Summary
Epigenetic and genetic alterations have long been thought of as two separate mechanisms participating in carcinogenesis. A recent outcome of whole exome sequencing of thousands of human cancers has been the unexpected discovery of many inactivating mutations in genes that control the epigenome. These mutations have the potential to disrupt DNA methylation patterns, histone modifications and nucleosome positioning and hence, gene expression. Genetic alteration of the epigenome therefore contributes to cancer just as epigenetic process can cause point mutations and disable DNA repair functions. This crosstalk between the genome and the epigenome offers new possibilities for therapy.
Cancer has traditionally been viewed as a set of diseases that are driven by the accumulation of genetic mutations which have been considered the major causes of neoplasia(Hanahan and Weinberg, 2011). However, this paradigm has now been expanded to incorporate the disruption of epigenetic regulatory mechanisms which are prevalent in cancer(Baylin and Jones, 2011; Sandoval and Esteller, 2012).
Both genetic and epigenetic views ultimately involve abnormal gene expression. The expression state of a particular gene is determined by the packaging of its DNA regulatory regions at promoters and/or enhancers and insulators in chromatin and by the presence of transcription factors (TFs) and chromatin modifying enzymes. The genetic path to cancer is relatively straightforward: mutation of tumor suppressors and/or oncogenes causes either loss or gain of function and abnormal expression. The epigenetic pathway to cancer is not as simple and is determined by chromatin structure including DNA methylation, histone variants and modifications, nucleosome remodeling as well as small non-coding regulatory RNAs(Sharma et al., 2010). During tumor initiation and progression, the epigenome goes through multiple alterations, including a genome-wide loss of DNA methylation (hypomethylation), frequent increases in promoter methylation of CpG islands, changes in nucleosome occupancy and modification profiles.
More recently, intriguing evidence has emerged that genetic and epigenetic mechanisms are not separate events in cancer; they intertwine and take advantage of each other during tumorigenesis. Alterations in epigenetic mechanisms can lead to genetic mutations and genetic mutations in epigenetic regulators leads to an altered epigenome. In this review, we will discuss the collusion between epigenetics and genetics in cancer.
How Epigenetics Affect Genetics
Epigenetic mechanisms help establish cellular identities and failure of the proper preservation of epigenetic marks can result in inappropriate activation or inhibition of various cellular signaling pathways leading to cancer. It is now generally accepted that human cancer cells harbor global epigenetic abnormalities and that epigenetic alterations may be the key to initiating tumorigenesis(Baylin and Jones, 2011; Sandoval and Esteller, 2012; Sharma et al., 2010). The cancer epigenome is characterized by substantial changes in various epigenetic regulatory layers; herein we introduce some important examples of epigenetic disruptions that cause mutation of key genes and/or alteration of signaling pathways in cancer development.
Epigenetic Silencing Causes the Loss of Function of Genes and Predisposes to Genetic Mutation
Promoter hypermethylation of classic tumor suppressor genes is commonly observed in cancers and cancer cells take advantage of this for tumorigenesis(Baylin and Jones, 2011). Genes controlling the cell cycle and DNA repair, such as RB, BRCA1/2 and PTEN have all been reported to be hypermethylated or mutated/deleted in cancer(Hatziapostolou and Iliopoulos, 2011). There are also several genes which are seldom mutated but are silenced in cancer; promoter hypermethylation is the predominant mechanism for the loss of their functions(Baylin and Jones, 2011). O6-methylguanine-DNA methyltransferase (MGMT), which encodes a DNA repair gene, Cyclin-dependent kinase inhibitor 2B (CDKN2B), which encodes a cell cycle regulator p15, and RASSF1A, which encodes a protein that binds to the RAS oncogene all belong to this category and they have been implicated with protective roles against tumorigenesis.
Several DNA repair genes are known to be subject to promoter methylation. MGMT removes carcinogen-induced O6-methylguanine adducts from DNA, which result in G to A transition mutations. Cancers with hypermethylated MGMT are susceptible to genetic mutation in critical genes such as p53 or KRAS(Baylin and Jones, 2011; Esteller, 2007). The mismatch-repair gene MLH1 plays an important role in genomic stability and the loss of function of this gene by promoter hypermethylation causes microsatellite instability, which is a key factor in several cancers including colorectal and endometrial cancers(Krivtsov and Armstrong, 2007). The MLH1 promoter is already hypermethylated in normal colonic epithelium of some colorectal cancer patients, suggesting this epigenetic change is early event of tumorigenesis and precedes downstream genetic mutation(Hitchins et al., 2011). Notably, single nucleotide variants (SNVs) of MLH1 5′UTR are correlated with the hypermethylation of its promoter, highlighting a close relationship between genetic and epigenetic disruption in cancer(Hitchins et al., 2011).
Epigenetic Silencing Facilitates the Selection of Mutations in Key Signaling Pathways
Direct evidence for a close epigenetic-genetic cooperation is apparent in the colon cancer cell line HCT116 in which one allele of MLH1 and CDKN2A is genetically mutated whereas the other allele is silenced by DNA methylation(Baylin and Ohm, 2006). The lack of functional expression of MLH1 and CDKN2A causes defects in DNA mismatch repair and cell cycle regulation. Another example of epigenetic–genetic cooperation is in the WNT signaling pathway(Schepers and Clevers, 2012). In normal cells, secreted frizzled-related proteins (SFRPs) antagonize WNT signaling. Epigenetic silencing of SFRPs induces abnormal activation of this signaling pathway, further promoting the expression of several genes whose products are responsible for cell proliferation. As a result of survival and proliferative advantages, these cells accumulate genetic mutations in other components of the WNT signaling pathway. There are also several examples where epigenetic silencing allows abnormal proliferation pathways and increases the likelihood for mutation in genetic gate keepers and increases cancer risk(Baylin and Jones, 2011).
More recent results from The Cancer Genome Atlas (TCGA) project provide an integrative view of ovarian carcinoma based on integrated genomic analyses (Network, 2011). The mutation spectrum is unexpectedly simple showing the predominance of p53 mutations and other low frequency mutations in nine genes including BRCA1, BRCA2 and RB. On the other hand, promoter hypermethylation is observed in 168 genes and those genes are epigenetically silenced and correlated with reduced expression. It is noteworthy that clustering of variable DNA methylation across tumors can identify subtypes. Indeed, the CpG island methylator phenotype (CIMP) is reported in colorectal cancer and glioblastoma and this subgroup shows distinctive characters such as genetic and clinical features(Hinoue et al., 2012; Noushmehr et al., 2010). A CIMP-high subgroup is strongly associated with MLH1 DNA hypermethylation and BRAF mutation, while a CIPM-low subgroup is related to KRAS mutation(Hinoue et al., 2012).
Role of 5-methylcytosine (5mC) in Generating Disease-causing Mutations
The methylation of cytosine residues in the germline has led to an approximately 75% decrease in the frequency of CpG methyl acceptor sites. This is thought to be due to the spontaneous hydrolytic deamination of 5mC to thymine rather than uracil, which is formed by deamination of cytosine. The resulting T:G mismatch is more difficult to repair and about a third of all disease causing familial mutations and single nucleotide polymorphisms or variants (SNPs or SNVs) occur at methylated CpG sites. What is often overlooked is that the presence of 5mC in the gene bodies and coding regions of genes such as p53 is responsible for generating inactivating C to T transition mutations causing hotspots in somatic cells(Rideout et al., 1990). For example, as many as 50% of p53 point mutations in colon cancer occur at such sites, clearly demonstrating that an epigenetic mark (5mC) directly causes somatic mutations.
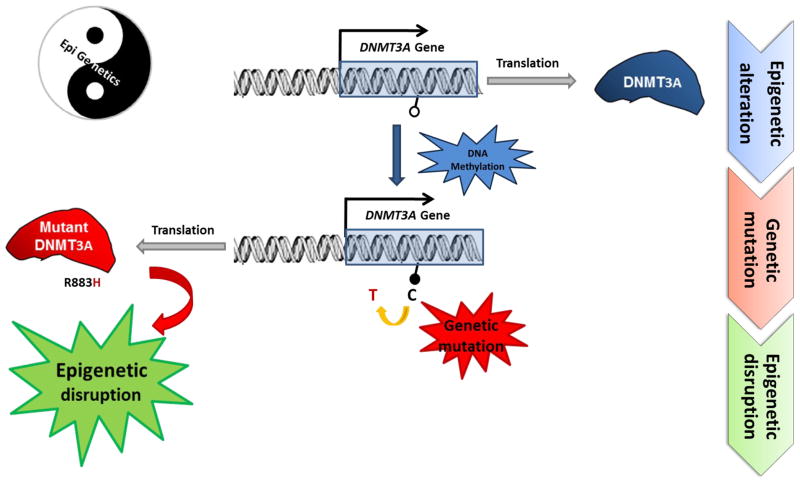
More interestingly, a somatic DNMT3A hotspot mutation in AML is caused by C to T transitions at a CpG site, possibly due to the methylation of its own exon by the enzyme (epigenetic alteration) and the subsequent deamination of 5mC (genetic mutation)(Ley et al., 2010) (Figure 1). The effect of the point mutation is not yet fully understood since methylation changes are not observed in the tumor. It is possible that this mutation alters DNMT3A function and/or activity and may further disrupt whole epigenetic regulation mechanism (Figure 1).
