Abstract
A ring species arises when a parental population expands around an area of unsuitable habitat in such a way that when the two fronts meet they behave as distinct species while still being connected through a series of intergrading populations. Ring species offer great possibilities for studying the forces causing species divergence (e.g. the nature of pre-zygotic or post-zygotic reproductive isolation) or helping to maintain species integrity (e.g. reinforcement). Yet, ring species are extremely rare, and have only been documented convincingly in animals. Here, we present phylogenetic analyses of two nuclear gene regions from the Caribbean slipper spurge (Euphorbia tithymaloides) species complex that provide evidence that this group forms a ring species. These data show that the species complex originated in the area where Mexico and Guatemala meet, and expanded around the Caribbean basin along two distinct fronts: one eastward through the Yucatan Peninsula and into the Greater Antilles (GA); one southeastward through northern South America and then northward to the Lesser Antilles and eastern GA. The two terminal forms co-occur in the Virgin Islands and appear to be morphologically and ecologically distinct. Thus, our results suggest that Euphorbia tithymaloides is the first compelling example of a ring species in plants.
Keywords: speciation, reproductive isolation, biogeography, phylogeography, phylogeny, nuclear gene
1. Introduction
Ring species are thought to arise most commonly when a population expands around an area of inhospitable habitat along two fronts that upon secondary contact behave as distinct species despite being the terminal forms of a single widespread population or a set of historically connected populations [1–3](figure 1). Evolutionary changes that accumulate independently in the two fronts can result in sufficient differentiation that the terminal forms can persist in sympatry without fusing.
Figure 1.
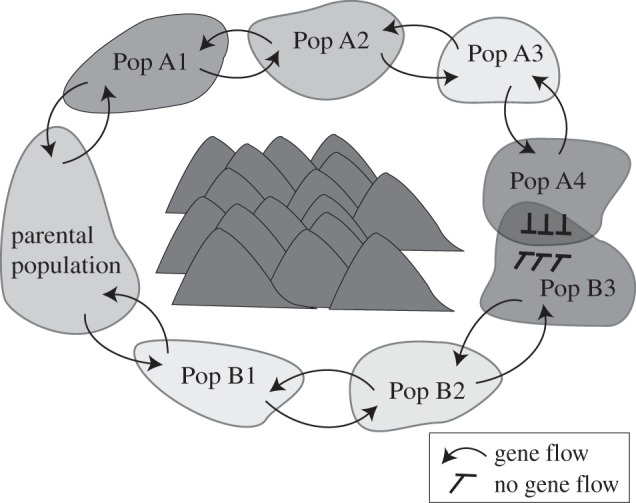
According to the ring-species model, a parental population diverges along two fronts around an area of unsuitable habitat so that its terminal forms behave as distinct species upon secondary contact.
Ernst Mayr [3] referred to ring species as ‘the perfect demonstrations of speciation’ because they spread out in space the process of species divergence. Ring species offer unique ‘real-time’ models for studying the genetic basis of pre- or post-zygotic isolation through comparative studies of different populations throughout the ring.
Conceptually, an unequivocal case of a ring-species system would have the following four features [1,4]: (i) a circular distribution that arose via historical range expansion of two diverging fronts (rather than, for example, hybridization of formerly distinct forms); (ii) the two terminal most divergent forms occur in sympatry such that they have the potential for directly exchanging genes; (iii) a chain of populations connects the two terminal forms such that there is the potential for indirect gene flow between the two termini; and, (iv) intrinsic reproductive isolation between the terminal forms such that there is no hybridization in sympatry. We would argue that when identifying putative ring species in nature, the first two criteria are essential, namely the historical range expansion along two fronts from an ancestral population, and the persistence of distinct forms in sympatry despite the opportunity for direct gene flow. While gaps in the distribution might be seen as problematic [1], strictly continuous clinal distributions hardly ever occur in naturally widespread species because of habitat heterogeneity, habitat loss or local extinction [4]. Likewise, in the same way that hybrid zones are common in traditional species, it does not seem essential that the reproductive isolation between the terminal forms be complete so long as there is sufficient reproductive isolation that allows the two forms to persist rather than fuse into a hybrid entity. Indeed, that reproductive barriers are semipermeable to gene flow and that species can differentiate despite ongoing interbreeding has been acknowledged as one of the most important new insights from modern speciation research already being integrated into species concepts [5].
