Sigrist S.J., Frohlich K.U., Madeo F. The Warburg effect suppresses oxidative stress induced apoptosis in a yeast model for cancer. PLoS One. 2009;4:e4592. doi: 10.1371/journal.pone.0004592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazurek S., Boschek C.B., Hugo F., Eigenbrodt E. Pyruvate kinase type M2 and its role in tumor growth and spreading. Semin. Cancer Biol. 2005;15:300–308. doi: 10.1016/j.semcancer.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 33.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- 34.Longo V.D., Nislow C., Fabrizio P. Endosomal protein sorting and autophagy genes contribute to the regulation of yeast life span. Autophagy. 2010;6:1227–1228. doi: 10.4161/auto.6.8.13850. [DOI] [PubMed] [Google Scholar]
- 35.Wei M., Fabrizio P., Madia F., Hu J., Ge H., Li L.M., Longo V.D. Tor1/Sch9-regulated carbon source substitution is as effective as calorie restriction in life span extension. PLoS Genet. 2009;5:e1000467. doi: 10.1371/journal.pgen.1000467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fabrizio P., Longo V.D. The chronological life span of Saccharomyces cerevisiae. Methods Mol. Biol. 2007;371:89–95. doi: 10.1007/978-1-59745-361-5_8. [DOI] [PubMed] [Google Scholar]
- 37.Weinberger M., Mesquita A., Caroll T., Marks L., Yang H., Zhang Z., Ludovico P., Burhans W.C. Growth signaling promotes chronological aging in budding yeast by inducing superoxide anions that inhibit quiescence. Aging (Albany NY) 2010;2:709–726. doi: 10.18632/aging.100215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baur J.A., Pearson K.J., Price N.L., Jamieson H.A., Lerin C., Kalra A., Prabhu V.V., Allard J.S., Lopez-Lluch G., Lewis K., Pistell P.J., Poosala S., Becker K.G., Boss O., Gwinn D., Wang M., Ramaswamy S., Fishbein K.W., Spencer R.G., Lakatta E.G., Le Couteur D., Shaw R.J., Navas P., Puigserver P., Ingram D.K., de Cabo R., Sinclair D.A. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harrison D.E., Strong R., Sharp Z.D., Nelson J.F., Astle C.M., Flurkey K., Nadon N.L., Wilkinson J.E., Frenkel K., Carter C.S., Pahor M., Javors M.A., Fernandez E., Miller R.A. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eisenberg T., Knauer H., Schauer A., Buttner S., Ruckenstuhl C., Carmona-Gutierrez D., Ring J., Schroeder S., Magnes C., Antonacci L., Fussi H., Deszcz L., Hartl R., Schraml E., Criollo A., Megalou E., Weiskopf D., Laun P., Heeren G., Breitenbach M., Grubeck-Loebenstein B., Herker E., Fahrenkrog B., Frohlich K.U., Sinner F., Tavernarakis N., Minois N., Kroemer G., Madeo F. Induction of autophagy by spermidine promotes longevity. Nat. Cell Biol. 2009;11:1305–1314. doi: 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- 41.Morselli E., Marino G., Bennetzen M.V., Eisenberg T., Megalou E., Schroeder S., Cabrera S., Benit P., Rustin P., Criollo A., Kepp O., Galluzzi L., Shen S., Malik S.A., Maiuri M.C., Horio Y., Lopez-Otin C., Andersen J.S., Tavernarakis N., Madeo F., Kroemer G. Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J. Cell Biol. 2011;192:615–629. doi: 10.1083/jcb.201008167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herker E., Jungwirth H., Lehmann K.A., Maldener C., Frohlich K.U., Wissing S., Buttner S., Fehr M., Sigrist S., Madeo F. Chronological aging leads to apoptosis in yeast. J. Cell Biol. 2004;164:501–507. doi: 10.1083/jcb.200310014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pan Y., Schroeder E.A., Ocampo A., Barrientos A., Shadel G.S. Regulation of yeast chronological life span by TORC1 via adaptive mitochondrial ROS signaling. Cell Metab. 2011;13:668–678. doi: 10.1016/j.cmet.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ristow M., Zarse K. How increased oxidative stress promotes longevity and metabolic health: the concept of mitochondrial hormesis (mitohormesis) Exp. Gerontol. 2010;45:410–418. doi: 10.1016/j.exger.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 45.Scherz-Shouval R., Shvets E., Fass E., Shorer H., Gil L., Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki S.W., Onodera J., Ohsumi Y. Starvation induced cell death in autophagy-defective yeast mutants is caused by mitochondria dysfunction. PLoS One. 2011;6:e17412. doi: 10.1371/journal.pone.0017412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fontana L., Partridge L., Longo V.D. Extending healthy life span—from yeast to humans. Science. 2010;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wullschleger S., Loewith R., Hall M.N. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 49.Wei M., Fabrizio P., Hu J., Ge H., Cheng C., Li L., Longo V.D. Life span extension by calorie restriction depends on Rim15 and transcription factors downstream of Ras/PKA, Tor, and Sch9. PLoS Genet. 2008;4:e13. doi: 10.1371/journal.pgen.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rubinsztein D.C., Marino G., Kroemer G. Autophagy and aging. Cell. 2011;146:682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 51.Ruckenstuhl C., Carmona-Gutierrez D., Madeo F. The sweet taste of death: glucose triggers apoptosis during yeast chronological aging. Aging (Albany NY) 2010;2:643–649. doi: 10.18632/aging.100223. [DOI] [PMC free article] [PubMed] [Google Scholar]