Figure 1. The Crosstalk between Cancer Genetics and Epigenetics.
The methylation of CpG sites located in DNMT3A exons (epigenetic alteration, represented as a black circle) potentially leads to genetic mutation in somatic cells by the hydrolytic deamination of 5mC to form a C to T transition mutation. Although it is not known whether DNMT3A directly methylates its own exon and the effect of this genetic mutation is not yet fully understood, it is possible that the C to T transition alters DNMT3A function and/or activity and thereby disrupts the epigenetic landscape. The Yin-Yang diagram emphasizes how epigenetic and genetic interactions are required to achieve perfect balance and suggests that disruption of the balance can lead to disease.
Role of MicroRNA in Tumorigenesis
MicroRNAs (miRNAs) are a class of small non-coding RNAs which play key roles in epigenetic regulation by controlling the translation and/or stability of mRNAs. There are over 1000 human miRNAs and, interestingly, these miRNAs frequently target regions related to cancer development(Ryan et al., 2010). They have been classified as oncogenic, tumor suppressive or context dependent miRNAs(Kasinski and Slack, 2011a). Indeed, oncogenic miRNAs such as miR-155 or miR-21 are frequently overexpressed and tumor suppressive miRNAs such as miR-146 or miR-15~16 are deleted in cancers(Kasinski and Slack, 2011a). Mutation in the miRNA can disrupt its recognition of binding targets and further result in oncogene activation and/or tumor suppressor repression. Additionally miRNAs including miR-101 and miR-29 target epigenetic modifiers such as EZH2(Friedman et al., 2009; Varambally et al., 2008) and DNMT3A/B(Fabbri et al., 2007) respectively. This can result in further widespread epigenetic alterations(Fabbri and Calin, 2010; Kasinski and Slack, 2011a) and might lead to the methylation of promoters of other miRNAs that target oncogenes. miR-127, which targets BCL6, is abnormally methylated and silenced in cancer(Saito et al., 2006) highlighting the reciprocal regulation of miRNAs, epigenetic modifiers and genetic defects in cancer.
Given the importance of epigenetic silencing in the development of cancer, distinguishing “drivers” and “passengers” is becoming an important priority for the field. Driver genes must be essential for cancer causation whereas passenger genes are not necessary(Kelly et al., 2010). With the improvement of technology, it may eventually be possible to specifically distinguish epigenetic disruptions of the driver genes(De Carvalho et al., 2012; Kalari and Pfeifer, 2010). Current evidence shows that epigenetic disruption plays a key role at every stage of tumorigenesis and has a significant impact on the underlying mechanisms of tumorigenesis and development of cancer therapy.
How Genetics Affect Epigenetics
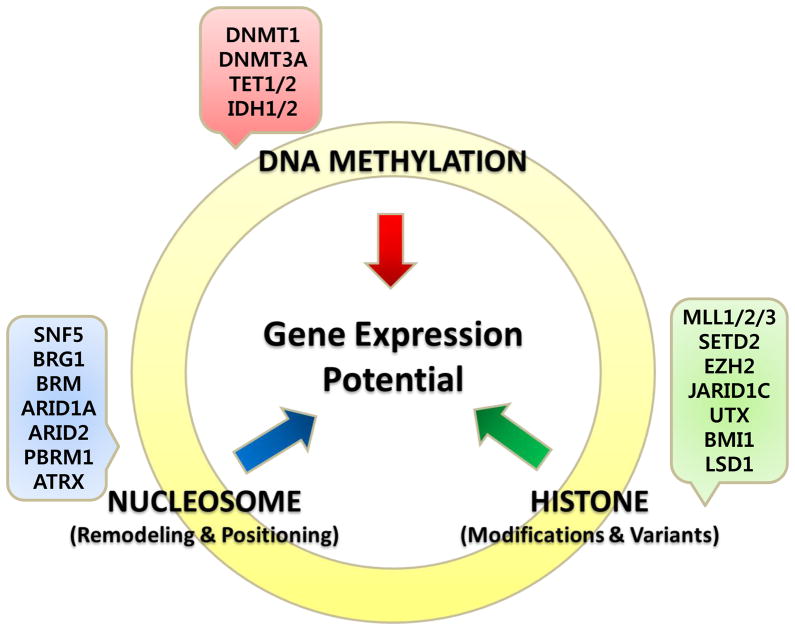
While epigenetics and genetics can cooperate in cancer initiation and progression, the interconnectedness between of these two processes is becoming increasingly apparent with the realization that several epigenetic modifiers are mutated in human cancers(Kasinski and Slack, 2011b; Rodriguez-Paredes and Esteller, 2011; Schuettengruber et al., 2011; Wilson and Roberts, 2011). Some examples of genetic mutations of epigenetic modifiers are shown in Table 1 and Figure 2. The mutation of epigenetic modifiers presumably leads to profound epigenetic changes, including aberrant DNA methylation, histone modifications and nucleosome positioning although this remains to be demonstrated. These epigenetic alterations can lead to abnormal gene expression and genomic instability, which may predispose to cancer(Rodriguez-Paredes and Esteller, 2011; Wilson and Roberts, 2011).
Table 1.
Epigenetic Modifiers in Cancer
| Gene | Function | Tumor Type | Alteration | |
|---|---|---|---|---|
| DNA Methylation | DNMT1 | DNA methyltransferase | Colorectal, Non-small cell lung, pancreatic, gastric, breast cancer | Mutation(Kanai et al., 2003) Overexpression(Wu et al., 2007) |
| DNMT3A | DNA methyltransferase | MDS; AML | Mutation(Ley et al., 2010; Yamashita et al., 2010; Yan et al., 2011) | |
| DNMT3B | DNA methyltransferase | ICF syndrome, SNPs in breast and lung adenoma | Mutation(Wijmenga et al., 2000) Mutation(Shen et al., 2002) |
|
| MBD1/2 | Methyl binding protein | Lung and breast cancer | Mutation(Sansom et al., 2007) | |
| TET1 | 5′methylcytosine hydroxylase | AML | Chromosome translocation(De Carvalho et al., 2010; Wu and Zhang, 2010) | |
| TET2 | 5′methylcytosine hydroxylase | MDS, myeloid malignancies (AML), Gliomas | Mutation/silencing(Tan and Manley, 2009) | |
| IDH1/2 | Isocitrate dehydrogenase | Glioma, AML | Mutation(Figueroa et al., 2010; Lu et al., 2012; Turcan et al., 2012) | |
| AID | 5′cytidine deaminase | CML | Aberrant expression(De Carvalho et al., 2010) | |
| Histone Modification | MLL1/2/3 | Histone methyltransferase H3K4 | Bladder TCC, ALL and AML, Non-Hodgkin lymphoma, B-Cell lymphoma, Prostate (primary) | Translocation, mutation, aberrant expression(Gui et al., 2011; Morin et al., 2011) |
| BRD4 | Bromodomain containing 4 | Nuclear protein in testis (NUT) midline carcinoma, Breast, Colon and AML | Translocation (fusion protein), aberrant expression(Filippakopoulos et al., 2010; Zuber et al., 2011) | |
| EZH2 | Histone methyltransferase H3K27 | breast, prostate, bladder, colon, pancreas, liver, gastric, uterine tumors, melanoma, lymphoma, myeloma, and Ewing’s sarcoma | Mutation, aberrant expression(Chase and Cross, 2011; Tsang and Cheng, 2010) | |
| ASXL | Enhancer of trithorax and polycomb group (EAP) Additional sex combs like 1 | MDS and AML, Bohring-Opitz syndrome | Mutation(Gelsi-Boyer et al., 2012; Hoischen et al., 2012) | |
| BMI-1 | PRC1 subunit | ovarian, mantle cell lymphomas and Merkel cell carcinomas | Overexpression(Jiang et al., 2009; Lukacs et al., 2011) | |
| G9a | Histone methyltransferase H3K9 | HCC, cervical, uterine, ovarian and breast cancer | Aberrant expression(Varier and Timmers, 2010) | |
| PRMT1/5 | Protein arginine methyltransferase | Breast/gastric | Aberrant expression(Miremadi et al., 2007) | |
| LSD1 | Histone demethylase H3K4/H3K9 | Prostate | Mutation(Rotili and Mai, 2011) | |
| UTX (KDM6A) | Histone demethylase H3K27 | Bladder, breast, kidney, lung, pancreas, oesophagus, colon, uterus, brain | Mutation(Rotili and Mai, 2011) | |
| JARID1B/C | Histone demethylase H3K4/H3K9 | Testicular and breast, RCCC | Overexpression(Rotili and Mai, 2011) | |
| EP300 | Histone deacetyltransferase | Breast, Colorectoal, pancreatic cancer | Mutation(Miremadi et al., 2007) | |
| CREBBP | Histone acetyltransferase | Gastric and colorectal, epithelial, ovarian, lung, esophageal cancer | Mutation, overexpression (Miremadi et al., 2007) | |
| PCAF | Histone acetyltransferase | Epithelial | Mutation(Miremadi et al., 2007) | |
| HDAC2 | Histone deacetyltransferase | Colonic, gastric, endometrial cancer | Mutation(Ropero et al., 2006) | |
| SIRT1, HDAC5/7A | Histone deacetyltransferase | Breast, colorectal, prostate cancer | Mutation, aberrant expression(Miremadi et al., 2007) | |