Despite the unique opportunity that ring species provide for the study of species divergence, ring species are rare [1,2,6]. There are only two biological systems for which compelling data support the phenomenon and its two core criteria (a historical chain of populations whose two termini meet and remain distinct): the salamander Ensatina eschscholtzii species complex in the mountains surrounding the Central Valley of California [7–9], and the greenish warblers (Phylloscopus trochiloides) around the Tibetan Plateau [10,11]. Previous to this study, only one of all proposed ring-species examples is a plant system: populations of Acacia karoo that surround the central massif of South Africa [12]. Unfortunately, the ring-species interpretation in that case received only equivocal support [12,13]. Here, we use a phylogenetic approach to ask whether a Caribbean plant species complex (Euphorbia tithymaloides L.) conforms to the ring-species model.
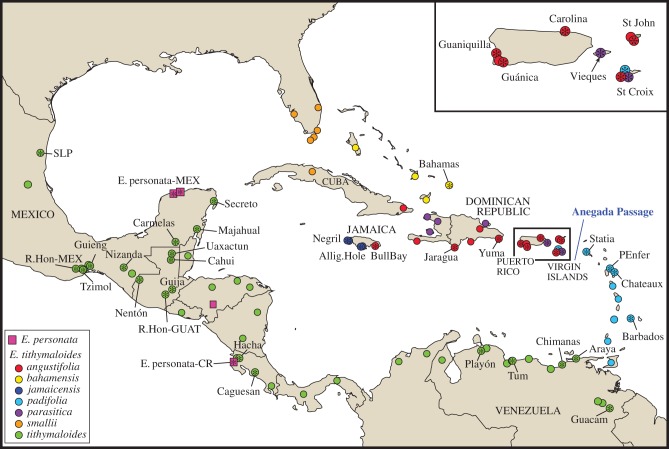
The slipper spurge E. tithymaloides is by far the most variable and widespread species of the Pedilanthus clade of Euphorbia, with a range that includes Mexico, Florida, Central America, northern South America, and most islands in the Caribbean (figure 2) [14,15].
Figure 2.
Documented populations of E. tithymaloides in the Caribbean are represented in full circles, colour-coded by subspecies. The 42 populations included in this study are marked with an asterisk and have been assigned names representative of their localities.
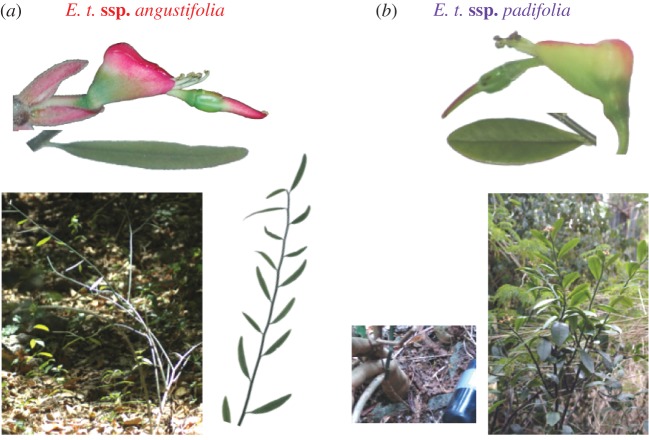
Eight subspecies of E. tithymaloides have been recognized based on geography and vegetative and reproductive traits (see electronic supplementary material, table S1). Owing to intergradation, subspecies assignation can be challenging, particularly among the Greater Antillean (GA) subspecies smallii, jamaicensis, bahamensis and parasitica, which bear close resemblance to the mainland populations traditionally assigned to subspecies tithymaloides. On the other hand, subspecies angustifolia (figure 3a) and padifolia (figure 3b) are more easily distinguishable since their features are fairly constant within the entity and clearly different from other subspecies. Individuals of subspecies angustifolia are viny shrubs about 1 m tall, with very thin, narrow and puberulent leaves and slender stems, and cyathia with short involucral tubes that occur in tropical deciduous forests in the eastern GA, including the Virgin Islands. In contrast, plants assigned to subspecies padifolia are stout shrubs up to 2 m in height with a thick main axis and have broad succulent and glabrous leaves with obtuse apices, and cyathia with relatively long involucral tubes. Individuals of the latter subspecies are found mainly in the Lesser Antilles (LA), with some records in the Virgin Islands only a few kilometres from populations of subspecies angustifolia [15]. Plants of subspecies parasitica, which have also been reported in the eastern GA, resemble those of subspecies padifolia in most regards but (like subspecies smallii, jamaicensis, bahamensis and tithymaloides) have acute leaf apices and round (rather than truncate) nectar spurs (see electronic supplementary material, table S1).