| Chromatin Remodeling | SNF5 (SMARCB1, INI1) | BAF subunit | Kidney malignant rhabdoid tumors, atypical rhabdoid/teratoid tumors (extrarenal), epithelioid sarcomas, small cell hepatoblastomas, extraskeletal myxoid chondrosarcomas, and undifferentiated sarcomas | Mutation, silencing, loss of expression(Wilson and Roberts, 2011) |
| BRG1 (SMARCA4) | ATPase of BAF | Lung, rhabdoid, medulloblastoma | Mutation, low expression(Wilson and Roberts, 2011) | |
| BRM (SMARCA2) | ATPase of BAF | Prostate, Basal cell carcinoma | Mutation, low expression(de Zwaan and Haass, 2010; Sun et al., 2007) | |
| ARID1A (BAF250A) | BAF subunit | Ovarian clear cell carcinomas, 30% of endometrioid carcinomas, endometrial carcinomas | Mutation, Genomic rearrangement, low expression(Guan et al., 2011; Jones et al., 2010) | |
| ARID2 (BAF200) | PBAF subunit | Primary pancreatic adenocarcinomas | Mutation(Li et al., 2011) | |
| BRD7 | PBAF subunit | Bladder TCC | Mutation(Drost et al., 2010) | |
| PBRM1 (BAF180) | PBAF subunit | Breast tumors | Mutation(Varela et al., 2011) | |
| SRCAP | ATPase of SWR1 | Prostate | Aberrant expression(Balakrishnan et al., 2007) | |
| P400/Tip60 | ATPase of SWR1, Acetylase of SWR1 | Colon, lymphomas, head-and-neck, breast | Mutation, aberrant expression(Mattera et al., 2009) | |
| CHD4/5 | ATPase of NURD | Colorectal and gastric cancer, ovarian, prostate, neuroblastoma | Mutation(Bagchi et al., 2007; Kim et al., 2011; Wang et al., 2011a) | |
| CHD7 | ATP-dependent helicase | gastric and colorectal | Mutation(Wessels et al., 2010) |
MDS: Myelodysplastic syndromes; AML: acute myeloid leukemia; TCC: transitional cell carcinoma; RCCC: renal clear cell carcinoma
Figure 2. Genetic Mutations in Epigenetic Modifiers in Cancer.
The drawing shows the interaction between epigenetic processes in specifying gene expression patterns. Recent whole exome sequencing studies show that mutations in the three classes of epigenetic modifiers is frequently observed in various types of cancers, further highlighting the crosstalk between genetics and epigenetics. Examples of some but not all of these mutations which are discussed in this review are shown. The mutations of epigenetic modifiers probably cause genome-wide epigenetic alterations in cancer but these have yet to be demonstrated in a genome wide scale. Understanding the relationship of genetic and the epigenetic changes in cancer will offer novel insights for cancer therapies.
DNA Methylation Machinery
While non-CpG methylation has been reported in pluripotent cells(Hawkins et al., 2010; Meissner et al., 2008), DNA methylation in mammals occurs predominantly at CpG dinucleotides, and methylation of CpG islands acts as a relatively stable gene silencing mechanism(Jones and Liang, 2009). The majority of the CpG islands, which represent over 50% of promoters, remain mostly unmethylated in somatic cells. DNA methylation is important for the regulation of non-CpG island as well as CpG island promoters and in repetitive sequences (LINE and/or SINE) to maintain genomic stability(De Carvalho et al., 2010; Jones and Liang, 2009). DNA methylation in mammalian cells is regulated by a family of DNA methyltransferases (DNMTs) which catalyze the transfer of methyl groups from S-adenosyl-L-methionine to the 5′ position of cytosine bases in the CpG dinucleotide. DNMT3A and DNMT3B, which are expressed throughout the cell cycle(Kinney and Pradhan, 2011), establish new DNA methylation patterns early in development. During replication, the original DNA methylation pattern is maintained largely by DNMT1 activity, which prefers hemi-methylated DNA over non-methylated DNA as a substrate and which is also supported by recent structure study(Song et al., 2011) and is therefore responsible for the maintenance of methylation patterns during cell division, with some participation by DNMT3A and DNMT3B(Jones and Liang, 2009; Sharma et al., 2010).
DNMT1 mutations have been described in colorectal cancer(Kanai et al., 2003) and as previously noted DNMT3A mutations are frequent in myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML)(Ley et al., 2010; Yamashita et al., 2010; Yan et al., 2011). Germ line mutations in DNMT3B underlie immunodeficiency-centromeric instability-facial anomalies (ICF) syndrome and chromosome instability(Wijmenga et al., 2000) and SNPs in DNMT3B have been suggested to be associated with risk of several cancers including breast and lung adenocarcinoma(Shen et al., 2002). In addition to the example in Figure1, other mutations of DNMT3A occur at several positions and generally represent a loss of function - similar to DNMT3B mutations that are associated with ICF syndrome(Ley et al., 2010). Recent studies uncovered a role of DNMT3A in silencing self-renewal genes in hematopoietic stem cells (HSCs) to permit efficient hematopoietic differentiation and its loss progressively impairs HSC differentiation(Challen et al., 2011; Trowbridge and Orkin, 2011). All known DNMT3A mutations are related to poor survival in AML(Ley et al., 2010; Yan et al., 2011), suggesting that these mutations prevent differentiation and have an important role in the progression of disease.
In addition to various mutations, DNMT1, DNMT3A and DNMT3B are often overexpressed in various cancers and possibly contribute to ectopic hypermethylation(Wu et al., 2007). However, careful studies should be done to understand the relationship between the expression of DNMTs and methylation disruption since the overexpression of DNMTs may be a reflection of increased cell proliferation.
Methyl-binding domain (MBD) proteins, including MeCP2, MBD1, MBD2 and MBD4, bind to methylated CpG sites and might be involved in mediating transcriptional repression(Bogdanovic and Veenstra, 2009). Genetic mutations in MBD1 and MBD2 increase the risk of lung and breast cancer, respectively(Sansom et al., 2007). MeCP2 and other MBD protein alterations have been reported in several cancers, however the mechanism is yet to be uncovered.
The field of DNA demethylation has been controversial(Ooi and Bestor, 2008) but recent evidence suggests that this demethylation can occur through two processes - active and passive(De Carvalho et al., 2010; Wu and Zhang, 2010). Passive DNA demethylation occurs when maintenance DNA methylation is impaired during DNA replication, resulting in loss of methylation of the newly synthesized DNA strand. In contrast, active DNA demethylation is dependent on the ability of one or more enzymes to hydroxylate, further oxidize or deaminate 5mC and can occur independent of DNA replication(Bhutani et al., 2011; Wu and Zhang, 2010). Recently, several proteins have been implicated to be erasers of DNA methylation including TET (Ten-eleven-translocation) and AID (Activation-induced cytidine deaminase)(De Carvalho et al., 2010; Ko et al., 2010; Wu and Zhang, 2010). Active DNA demethylation is currently thought of as being a stepwise process - hydroxylation of 5mC (5hmC) by TET proteins followed by deamination by AID/APOBEC protein or carboxylation and entry in to the subsequent base excision repair (BER) pathway(Bhutani et al., 2011). Alternatively, 5hmC is not recognized by DNMT1(Lao et al., 2010), replication of DNA containing this base would lead to loss of the 5mC mark in the subsequent S phase.
Three TET family members (TET1, TET2 and TET3) have been reported so far and each protein seems to have a distinct function in different cellular contexts(Cimmino et al., 2011). Mutations in TET2 including frame shift, nonsense and missense mutations, have been found in myelodysplastic syndrome and in myeloproliferative neoplasms(Tan and Manley, 2009). Notably, TET2 loss-of-function mutations were mutually exclusive of mutations in IDH1 (isocitrate dehydrogenase1) and IDH2, which are known to induce DNA hypermethylation and impair differentiation in hematopoietic cells (Figueroa et al., 2010). IDH1/2 mutations in glioma and AML cause accumulation of 2-hydoxyglutarate which is called a “oncometabolite” and further impairs the DNA demethylation process and causes hypermethylation in glioma(Turcan et al., 2012). Remarkably, IDH1/2 mutations also disrupt histone demethylation and block cell differentiation in non-transformed cells(Lu et al., 2012).
Considering that DNMTs/MBD proteins and enzymes involved in DNA demethylation contribute directly to the level of DNA methylation but also to nucleosome occupancy patterns, the alteration of these machineries in cancer development could be broader than previously realized.
Histone Modification Machinery
Nucleosomes, which are the basic building blocks of chromatin, contain DNA wrapped around histones (Luger et al., 1997). Histones are regulators of chromatin dynamics either by changing chromatic structure by altering electrostatic charge or providing protein recognition sites by specific modifications(Mills, 2010; Suganuma and Workman, 2011). Histone modifications at specific residues characterize genomic regulatory regions, such as active promoter regions which are enriched in trimethylated H3 at lysine 4 (H3K4me3), inactive promoters which are enriched in trimethylated H3 at lysine 27 (H3K27me3) or trimethylated H3 at lysine 9 (H3K9me3), and regulatory enhancers that are enriched in monomethylated H3 at lysine 4 (H3K4me1) and/or acetylated H3 at lysine 27 (H3K27ac)(Hawkins et al., 2011; Hon et al., 2009; Mills, 2010). These histone modification patterns are regulated by enzymes including histone acetyltransferases (HATs) and deacetylases (HDACs), which introduce and remove acetyl groups respectively. Histone methyltransferases (HMTs) and demethylases (HDMs), on the other hand, introduce and remove methyl groups. During tumorigenesis, cells undergo global changes in histone modifications and in the distribution of histone variants such as H2A.Z(Conerly et al., 2010) which may affect the recruitment of transcription factors and often components of the transcription machinery, thereby contributing to aberrant gene expression(Mills, 2010; Sharma et al., 2010).
The acetylation of lysine residues on histones is generally associated with active gene transcription. HATs can be grouped in to three categories based on their sequence similarities: Gcn5/PCAF, p300/CBP and the MYST families(Yang, 2004). Mutations or translocations of these genes are observed in colon, uterine, lung tumors and in leukemias(Esteller, 2007). Further these HATs (p300, CBP and MYST4) are commonly involved in chromosomal translocations in hematological cancers rather than in solid tumors(Iyer et al., 2004). For example, AML1-ETO, the fusion protein generated by the t(8;21) translocation which is also the most common fusion protein in AML, requires its acetylation mediated by p300 for oncogenic activity(Wang et al., 2011b). HDACs remove acetyl groups from histone tails and at least 18 HDAC genes have been identified in the human genome. HDACs as well as HATs function as part of large multi-protein complexes(Marks et al., 2001). HDACs have been implicated in cancer due to their aberrant binding and consequent silencing of tumor suppressor genes. For example, hypoacetylation of the p21waf1/cif1 (CDKN1A) promoter results in its silencing and can be reversed by HDAC inhibitors(Ocker and Schneider-Stock, 2007). Germline mutations of HDACs increase the risk of breast and lung cancers and abnormal HDAC overexpression has also been observed in various cancers(Miremadi et al., 2007). As a result, HDAC inhibitors have been developed as anti-cancer drugs(Shankar and Srivastava, 2008). Several independent reports have identified truncation mutations in HDAC2 in epithelial, colonic, gastric and endometrial cancers and these mutations confer resistance to HDAC inhibitors(Smith and Workman, 2009). Screening for these mutations may improve the efficacy of HDAC inhibitors. Conversely, there is evidence that HDACs may function as tumor suppressors by maintaining proper chromatin structure and further stabilizing the genome(Bhaskara et al., 2010). Potentially, either loss or gain of function mutations of HDACs could contribute to tumorigenesis.
In addition to chromatin modifying enzymes, chromatin binding proteins or so-called epigenetic “readers” such as the bromodomain proteins which read lysine acetylation marks can also play an important during tumorigenesis. For example, the fusion of the bromodomain protein Brd4 with nuclear protein in testis (NUT) results in the development aggressive NUT midline carcinoma(Filippakopoulos et al., 2010). Aberrant regulation of Brd4 has also been reported in other cancers such as colon and breast, suggesting that the selective inhibitors which target these kinds of epigenetic readers may give us a novel clue for cancer therapy(Filippakopoulos et al., 2010; Zuber et al., 2011).
Methylation of arginine and lysine residues on histones or non-histone proteins such as transcription factors regulate chromatin structure and therefore gene expression(Greer and Shi, 2012). The best-known example of alterations in HMTs during tumorigenesis may be in the mixed lineage leukemia (MLL) protein which introduces the active H3K4me3 mark and plays important roles in development. MLL is located on chromosome 11q23, which is a common region of chromosomal translocation in AML and ALL(Slany, 2009). Translocations of MLL with multiple different partners can result in the generation of fusion proteins that are frequently associated with tumorigenesis and poor prognosis by generating abnormal patterns of H3K4me3 and/or recruiting other epigenetic modifiers(Balgobind et al., 2011). These MLL fusion proteins have close relationships with other epigenetic modifiers and cause altered epigenetic programs in cancer. For example, the aberrant H3K79 methylation pattern mediated by DOT1L is required for the maintenance of the MLL translocation-associated oncogenic program(Bernt et al., 2011). Inhibition of DOT1L activities decreases expression of MLL fusion-driven transcriptional programs and might have profound therapeutic implications (detailed discussion in therapeutic perspective section in this review). In addition, alternative splicing and mutations in MLL1, MLL2 and MLL3 genes have been identified in bladder, breast, pancreatic cancers and in glioblastoma(Gui et al., 2011; Morin et al., 2011).
The Polycomb group (PcG) of repressor proteins controls the accessibility of gene regulatory elements to the transcription machinery(Mills, 2010), This group is crucial for early development and often becomes deregulated in cancer. EZH2, together with SUZ12 and EED, form the polycomb repressive complex 2 (PRC2), which methylates H3K27. Overexpression of EZH2 has been reported in several cancers such as prostate, breast, lung and bladder and seems to result in an increase in H3K27me3(Chase and Cross, 2011). However, other studies show that there is no association between EZH2 and H3K27me3 in ovarian and pancreatic cancers(Fullgrabe et al., 2011). Down-regulation of microRNA-101, a negative regulator of EZH2, has been described as a cause of overexpression of EZH2 in bladder and prostate cancers(Friedman et al., 2009; Varambally et al., 2008) and EZH2 mutations have been reported in lymphoma and myeloid neoplasm(Chase and Cross, 2011). In lymphoma, a heterozygous missense mutation at amino acid Y641, within the SET domain, results in a gain of function showing enhanced catalytic activity. The EZH2 mutations in myeloid neoplasms are associated with poor prognosis and the mutations frequently result in loss of function of HMT. Although the mechanism of action of EZH2 in cancer is not yet clear, it appears to play a role in growth control(Tsang and Cheng, 2010).
BMI-1, a component of PRC1, is indispensable for the regulation of self-renewal of normal and leukemic stem cells and for the differentiation of T cells (Nakayama and Yamashita, 2009; Sauvageau and Sauvageau, 2011). BMI-1 has been considered as a key regulator of self-renewal in cancer stem cells(Jiang et al., 2009). More recently, overexpression of BMI-1 has been observed in solid tumors such as prostate cancer(Lukacs et al., 2011; Yang et al., 2010).
Subgroups of genes which are normally repressed by H3K27me3 in early development often acquire abnormal DNA methylation in cancers, a process which we have called “epigenetic switching.”(Sharma et al., 2010). The differentiation of stem cells begins by turning off master regulators which define “stemness” (e.g. OCT4 in embryonic stem cells) followed by the expression of lineage specific genes resulting in the acquisition of particular phenotypes (e.g. MYOD1 in muscle and NEUROG1 in neurons)(Young, 2011). Progress through these steps is often, but not always, controlled by PcG and does not involve DNA methylation. Once these key regulators become methylated, they become locked in a repressed state and this prevents switching from one phenotype to another. The outcome of the “epigenetic switch” may therefore be an increase in the number of cancer initiating cells(Baylin and Jones, 2011). Full understanding of this mechanism remains to be clarified.