Figure 3.
Two morphologically and ecologically distinct forms of E. tithymaloides coexist in the Greater Antilles. (a) E. t. ssp. angustifolia; (b) E. t. ssp. padifolia.
Based on his careful morphological study of these organisms, Robert Dressler [15] suggested that angustifolia and parasitica (p.137): ‘ … are broadly sympatric and appear to behave as distinct species […], each closely tied in to the mainland population by a different chain of taxa.’ Although Dressler's formulation did not specifically hypothesize that E. tithymaloides might be a ring species, such a scenario seems a priori plausible in this case given (i) the circular distribution of E. tithymaloides around the rim of the Caribbean, and (ii) the oceanic currents within the Caribbean Sea [16–18] which would tend to promote dispersal to adjacent islands and inhibit direct migration across the Caribbean Sea.
We use two low-copy nuclear regions and a molecular phylogenetic approach to test the hypothesis that the E. tithymaloides species complex in the Caribbean fits the ring-species model. Under a ring-species scenario, we would expect statistical support for a phylogenetic pattern involving two colonization fronts of the Caribbean whose terminal forms remain distinguishable despite co-occurring and show reduced or no gene flow. Examples of alternative scenarios would be that the several forms in the Caribbean are derived from a single invasion (and thus sister to each other) rather than conforming to two colonization fronts whose termini meet, or that the Caribbean populations originated from multiple long-distance dispersal events directly from mainland populations.
2. Material and methods
Sampling, laboratory methods and analyses are detailed in the electronic supplementary material.
(a). Taxon and gene sampling
We sequenced two nuclear regions for 1–16 individuals for each of 40 natural populations of E. tithymaloides spanning as much of the taxonomic and geographical ranges of the species complex as possible, and two populations of its sister species, Euphorbia personata. For brevity, we will refer to these two regions as SGN (region based on Solanum Gene Network Unigene SGN-U342009), and G3p (GAPC-2 locus of the glyceraldehyde 3-phosphate dehydrogenase subunit C gene) [14].
The subspecies retusa is only known from very few scattered locations in the Peruvian and Brazilian Amazon and lacks any compelling differences from E. t. ssp. tithymaloides from Venezuela [15]. Given that E. t. ssp. tithymaloides is commonly cultivated and used in folk medicine in the area, it is likely that subspecies retusa is an escape from cultivation of plants that originated from Venezuela [15]. Based on these issues, we decided to exclude the subspecies retusa from the present study.
Despite intensive field and herbarium work in Florida, we were unable to locate populations of E. t. ssp. smallii. Areas where healthy and thriving populations were reported as late as 1982 have been destroyed by real estate development. We thus fear that E. t. ssp. smallii might be now extinct. However, since this subspecies is only marginally insular and very similar to E. t. ssp. tithymaloides [15], it is unlikely that its inclusion in our study would have substantially changed our conclusions.
A combination of single-stranded conformation polymorphism (SSCP) gel separation of PCR products and cloning was used to isolate all alleles present in each of the individuals analysed. Thus, the resulting data matrix has no missing data. Briefly, once the procedures were standardized, the approach for allele separation was as follows. After genomic PCR, direct sequencing was performed. Those PCR products that yielded overlapping sequence reads or double peaks were ran in SSCP gels for allele separation [19–21]. If SSCP allele separation failed owing to co-migration of different sequences, cloning was used to recover both alleles. Allele separation by SSCP was successful in 40 per cent of the heterozygotes for SGN and 58.5 per cent for G3p.
Data analysis—sequences were easily aligned manually and analysed in maximum parsimony (MP), maximum likelihood (ML) and Bayesian Metropolis-coupled-Markov-chain Monte Carlo (BA) frameworks. Haplotype networks were estimated at a 95 per cent reconnection limit treating gaps as missing data in TCS v. 1.21 [22]. Observed heterozygosity was calculated as the proportion of the individuals sampled that were heterozygous at a given locus [23].
3. Results
We sequenced and performed phylogenetic analyses on two nuclear regions (SGN and G3p) for individuals representing 40 populations spanning as much as possible of the range of E. tithymaloides. Details on data matrices, diagnostic statistics, model selection, inferred trees and topology tests are presented in electronic supplementary material, tables S2 and S3.
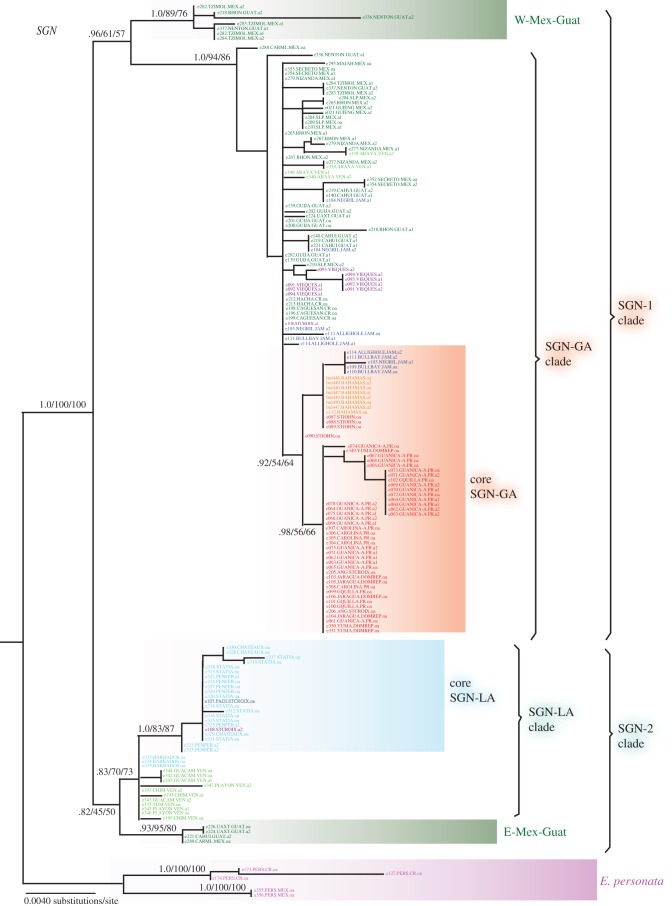
Figure 4 and electronic supplementary material, figure S1 show the ML trees with support values (bootstrap, BS; posterior probability, PP) for SGN and G3p, respectively. MP and BA consensus topologies are very similar to the ML trees; most differences being due to a lack of resolution of weakly supported clades. Haplotype networks for each marker are shown in electronic supplementary material, figures S2 and S3.
Figure 4.
ML allele tree for SGN. Tips represent alleles; only one allele is depicted for those individuals that are homozygous (oa), and both alleles (a1, a2) for heterozygotes (individuals of E. tithymaloides are diploid [15]). Support values correspond to posterior probability/ML bootstrap/MP bootstrap. Tips are colour coded by subspecies and abbreviations for the population name correspond to those on figure 2.
The SGN tree (figure 4) shows clear support for the monophyly of E. tithymaloides (PP = 1.0/ML-BS = 100/MP-BS = 100), as does the haplotype network (see electronic supplementary material, figure S2). Within E. tithymaloides, two main clades are suggested, albeit with weak support: SGN-1 (0.96/61/57) and SGN-2 (0.82/45/50). Each of these clades consists of one clade containing either the GA alleles (SGN-GA) or the LA ones (SGN-LA) that is sister to a clade of alleles restricted to Mexico/Guatemala (W-Mex-Guat or E-Mex-Guat, respectively). This pattern is consistent with a two-fronted invasion of the Caribbean from Mexico/Guatemala, which would be characteristic of a ring-species scenario.
Similarly for G3p (electronic supplementary material, figure S1), the two clades G3p-GA and G3p-LA contain most of the alleles from the GA and the LA, respectively. While the monophyly of E. tithymaloides is not clearly supported by this gene, neither is it significantly rejected (Wilcoxon sign-rank p-value = 0.655; PP of monophyly = 0.181; electronic supplementary material, table S3). Patterns of allele evolution are also consistent with a two-fronted invasion (see §4). A more detailed description of each clade and its composition is presented in the electronic supplementary material.
We found no examples of LA plants that harboured alleles from the GA front, suggesting that the GA front has not crossed the Anegada Passage eastward into the LA. On the other hand, there is evidence that the LA front has successfully crossed the Anegada Passage westward into the GA (see §4).