Other lysine HMTs (NSD1, SMYD3 and G9a) are aberrantly expressed in several cancers(Varier and Timmers, 2010). Evidence for the role of arginine HMTs (PRMTs) in tumorigenesis has not been as well established as that of lysine HMTs, although alteration of expression of PRMT1 in breast cancer and PRMT5 in gastric cancer has been reported(Lee and Stallcup, 2009).
Two distinct classes of HDMs have been defined based on their mechanism of action(Mosammaparast and Shi, 2010). LSD1 (lysine-specific histone demethylase1), KDM6A/UTX (lysine-specific demethylase 6A) and JARID1A-D (jumonji C-domain containing proteins) have all been implicated in tumorigensis. Mutations in LSD1 (prostate cancer) and KDM6A/UTX (various cancers including bladder, breast, kidney and colon) have been reported(Rotili and Mai, 2011). Reintroduction of KDM6A/UTX in the UTX mutant cancer cells results in the slowing of proliferation, suggesting that genetic mutations of these enzymes reinforce the epigenetic deregulation in cancers.
The exact mechanism by which these histone modifying enzymes affect tumorigenesis remains to be elucidated; altered expression of histone modifiers caused by mutations may disrupt whole epigenetic regulation mechanisms and result in aberrant gene expression patterns. Indeed, the disruption of histone modifications has been linked to all the hallmarks of cancer and it is important to be aware that a precise balance between the enzymes that write, read and erase histone marks is crucial in preventing tumorigenesis.
Chromatin Remodeling Complexes
Nucleosome occupancy is a key mechanism for gene expression and it has been known for some time that chromatin remodelers are responsible for regulating this process(Clapier and Cairns, 2009; Segal and Widom, 2009; Valouev et al., 2011). ATP dependent chromatin remodelers are generally divided into 4 main families: SWI/SNF (switch/sucrose non-fermenting), ISWI (imitation SWI), INO80 (inositol requiring 80) and NURD (nucleosome remodeling and deacetylation)/Mi2/CHD (chromatin helicase DNA binding) complexes(Ho and Crabtree, 2010). Although the ATPase domains are highly similar, the distinct chromatin interacting domains carry out specific roles and can be selectively targeted. These ATPase dependent remodelers act in the context of multisubunit complexes and have dual roles as activators and repressors of gene expression. The importance of chromatin remodeling machines is becoming apparent with the realization that many of them are mutated in several types of cancer (Hargreaves and Crabtree, 2011; Wilson and Roberts, 2011).
SWI/SNF is a large complex with 9 to 12 subunits including ATPases (BRG1 or BRM), core subunits (SNF5, BAF155 and BAF 170) and other accessory subunits(Ho and Crabtree, 2010). The variety of subunits allows for combinatorial assembly that leads to functional diversity as evidenced by the cellular stage-specific composition of SWI/SNF complexes(Hargreaves and Crabtree, 2011). SWI/SNF complexes remodel chromatin by changing nucleosome occupancy pattern thereby contributing to either transcriptional activation or repression(Reisman et al., 2009; Wilson and Roberts, 2011).
SNF5 of the SWI/SNF core subunit is at the nexus of the link between chromatin remodeling and tumorigenesis and many rhabdoid tumors contain inactivating mutations in this gene. Loss of SNF5 is also observed in renal carcinomas and melanomas, where it is correlated with poor survival rates(Lin et al., 2009). SNF5 loss affects expression of genes associated with cell proliferation and cell cycle such as RB or p53 and Hedgehog-Gli, a key signaling pathway in early development and cancer. Antagonism between EZH2 and SNF5 has also been reported during tumorigenesis(Jagani et al., 2010; Wilson and Roberts, 2011) and there is accumulating evident that SNF5 deletion plays a role in tumorigenesis, but the exact mechanism of SNF5 loss in tumorigenesis remains to be elucidated.
ARID1A/BAF250a mutations have been frequently observed in ovarian clear cell carcinoma (50%) and endometrioid carcinomas (30%)(Guan et al., 2011; Jones et al., 2010; Wiegand et al., 2010). More recently, ARID1A/BAF250a mutations have been observed in primary pancreatic adenocarcinomas and transitional cell carcinoma and low ARID1A expression was found to be significantly associated with a specific subgroup of breast cancers (ER−/PR−/HER2−)(Zhang et al., 2011). In mice, ARID1B/BAF250b containing complexes, which include components of an E3 ubiquitin ligase and are mutually exclusive of ARID1A, have also been shown to play a role in the control of cell cycle and differentiation. Mutations in human ARID1B/BAF250b have been reported very recently as a cause of Coffin-Siris syndrome(Santen et al., 2012; Tsurusaki et al., 2012).
The PBRM1/BAF180, BAF200 and BRD7 subunits belong to polybromo BRG1 associated factor (PBAF) complexes and facilitate transcriptional activation by nuclear receptors(Wilson and Roberts, 2011). Mutation of PBRM/BAF180 has been identified in 41% renal cell carcinomas and in breast cancers and this mutation affects senescence in human cells. Mutation in another PBAF specific subunit, BRD7, has been reported in breast cancers. Since BRD7 has a variety of binding partners including p53 and BRCA1, mutations in it may be important in tumorigenesis.
Mutations in SWI/SNF ATPase subunits BRG1 or BRM have been reported in several cancers including lung, medulloblastoma, rhabdoid and prostate tumors(Wilson and Roberts, 2011). Although BRG1 and BRM show some redundancy in vivo and in vitro, they seem to be mutually exclusive and have distinctive roles based on their expression changes during early development. Tumor suppressor properties of BRG1 and BRM have been reported in lung, breast and prostate cancer cell lines(Roberts and Orkin, 2004) and, in vitro, BRG1 and BRM have been observed to interact with several tumor suppressors including BRCA1(Wang et al., 2007).
Mutation of BAF complexes is a frequent event in various cancers; however the dependency between the subunits and whether mutation of one subunit results in a modification of the activity of the complex is not clear. In addition, mutations of BAF complex components frequently coexist with those of canonical oncogenes or tumor suppressors such as KRAS, CDKN2A or p53, suggesting a synergistic effect on tumorigenesis(Wilson and Roberts, 2011).
In addition to SWI/SNF complexes, mutations of other ATP dependent chromatin remodelers are beginning to be identified in several cancers(Clapier and Cairns, 2009; Hargreaves and Crabtree, 2011; Ho and Crabtree, 2010). Despite emerging evidence that closely connects these ATPase remodelers in tumorigenesis, the direct causality and/or mechanism still remains to be explicated.
The Role of SNPs on Epigenetic Regulation and Cancer
GWAS (genome-wide association studies) have identified a large number of single nucleotide polymorphisms (SNPs) associated with an increased risk of a variety of diseases including several cancers. Surprisingly, cancer-associated SNPs are significantly enriched at regions defined as functional enhancers in ES cells(Teng et al., 2011) and might confer cancer susceptibility by altering the chromatin landscape. Further, several genome-wide expression quantitative trait loci (eQTLs) studies in humans have demonstrated a link between genetic variation and changes in gene regulation(Nica et al., 2010; Nicolae et al., 2010). More recently, these genetic variants were shown to modify the chromatin accessibility of TF binding sites, thereby leading to gene expression differences(Degner et al., 2012). Although allele-specific DNA methylation and allele-specific gene expression have been well studied in imprinting and X chromosome inactivation, recent studies show that these allele-specific phenomena are more pervasive to other cellular activities(Tycko, 2010). Notably, most of the allele-specific DNA methylation outside of imprinted genes shows a strong correlation with SNP genotypes that affect TF binding insulators and long-range chromosome structure. Conversely, SNPs can create or delete CpGs (termed as CpG SNPs) thereby influencing the binding of specific TFs(Tycko, 2010). Future studies aimed at understanding functional associations among epigenetic variation (epigenotype), genetic variation (genotype) and trait or disease (phenotype) may help us to determine the causality of diseases.