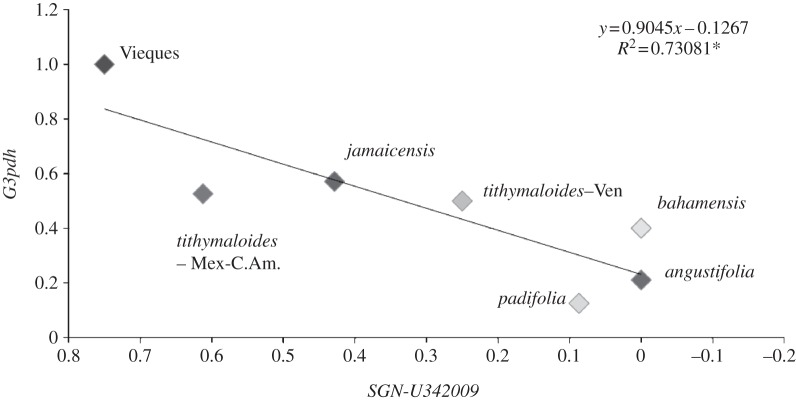
Taking subspecies and geographical areas into consideration for population assignation, heterozygosity at SGN and G3p were significantly positively correlated (figure 5 and electronic supplementary material, table S4). Leaving the Vieques population aside (see §4), heterozygosity is highest in Mexican/Central American populations of subspecies tithymaloides and decreases with distance towards the Anegada Passage, achieving its lowest frequency in subspecies bahamensis, angustifolia and padifolia.
Figure 5.
Observed heterozygosity for subspecies of E. tithymaloides in the Caribbean. Subspecies tithymaloides Mex-C.Am includes Guanica (see the electronic supplementary material); padifolia includes St Croix. Correlation is significant at α = 0.05 (p = 0.01423). Excluding Vieques from the analysis yields a model significant at α = 0.1 (y = 1.0066x − 0.1618, R2 = 0.5391, p = 0.09656).
4. Discussion
(a). Euphorbia tithymaloides colonized the Caribbean along two fronts from Mexico/Guatemala
Evidence for a historical two-fronted range expansion from Mexico/Guatemala proceeding either through the GA or the LA towards the Anegada Passage is seen at both loci (see the electronic supplementary material).
The easternmost GA alleles form the Core-SGN-GA clade, which is nested within a larger clade (SGN-GA) whose basal grade is restricted to Guatemala and Mexico. Moreover, the clade sister to SGN-GA is composed of alleles also restricted to Mexico and Guatemala (W-Mex-Guat). This topology clearly supports an eastwards migration of E. tithymaloides from Mexico/Guatemala into the GA giving rise to subspecies jamaicensis, bahamensis and angustifolia, which we refer to as the GA front. The G3p tree (electronic supplementary material, figure S1) also supports a GA front extending eastwards from Mexico/Guatemala: the GA alleles are concentrated in the clade G3p-GA whose easternmost alleles appear to be of most recent origin, and whose basal split involves a Mexican allele.
The second colonization front (LA front) has reached the LA through Venezuela. On the SGN tree (figure 4), alleles from E. t. ssp. padifolia from the northernmost LA (Guadeloupe and St Eustatius) form the clade Core-SGN-LA, which together with alleles from Barbados and Venezuela forms the SGN-LA clade. The SGN-LA clade is then sister to a Mexican/Guatemalan clade of E. t. ssp. tithymaloides (E-Mex-Guat). This pattern suggests a migration out of Mexico/Guatemala, first into Venezuela (with loss of much ancestral variation) and thence northwards along the LA island chain. While consistent with a migration through Venezuela into the LA, the G3p tree lacks resolution: most individuals from the LA share an ancestral allele (as shown by zero-length branches in the G3p-LA clade) that also occurs in Venezuela and Central America.
Our sampling of Central American populations is somewhat limited, and Cuba represents a gap as well. It is likely that including additional populations (e.g. Honduras, Salvador, Panamá, Colombia) would result in further resolution in the LA front. Nonetheless, even with the current sampling, the concordance of the two nuclear regions, and the patterns of heterozygosity and morphological variation observed around the ring provide compelling evidence of two invasion fronts that have converged in the GA.
(b). The Greater Antilles and Lesser Antilles fronts have come into contact in Vieques and the Virgin Islands
Our data provide evidence of only limited gene flow from east to west across the Anegada Passage, characterized by a relatively high inter-island distance (150+ km) and moderate to strong currents [17] compared with other Caribbean passages. Some alleles from individuals sampled in the Virgin Island of St Croix (GA) are embedded in the LA clades for both gene trees. This is also true for most G3p alleles from a population sampled in Vieques. This finding can only readily be explained by gene flow into the GA from the east.
The population on the island of Vieques seems to be greatly admixed: the SGN alleles belong to the SGN-GA clade (Mexican–Jamaican grade portion), whereas the G3p alleles seem to be of LA origin (with the exception of one individual with an allele that is otherwise unique to angustifolia). In our view, the most plausible interpretation of these data is that at some point in the past, before a significant barrier to gene flow existed (aside from geography), one or a few migrants from the LA front arrived on the island of Vieques (most probably as plant fragments—E. tithymaloides can root with relative ease), and hybridized with plants occupying the island at the time (which possibly resembled subspecies jamaicensis). The resulting introgression yielded a genetically diverse population with alleles from both the LA and GA fronts. Subsequent genetic drift or selection would explain why different loci might be fixed (or almost so) for LA or GA front-derived alleles. The allele that is otherwise restricted to angustifolia seems to be of recent origin, suggesting that this might be the result of a more recent genetic exchange. Finding new populations of parasitica (e.g. on Hispaniola) should be a priority for future work; they would allow to test additional specific hypotheses regarding the origin of the Vieques population (e.g. its relationship with the admixed e108) and shed light on parasitica as a whole, which has been dubbed a ‘mysterious entity’ [15]. Nonetheless, even without additional sampling, the population in Vieques provides evidence of a lack of complete reproductive isolation between the GA and LA fronts as represented by ‘padifolia’-type and ‘jamaicensis’-type plants. It also shows that such introgression (presence of LA alleles in GA plants), while surviving in this potentially hybrid population is rather ‘contained’, having not spread to a significant number of populations. Thus, the admixture represented by the Vieques population has so far been insufficient to breakdown the distinction between the two invasion fronts.
On St Croix, we sampled individuals of three distinct populations. One is pure angustifolia (e205, e206); its morphology and alleles match those from subspecies angustifolia on other GA islands. The other two populations are 0.5 km from each other and within 21 km of the angustifolia population. The population represented by the individual e107 bears strong morphological similarities to padifolia from the LA and harbours alleles at both loci that are nested within the LA clade, thus appearing to be a padifolia individual that arrived in St Croix by westward migration across the Anegada Passage. The population represented by individual e108 is also predominantly composed of padifolia alleles, and morphologically resembles padifolia, but also contains a single allele from the SGN-Mexican-Jamaican grade. There is no evidence of either population of padifolia in St Croix capturing alleles from the morphologically and molecularly more cohesive (and putatively younger; see the electronic supplementary material) angustifolia.
There seems to be no reason to doubt that the presence of both subspecies (padifolia and angustifolia) on St Croix is natural. Records of angustifolia populations are on a protected portion of the island, and herbarium records document that the padifolia populations are at least 50 years old and that they used to be numerous before significant development had occurred on St Croix. Thus, our data show that angustifolia and padifolia, two morphologically [15] and genetically distinct forms co-occur on St Croix and are persisting as separate entities. Moreover, these two forms represent the termini of two distinct chains of populations that connect Mexico/Guatemala to the GA. Euphorbia tithymaloides thus satisfies the core criteria to be considered a ring species, becoming the first known plant example of the phenomenon.
(c). The ring-species Euphorbia tithymaloides provides an opportunity to study speciation in real time
Here, we document colonization of the Caribbean by two fronts advancing eastwards, one through the GA (GA front), and a second through northern South America and then into the LA (LA front). The GA front appears never to have crossed the Anegada Passage, so contact between these two fronts has only taken place when alleles (most probably as plant fragments but also possible via pollen or seeds) have crossed the Anegada Passage from the LA into the easternmost GA.
Our data suggest that the LA front has crossed the Anegada Passage on at least two occasions with quite different outcomes. In one instance, plants that were genetically similar to today's padifolia came into contact with plants from the GA front in (or near) the island of Vieques. Interbreeding occurred to yield a hybrid population that contains approximately equal representation of alleles from the GA and LA fronts, with evidence of minimal (and recent) gene flow from angustifolia. In what would be a second event of contact, plants of padifolia and angustifolia became established in St Croix within 21 km of each other. Despite this proximity, there is no evidence of any introgression with angustifolia. The different fates of the contact between the GA and LA fronts in Vieques and St Croix could reflect ecogeographical differences between these two areas, but also suggest the hypothesis that specific features of angustifolia limit the potential for gene flow with populations originating from the LA front.