Therapeutic Perspective
An increasing number of nucleoside analogues/small molecules are being studied as anti-cancer drugs. Inhibitiors of DNMTs 5-Azacytidine (5-Aza-CR; Vidaza; azacitidine) and 5-Aza-2-deoxycytidine (5-Aza-CdR; Dacogen; decitabine) or HDACs by SAHA or Rhomidepsin have been approved for cancer treatment by the FDA and proven to have therapeutic efficacy in a variety of malignancies(Kelly et al., 2010). Recently, several novel compounds have been reported to target epigenetic components and have therapeutic effects in the presence of specific genetic defects. The DOT1L inhibitor (EPZ004777) inhibits H3K79 methylation, prevents transcription of genes which are involved in leukemogenesis and kills cancer cells bearing MLL translocations(Daigle et al., 2011). Selective bromodomain inhibitors (JQ1 or GSK525762)(Filippakopoulos et al., 2010; Nicodeme et al., 2010) inhibit transcription by MYC which is overexpressed in a majority of cancers(Delmore et al., 2011). The presence of multiple genetic and epigenetic aberrations within a cancer suggests that effective cancer therapies will be most beneficial when combined with epigenetic and/or other anti-cancer strategies such as standard chemotherapy(Juergens et al., 2011; Matei and Nephew, 2010).
Conclusions
Recent whole exome sequencing of thousands of human cancers have come up with the unexpected results that mutations in genes which control the epigenome are surprisingly common in human cancers. The presence of these mutations was unknown and overlooked which is surprising in view of the fact that were almost 1,000 cell lines recently analyzed by whole exome sequencing contain a large number of potential mutations in epigenetic modifiers(Barretina et al., 2012). The fact that the epigenome acts at the pinnacle of the hierarchy of gene control mechanisms means that the mutations probably have effects on multiple pathways relevant to the cancer phenotype and so a single mutation could cause wide scale misregulation. This realization opens the door to further drug development, since it might be possible to correct several pathways by altering or inhibiting one enzyme. These data also show a much closer Yin-Yang relationship between the genome and the epigenome as indicated in this review. This has heralded the dawn of a new era in cancer research in which the way the genes are organized and controlled is being recognized as a major relevant factor for human carcinogenesis. Traditionally cancer is diagnosed by pathologists using light microscopes to analyze the morphology of the nucleus among other cellular features. Understanding how epigenetic modifiers communicate with each other and alter nuclear architecture and therefore gene expression is a major challenge for the future but one which should yield better options for patients.
Acknowledgments
We thank Jones lab members and Gerry Coetzee for helpful discussion of the manuscript. We apologize to those whose work has not been included because of space constraints. Funding for this work to P.A.J was provided by NIH R37 CA-082422-12.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bagchi A, Papazoglu C, Wu Y, Capurso D, Brodt M, Francis D, Bredel M, Vogel H, Mills AA. CHD5 is a tumor suppressor at human 1p36. Cell. 2007;128:459–475. doi: 10.1016/j.cell.2006.11.052. [DOI] [PubMed] [Google Scholar]
- Balakrishnan A, Bleeker FE, Lamba S, Rodolfo M, Daniotti M, Scarpa A, van Tilborg AA, Leenstra S, Zanon C, Bardelli A. Novel somatic and germline mutations in cancer candidate genes in glioblastoma, melanoma, and pancreatic carcinoma. Cancer research. 2007;67:3545–3550. doi: 10.1158/0008-5472.CAN-07-0065. [DOI] [PubMed] [Google Scholar]
- Balgobind BV, Zwaan CM, Pieters R, Van den Heuvel-Eibrink MM. The heterogeneity of pediatric MLL-rearranged acute myeloid leukemia. Leukemia. 2011;25:1239–1248. doi: 10.1038/leu.2011.90. [DOI] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, Feng Z, Punt N, Daigle A, Bullinger L, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer cell. 2011;20:66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskara S, Knutson SK, Jiang G, Chandrasekharan MB, Wilson AJ, Zheng S, Yenamandra A, Locke K, Yuan JL, Bonine-Summers AR, et al. Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer cell. 2010;18:436–447. doi: 10.1016/j.ccr.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutani N, Burns DM, Blau HM. DNA demethylation dynamics. Cell. 2011;146:866–872. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanovic O, Veenstra GJ. DNA methylation and methyl-CpG binding proteins: developmental requirements and function. Chromosoma. 2009;118:549–565. doi: 10.1007/s00412-009-0221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challen GA, Sun D, Jeong M, Luo M, Jelinek J, Berg JS, Bock C, Vasanthakumar A, Gu H, Xi Y, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet. 2011;44:23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase A, Cross NC. Aberrations of EZH2 in cancer. Clin Cancer Res. 2011;17:2613–2618. doi: 10.1158/1078-0432.CCR-10-2156. [DOI] [PubMed] [Google Scholar]
- Cimmino L, Abdel-Wahab O, Levine RL, Aifantis I. TET family proteins and their role in stem cell differentiation and transformation. Cell Stem Cell. 2011;9:193–204. doi: 10.1016/j.stem.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annual review of biochemistry. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Conerly ML, Teves SS, Diolaiti D, Ulrich M, Eisenman RN, Henikoff S. Changes in H2A.Z occupancy and DNA methylation during B-cell lymphomagenesis. Genome research. 2010;20:1383–1390. doi: 10.1101/gr.106542.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J, Johnston LD, Scott MP, Smith JJ, Xiao Y, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer cell. 2011;20:53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Carvalho DD, Sharma S, You JS, Su SF, Taberlay PC, Kelly TK, Yang X, Liang G, Jones PA. DNA methylation screening identifies driver epigenetic events of cancer cell survival. Cancer cell. 2012;21:655–667. doi: 10.1016/j.ccr.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Carvalho DD, You JS, Jones PA. DNA methylation and cellular reprogramming. Trends in cell biology. 2010;20:609–617. doi: 10.1016/j.tcb.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Zwaan SE, Haass NK. Genetics of basal cell carcinoma. The Australasian journal of dermatology. 2010;51:81–92. doi: 10.1111/j.1440-0960.2009.00579.x. quiz 93–84. [DOI] [PubMed] [Google Scholar]
- Degner JF, Pai AA, Pique-Regi R, Veyrieras JB, Gaffney DJ, Pickrell JK, De Leon S, Michelini K, Lewellen N, Crawford GE, et al. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature. 2012;482:390–394. doi: 10.1038/nature10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost J, Mantovani F, Tocco F, Elkon R, Comel A, Holstege H, Kerkhoven R, Jonkers J, Voorhoeve PM, Agami R, Del Sal G. BRD7 is a candidate tumour suppressor gene required for p53 function. Nature cell biology. 2010;12:380–389. doi: 10.1038/ncb2038. [DOI] [PubMed] [Google Scholar]
- Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- Fabbri M, Calin GA. Epigenetics and miRNAs in human cancer. Adv Genet. 2010;70:87–99. doi: 10.1016/B978-0-12-380866-0.60004-6. [DOI] [PubMed] [Google Scholar]
- Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JM, Liang G, Liu CC, Wolff EM, Tsai YC, Ye W, Zhou X, Jones PA. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer research. 2009;69:2623–2629. doi: 10.1158/0008-5472.CAN-08-3114. [DOI] [PubMed] [Google Scholar]
- Fullgrabe J, Kavanagh E, Joseph B. Histone onco-modifications. Oncogene. 2011;30:3391–3403. doi: 10.1038/onc.2011.121. [DOI] [PubMed] [Google Scholar]