The small geographical area of St Croix makes it a natural laboratory for exploring lineage divergence to a point where the long-term coexistence of sister lineages becomes possible. It is not yet clear if angustifolia and padifolia on St Croix will persist as different entities in the longer term. It is possible that one or other will go extinct, or that they will fuse to yield a hybrid taxon as seems to have occurred on Vieques. To evaluate these outcomes, field and greenhouse research is needed to explore factors that might be acting as pre-mating or post-mating barriers to the reproduction of these two termini (e.g. evolution of niche differentiation, barriers to pollen transfer, or reduced fitness of F1s). The great value of a ring-species system is that the process of divergence can be studied by addressing such questions with plants from different populations throughout the ring. Additionally, it will be important to thoroughly survey islands in the eastern GA, including St Croix, to determine if there are additional cases of contact between the GA and LA fronts and, if so, what the fate of the resulting populations has been. Through such follow-up work, E. tithymaloides may emerge as a unique system for studying ecological and reproductive aspects of the process of speciation in plants.
Acknowledgements
Paul Berry, Kenneth Sytsma, Bret Larget, Johanne Brunet, Flor Rodríguez, Bryan Drew, Talline Martins, and Patrick McIntyre provided helpful discussion. Fieldwork and permits were facilitated by: Bahamas Turks & Caicos National Trust (N. Manco); INBio-Costa Rica (F. Morales, R. Espinoza, J. Hernández); JBSD-Dominican Republic (T. Clase); Guatemala: CECON-USCG (M. Pérez, A. Cobar, J. Morales), UVAL (A. MacVean); Institute of Jamaica (G. Proctor, T. Commock); MEXU-UNAM-Mexico (C. Hinostrosa, G. Salazar, B. Drew); UW-Madison (M. Wetter); Puerto Rico: UPR-RP (M. Caraballo, E. Santiago), DNR-Guánica (M. Canals); STENAPA-St Eustatius (H. Leslie, A. Herrera); US-Florida: FTG (L. Woodbury), IRC (S. Woodmansee), Everglades-NP (J. Sadle); Virgin Islands: USVI-NP-St Croix (R. O'Reilly), St John (G. Gray, E. Gibny); Venezuela: O. Huber, FIBV-VEN (L. Rodríguez, O. Hotchke, C. Reyes), MY (D. Conde, N. Díaz), GUYN (W. Díaz), CORO (R. Winfield). NSF grants DDIG (DEB-0608428) and PBI (DEB-0616533), and awards from The Garden Club of America, the ASPT and UW-Madison (LACIS, Graduate School, and Botany—Davis, J. Croxdale and ON Allen funds) funded this study.
References
- 1.Coyne J., Orr H. 2004. Speciation. Sunderland, MA: Sinauer Associates Inc [Google Scholar]
- 2.Irwin D., Irwin J., Price T. 2001. Ring species as bridges between microevolution and speciation. Genetica 112, 223–243 10.1023/A:1013319217703 (doi:10.1023/A:1013319217703) [DOI] [PubMed] [Google Scholar]
- 3.Mayr E. 1942. Systematics and the origin of species. New York, NY: Columbia University Press [Google Scholar]
- 4.Irwin D. E., Irwin J. H. 2002. Circular overlaps: rare demonstrations of speciation. Auk 119, 596–602 10.1642/0004-8038(2002)119[0596:CORDOS]2.0.CO;2 (doi:10.1642/0004-8038(2002)119[0596:CORDOS]2.0.CO;2) [DOI] [Google Scholar]
- 5.Hausdorf B. 2011. Progress toward a general species concept. Evol. Int. J. Org. Evol. 65, 923–931 10.1111/j.1558-5646.2011.01231.x (doi:10.1111/j.1558-5646.2011.01231.x) [DOI] [PubMed] [Google Scholar]
- 6.Ashlock D., Clare E. L., von Königslöw T. E., Ashlock W. 2010. Evolution and instability in ring species complexes: an in silico approach to the study of speciation. J. Theor. Biol. 264, 1202–1213 10.1016/j.jtbi.2010.03.017 (doi:10.1016/j.jtbi.2010.03.017) [DOI] [PubMed] [Google Scholar]
- 7.Moritz C., Schneider C., Wake D. 1992. Evolutionary relationships within the Ensatina eschscholtzii complex confirm the ring species interpretation. Syst. Biol. 41, 273–291 [Google Scholar]
- 8.Stebbins R. C. 1957. Intraspecific sympatry in the lungless salamander Ensatina eschscholtzii. Evolution 11, 265–270 10.2307/2405789 (doi:10.2307/2405789) [DOI] [Google Scholar]
- 9.Wake D. B., Yanev K. P., Brown C. W. 1986. Intraspecific sympatry in a ring species, the plethodontid salamander Ensatina eschscholtzii, in southern California. Evolution 40, 866–868 10.2307/2408473 (doi:10.2307/2408473) [DOI] [PubMed] [Google Scholar]
- 10.Irwin D., Bensch S., Irwin J., Price T. 2005. Speciation by distance in a ring species. Science 307, 414–416 10.1126/science.1105201 (doi:10.1126/science.1105201) [DOI] [PubMed] [Google Scholar]
- 11.Irwin D., Bensch S., Price T. 2001. Speciation in a ring. Nature 409, 333–337 10.1038/35053059 (doi:10.1038/35053059) [DOI] [PubMed] [Google Scholar]
- 12.Brain P. 1989. Genetic races in a ring species, Acacia karroo. S. Afr. J. Sci. 85, 181–185 [Google Scholar]
- 13.Ward D. 2011. Population differentiation in a purported ring species, Acacia karroo (Mimosoideae). Biol. J. Linn. Soc. 104, 748–755 10.1111/j.1095-8312.2011.01757.x (doi:10.1111/j.1095-8312.2011.01757.x) [DOI] [Google Scholar]
- 14.Cacho N. I., Berry P. E., Olson M. E., Steinmann V. W., Baum D. A. 2010. Are spurred cyathia a key innovation? Molecular systematics and trait evolution in the slipper spurges (Pedilanthus clade: Euphorbia, Euphorbiaceae). Am. J. Bot. 97, 493–510 10.3732/ajb.0900090 (doi:10.3732/ajb.0900090) [DOI] [PubMed] [Google Scholar]
- 15.Dressler R. 1957. The genus Pedilanthus (Euphorbiaceae). Contributions from the Gray Herbarium of Harvard University, vol. 182, pp. 1–188. Cambridge, MA: Harvard University Press [Google Scholar]
- 16.Gordon A. L. 1967. Circulation of caribbean sea. J. Geophys. Res. 72, 6207–6223 10.1029/JZ072i024p06207 (doi:10.1029/JZ072i024p06207) [DOI] [Google Scholar]
- 17.Johns W. E., Townsend T. L., Fratantoni D. M., Wilson W. D. 2002. On the Atlantic inflow to the Caribbean Sea. Deep Sea Res. Part I Oceanogr. Res. Papers 49, 211–243 10.1016/S0967-0637(01)00041-3 (doi:10.1016/S0967-0637(01)00041-3) [DOI] [Google Scholar]
- 18.Thruman H. 1985. Introductory oceanography. Columbus, OH: Merrill Publisher [Google Scholar]
- 19.Gasser R. B., Hu M., Chilton N. B., Campbell B. E., Jex A. J., Otranto D., Cafarchia C., Beveridge I., Zhu X. 2006. Single-strand conformation polymorphism (SSCP) for the analysis of genetic variation. Nat. Protoc. 1, 3121–3128 10.1038/nprot.2006.485 (doi:10.1038/nprot.2006.485) [DOI] [PubMed] [Google Scholar]
- 20.Koopman M. M., Baum D. A. 2010. Isolating nuclear genes and identifying lineages without monophyly: an example of closely related species from southern Madagascar. Int. J. Plant Sci. 171, 761–771 10.1086/654847 (doi:10.1086/654847) [DOI] [Google Scholar]
- 21.Ortí G., Hare M. P., Avise J. C. 1997. Detection and isolation of nuclear haplotypes by PCR-SSCP. Mol. Ecol. 6, 575–580 10.1046/j.1365-294X.1997.00212.x (doi:10.1046/j.1365-294X.1997.00212.x) [DOI] [PubMed] [Google Scholar]
- 22.Clement M., Posada D., Crandall K. A. 2000. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9, 1657–1659 10.1046/j.1365-294x.2000.01020.x (doi:10.1046/j.1365-294x.2000.01020.x) [DOI] [PubMed] [Google Scholar]
- 23.Allendorf F. W. 1986. Genetic drift and the loss of alleles versus heterozygosity. Zoo. Biol. 5, 181–190 10.1002/zoo.1430050212 (doi:10.1002/zoo.1430050212) [DOI] [Google Scholar]