- Gelsi-Boyer V, Brecqueville M, Devillier R, Murati A, Mozziconacci MJ, Birnbaum D. Mutations in ASXL1 are associated with poor prognosis across the spectrum of malignant myeloid diseases. J Hematol Oncol. 2012;5:12. doi: 10.1186/1756-8722-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012 doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan B, Mao TL, Panuganti PK, Kuhn E, Kurman RJ, Maeda D, Chen E, Jeng YM, Wang TL, Shih Ie M. Mutation and loss of expression of ARID1A in uterine low-grade endometrioid carcinoma. The American journal of surgical pathology. 2011;35:625–632. doi: 10.1097/PAS.0b013e318212782a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui Y, Guo G, Huang Y, Hu X, Tang A, Gao S, Wu R, Chen C, Li X, Zhou L, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43:875–878. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg R. Hallmarks of Cancer: The Next Generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell research. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatziapostolou M, Iliopoulos D. Epigenetic aberrations during oncogenesis. Cell Mol Life Sci. 2011;68:1681–1702. doi: 10.1007/s00018-010-0624-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Yang C, Antosiewicz-Bourget JE, Lee LK, Ngo QM, Klugman S, Ching KA, Edsall LE, Ye Z, et al. Dynamic chromatin states in human ES cells reveal potential regulatory sequences and genes involved in pluripotency. Cell research. 2011;21:1393–1409. doi: 10.1038/cr.2011.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinoue T, Weisenberger DJ, Lange CP, Shen H, Byun HM, Van Den Berg D, Malik S, Pan F, Noushmehr H, van Dijk CM, et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome research. 2012;22:271–282. doi: 10.1101/gr.117523.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchins MP, Rapkins RW, Kwok CT, Srivastava S, Wong JJ, Khachigian LM, Polly P, Goldblatt J, Ward RL. Dominantly inherited constitutional epigenetic silencing of MLH1 in a cancer-affected family is linked to a single nucleotide variant within the 5′UTR. Cancer cell. 2011;20:200–213. doi: 10.1016/j.ccr.2011.07.003. [DOI] [PubMed] [Google Scholar]
- Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoischen A, van Bon BW, Rodriguez-Santiago B, Gilissen C, Vissers LE, de Vries P, Janssen I, van Lier B, Hastings R, Smithson SF, et al. De novo nonsense mutations in ASXL1 cause Bohring-Opitz syndrome. Nat Genet. 2012;43:729–731. doi: 10.1038/ng.868. [DOI] [PubMed] [Google Scholar]
- Hon GC, Hawkins RD, Ren B. Predictive chromatin signatures in the mammalian genome. Hum Mol Genet. 2009;18:R195–201. doi: 10.1093/hmg/ddp409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer NG, Ozdag H, Caldas C. p300//CBP and cancer. Oncogene. 2004;23:4225–4231. doi: 10.1038/sj.onc.1207118. [DOI] [PubMed] [Google Scholar]
- Jagani Z, Mora-Blanco EL, Sansam CG, McKenna ES, Wilson B, Chen D, Klekota J, Tamayo P, Nguyen PT, Tolstorukov M, et al. Loss of the tumor suppressor Snf5 leads to aberrant activation of the Hedgehog-Gli pathway. Nature medicine. 2010;16:1429–1433. doi: 10.1038/nm.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Li J, Song L. Bmi-1, stem cells and cancer. Acta Biochim Biophys Sin (Shanghai) 2009;41:527–534. doi: 10.1093/abbs/gmp040. [DOI] [PubMed] [Google Scholar]
- Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet. 2009;10:805–811. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Wang TL, Shih Ie M, Mao TL, Nakayama K, Roden R, Glas R, Slamon D, Diaz LA, Jr, Vogelstein B, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juergens RA, Wrangle J, Vendetti FP, Murphy SC, Zhao M, Coleman B, Sebree R, Rodgers K, Hooker CM, Franco N, et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov. 2011;1:598–607. doi: 10.1158/2159-8290.CD-11-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalari S, Pfeifer GP. Identification of driver and passenger DNA methylation in cancer by epigenomic analysis. Adv Genet. 2010;70:277–308. doi: 10.1016/B978-0-12-380866-0.60010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Ushijima S, Nakanishi Y, Sakamoto M, Hirohashi S. Mutation of the DNA methyltransferase (DNMT) 1 gene in human colorectal cancers. Cancer letters. 2003;192:75–82. doi: 10.1016/s0304-3835(02)00689-4. [DOI] [PubMed] [Google Scholar]
- Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011a;11:849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasinski AL, Slack FJ. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011b;11:849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly TK, De Carvalho DD, Jones PA. Epigenetic modifications as therapeutic targets. Nature biotechnology. 2010;28:1069–1078. doi: 10.1038/nbt.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Chung NG, Kang MR, Yoo NJ, Lee SH. Genetic and expressional alterations of CHD genes in gastric and colorectal cancers. Histopathology. 2011;58:660–668. doi: 10.1111/j.1365-2559.2011.03819.x. [DOI] [PubMed] [Google Scholar]
- Kinney SR, Pradhan S. Regulation of expression and activity of DNA (cytosine-5) methyltransferases in mammalian cells. Prog Mol Biol Transl Sci. 2011;101:311–333. doi: 10.1016/B978-0-12-387685-0.00009-3. [DOI] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- Lao VV, Darwanto A, Sowers LC. Impact of base analogues within a CpG dinucleotide on the binding of DNA by the methyl-binding domain of MeCP2 and methylation by DNMT1. Biochemistry. 2010;49:10228–10236. doi: 10.1021/bi1011942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Stallcup MR. Minireview: protein arginine methylation of nonhistone proteins in transcriptional regulation. Mol Endocrinol. 2009;23:425–433. doi: 10.1210/me.2008-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Zhao H, Zhang X, Wood LD, Anders RA, Choti MA, Pawlik TM, Daniel HD, Kannangai R, Offerhaus GJ, et al. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat Genet. 2011;43:828–829. doi: 10.1038/ng.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Wong RP, Martinka M, Li G. Loss of SNF5 expression correlates with poor patient survival in melanoma. Clin Cancer Res. 2009;15:6404–6411. doi: 10.1158/1078-0432.CCR-09-1135. [DOI] [PubMed] [Google Scholar]
- Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012:483. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Lukacs RU, Memarzadeh S, Wu H, Witte ON. Bmi-1 is a crucial regulator of prostate stem cell self-renewal and malignant transformation. Cell Stem Cell. 2011;7:682–693. doi: 10.1016/j.stem.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks PA, Rifkind RA, Richon VM, Breslow R, Miller T, Kelly WK. Histone deacetylases and cancer: causes and therapies. Nat Rev Cancer. 2001;1:194–202. doi: 10.1038/35106079. [DOI] [PubMed] [Google Scholar]
- Matei DE, Nephew KP. Epigenetic therapies for chemoresensitization of epithelial ovarian cancer. Gynecologic oncology. 2010;116:195–201. doi: 10.1016/j.ygyno.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattera L, Escaffit F, Pillaire MJ, Selves J, Tyteca S, Hoffmann JS, Gourraud PA, Chevillard-Briet M, Cazaux C, Trouche D. The p400/Tip60 ratio is critical for colorectal cancer cell proliferation through DNA damage response pathways. Oncogene. 2009;28:1506–1517. doi: 10.1038/onc.2008.499. [DOI] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills AA. Throwing the cancer switch: reciprocal roles of polycomb and trithorax proteins. Nat Rev Cancer. 2010;10:669–682. doi: 10.1038/nrc2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miremadi A, Oestergaard MZ, Pharoah PD, Caldas C. Cancer genetics of epigenetic genes. Hum Mol Genet. 2007;16(Spec No 1):R28–49. doi: 10.1093/hmg/ddm021. [DOI] [PubMed] [Google Scholar]
- Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, Johnson NA, Severson TM, Chiu R, Field M, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosammaparast N, Shi Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annual review of biochemistry. 2010;79:155–179. doi: 10.1146/annurev.biochem.78.070907.103946. [DOI] [PubMed] [Google Scholar]
- Nakayama T, Yamashita M. Critical role of the Polycomb and Trithorax complexes in the maintenance of CD4 T cell memory. Semin Immunol. 2009;21:78–83. doi: 10.1016/j.smim.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Network CGAR. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nica AC, Montgomery SB, Dimas AS, Stranger BE, Beazley C, Barroso I, Dermitzakis ET. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS genetics. 2010;6:e1000895. doi: 10.1371/journal.pgen.1000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, Chandwani R, Marazzi I, Wilson P, Coste H, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS genetics. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocker M, Schneider-Stock R. Histone deacetylase inhibitors: signalling towards p21cip1/waf1. Int J Biochem Cell Biol. 2007;39:1367–1374. doi: 10.1016/j.biocel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Reisman D, Glaros S, Thompson EA. The SWI/SNF complex and cancer. Oncogene. 2009;28:1653–1668. doi: 10.1038/onc.2009.4. [DOI] [PubMed] [Google Scholar]
- Rideout WM, 3rd, Coetzee GA, Olumi AF, Jones PA. 5-Methylcytosine as an endogenous mutagen in the human LDL receptor and p53 genes. Science. 1990;249:1288–1290. doi: 10.1126/science.1697983. [DOI] [PubMed] [Google Scholar]
- Roberts CW, Orkin SH. The SWI/SNF complex--chromatin and cancer. Nat Rev Cancer. 2004;4:133–142. doi: 10.1038/nrc1273. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nature medicine. 2011;17:330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- Ropero S, Fraga MF, Ballestar E, Hamelin R, Yamamoto H, Boix-Chornet M, Caballero R, Alaminos M, Setien F, Paz MF, et al. A truncating mutation of HDAC2 in human cancers confers resistance to histone deacetylase inhibition. Nat Genet. 2006;38:566–569. doi: 10.1038/ng1773. [DOI] [PubMed] [Google Scholar]
- Rotili D, Mai A. Targeting Histone Demethylases: A New Avenue for the Fight against Cancer. Genes Cancer. 2011;2:663–679. doi: 10.1177/1947601911417976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10:389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, Jones PA. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Sandoval J, Esteller M. Cancer epigenomics: beyond genomics. Curr Opin Genet Dev. 2012;22:50–55. doi: 10.1016/j.gde.2012.02.008. [DOI] [PubMed] [Google Scholar]
- Sansom OJ, Maddison K, Clarke AR. Mechanisms of disease: methyl-binding domain proteins as potential therapeutic targets in cancer. Nat Clin Pract Oncol. 2007;4:305–315. doi: 10.1038/ncponc0812. [DOI] [PubMed] [Google Scholar]
- Santen GW, Aten E, Sun Y, Almomani R, Gilissen C, Nielsen M, Kant SG, Snoeck IN, Peeters EA, Hilhorst-Hofstee Y, et al. Mutations in SWI/SNF chromatin remodeling complex gene ARID1B cause Coffin-Siris syndrome. Nat Genet. 2012 doi: 10.1038/ng.2217. [DOI] [PubMed] [Google Scholar]
- Sauvageau M, Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2011;7:299–313. doi: 10.1016/j.stem.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers A, Clevers H. Wnt signaling, stem cells, and cancer of the gastrointestinal tract. Cold Spring Harb Perspect Biol. 2012:4. doi: 10.1101/cshperspect.a007989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Martinez AM, Iovino N, Cavalli G. Trithorax group proteins: switching genes on and keeping them active. Nat Rev Mol Cell Biol. 2011;12:799–814. doi: 10.1038/nrm3230. [DOI] [PubMed] [Google Scholar]
- Segal E, Widom J. What controls nucleosome positions? Trends Genet. 2009;25:335–343. doi: 10.1016/j.tig.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar S, Srivastava RK. Histone deacetylase inhibitors: mechanisms and clinical significance in cancer: HDAC inhibitor-induced apoptosis. Adv Exp Med Biol. 2008;615:261–298. doi: 10.1007/978-1-4020-6554-5_13. [DOI] [PubMed] [Google Scholar]
- Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Wang L, Spitz MR, Hong WK, Mao L, Wei Q. A novel polymorphism in human cytosine DNA-methyltransferase-3B promoter is associated with an increased risk of lung cancer. Cancer research. 2002;62:4992–4995. [PubMed] [Google Scholar]
- Slany RK. The molecular biology of mixed lineage leukemia. Haematologica. 2009;94:984–993. doi: 10.3324/haematol.2008.002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KT, Workman JL. Histone deacetylase inhibitors: anticancer compounds. Int J Biochem Cell Biol. 2009;41:21–25. doi: 10.1016/j.biocel.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Song J, Teplova M, Ishibe-Murakami S, Patel DJ. Structure-based mechanistic insights into DNMT1-mediated maintenance DNA methylation. Science. 2011;335:709–712. doi: 10.1126/science.1214453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganuma T, Workman JL. Signals and combinatorial functions of histone modifications. Annual review of biochemistry. 2011;80:473–499. doi: 10.1146/annurev-biochem-061809-175347. [DOI] [PubMed] [Google Scholar]
- Sun A, Tawfik O, Gayed B, Thrasher JB, Hoestje S, Li C, Li B. Aberrant expression of SWI/SNF catalytic subunits BRG1/BRM is associated with tumor development and increased invasiveness in prostate cancers. The Prostate. 2007;67:203–213. doi: 10.1002/pros.20521. [DOI] [PubMed] [Google Scholar]
- Tan AY, Manley JL. The TET family of proteins: functions and roles in disease. Journal of molecular cell biology. 2009;1:82–92. doi: 10.1093/jmcb/mjp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng L, Firpi HA, Tan K. Enhancers in embryonic stem cells are enriched for transposable elements and genetic variations associated with cancers. Nucleic acids research. 2011;39:7371–7379. doi: 10.1093/nar/gkr476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowbridge JJ, Orkin SH. Dnmt3a silences hematopoietic stem cell self-renewal. Nat Genet. 2011;44:13–14. doi: 10.1038/ng.1043. [DOI] [PubMed] [Google Scholar]
- Tsang DP, Cheng AS. Epigenetic regulation of signaling pathways in cancer: role of the histone methyltransferase EZH2. Journal of gastroenterology and hepatology. 2010;26:19–27. doi: 10.1111/j.1440-1746.2010.06447.x. [DOI] [PubMed] [Google Scholar]
- Tsurusaki Y, Okamoto N, Ohashi H, Kosho T, Imai Y, Hibi-Ko Y, Kaname T, Naritomi K, Kawame H, Wakui K, et al. Mutations affecting components of the SWI/SNF complex cause Coffin-Siris syndrome. Nat Genet. 2012 doi: 10.1038/ng.2219. [DOI] [PubMed] [Google Scholar]
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012:483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycko B. Allele-specific DNA methylation: beyond imprinting. Hum Mol Genet. 2010;19:R210–220. doi: 10.1093/hmg/ddq376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev A, Johnson SM, Boyd SD, Smith CL, Fire AZ, Sidow A. Determinants of nucleosome organization in primary human cells. Nature. 2011;474:516–520. doi: 10.1038/nature10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, Laxman B, Cao X, Jing X, Ramnarayanan K, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–1699. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, Davies H, Jones D, Lin ML, Teague J, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varier RA, Timmers HT. Histone lysine methylation and demethylation pathways in cancer. Biochimica et biophysica acta. 2010;1815:75–89. doi: 10.1016/j.bbcan.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Wang GG, Allis CD, Chi P. Chromatin remodeling and cancer, Part II: ATP-dependent chromatin remodeling. Trends Mol Med. 2007;13:373–380. doi: 10.1016/j.molmed.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Chen H, Fu S, Xu ZM, Sun KL, Fu WN. The involvement of CHD5 hypermethylation in laryngeal squamous cell carcinoma. Oral oncology. 2011a;47:601–608. doi: 10.1016/j.oraloncology.2011.05.003. [DOI] [PubMed] [Google Scholar]
- Wang L, Gural A, Sun XJ, Zhao X, Perna F, Huang G, Hatlen MA, Vu L, Liu F, Xu H, et al. The leukemogenicity of AML1-ETO is dependent on site-specific lysine acetylation. Science. 2011b;333:765–769. doi: 10.1126/science.1201662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels K, Bohnhorst B, Luhmer I, Morlot S, Bohring A, Jonasson J, Epplen JT, Gadzicki D, Glaser S, Gohring G, et al. Novel CHD7 mutations contributing to the mutation spectrum in patients with CHARGE syndrome. European journal of medical genetics. 2010;53:280–285. doi: 10.1016/j.ejmg.2010.07.002. [DOI] [PubMed] [Google Scholar]
- Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, Senz J, McConechy MK, Anglesio MS, Kalloger SE, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijmenga C, Hansen RS, Gimelli G, Bjorck EJ, Davies EG, Valentine D, Belohradsky BH, van Dongen JJ, Smeets DF, van den Heuvel LP, et al. Genetic variation in ICF syndrome: evidence for genetic heterogeneity. Hum Mutat. 2000;16:509–517. doi: 10.1002/1098-1004(200012)16:6<509::AID-HUMU8>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11:481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Strawn E, Basir Z, Halverson G, Guo SW. Aberrant expression of deoxyribonucleic acid methyltransferases DNMT1, DNMT3A, and DNMT3B in women with endometriosis. Fertil Steril. 2007;87:24–32. doi: 10.1016/j.fertnstert.2006.05.077. [DOI] [PubMed] [Google Scholar]
- Yamashita Y, Yuan J, Suetake I, Suzuki H, Ishikawa Y, Choi YL, Ueno T, Soda M, Hamada T, Haruta H, et al. Array-based genomic resequencing of human leukemia. Oncogene. 2010;29:3723–3731. doi: 10.1038/onc.2010.117. [DOI] [PubMed] [Google Scholar]
- Yan XJ, Xu J, Gu ZH, Pan CM, Lu G, Shen Y, Shi JY, Zhu YM, Tang L, Zhang XW, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet. 2011;43:309–315. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]
- Yang GF, He WP, Cai MY, He LR, Luo JH, Deng HX, Guan XY, Zeng MS, Zeng YX, Xie D. Intensive expression of Bmi-1 is a new independent predictor of poor outcome in patients with ovarian carcinoma. BMC cancer. 2010;10:133. doi: 10.1186/1471-2407-10-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic acids research. 2004;32:959–976. doi: 10.1093/nar/gkh252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. Control of the Embryonic Stem Cell State. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhang Y, Yang Y, Niu M, Sun S, Ji H, Ma Y, Yao G, Jiang Y, Shan M, et al. Frequent low expression of chromatin remodeling gene ARID1A in breast cancer and its clinical significance. Cancer epidemiology. 2011 doi: 10.1016/j.canep.2011.07.006. [DOI] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]