Abstract
The origins of tropical southwest Pacific diversity are traditionally attributed to southeast Asia or Australia. Oceanic and fragment islands are typically colonized by lineages from adjacent continental margins, resulting in attrition of diversity with distance from the mainland. Here, we show that an exceptional tropical family of harvestmen with a trans-Pacific disjunct distribution has its origin in the Neotropics. We found in a multi-locus phylogenetic analysis that the opilionid family Zalmoxidae, which is distributed in tropical forests on both sides of the Pacific, is a monophyletic entity with basal lineages endemic to Amazonia and Mesoamerica. Indo-Pacific Zalmoxidae constitute a nested clade, indicating a single colonization event. Lineages endemic to putative source regions, including Australia and New Guinea, constitute derived groups. Divergence time estimates and probabilistic ancestral area reconstructions support a Neotropical origin of the group, and a Late Cretaceous (ca 82 Ma) colonization of Australasia out of the Fiji Islands and/or Borneo, which are consistent with a transoceanic dispersal event. Our results suggest that the endemic diversity within traditionally defined zoogeographic boundaries might have more complex evolutionary origins than previously envisioned.
Keywords: biogeography, dispersal, disjunction, Arthropods, islands
1. Introduction
A growing body of evidence supports a role for transoceanic dispersal in historical biogeography [1–4]. One compelling biogeographic theatre that demonstrates the importance of transoceanic dispersal is the tropical South Pacific. The juxtaposition of a plethora of continental terranes and oceanic islands with diverse geological histories in this region has resulted in unique assemblages of biota found nowhere else in the world [5,6], with nearly all landmasses of the South Pacific categorized as biodiversity hotspots for conservation priority [7]. Biogeographic studies based on molecular phylogenies have implicated multiple source areas of South Pacific diversity, principally the Indo-Malay Archipelago, Australia, New Zealand, New Caledonia and the Neotropics (often via the Hawaiian Archipelago [4,8,9]).
A curious phenomenon that is periodically encountered in the biogeography of tropical Pacific lineages is the case of trans-Pacific disjunctions. These distributions typically include the Neotropics, continental parts of Australasia (e.g. Australia, New Guinea, the Thai–Malay Peninsula), and oceanic islands in the tropical belt (e.g. Fiji, Polynesia, Micronesia). Trans-Pacific disjunct distributions are generally attributed to one of three phenomena: taxonomic oversight (distant unrelated lineages are erroneously assigned to the same taxon), relictualism (range contraction of a previously broadly distributed taxon) and transoceanic dispersal. The last of these is considered uncommon owing to the dimensions of the barrier to dispersal: the breadth of the Pacific Ocean.
Most studied examples of trans-Pacific disjunct distributions resulting from oceanic dispersal consist of plant lineages [10–14]. However, trans-Pacific disjunctions are difficult to account for when the taxon of interest does not have adaptations for long distance oceanic dispersal, such as winged seeds [11] or floating fruit [12]. Apropos, fewer studied examples of trans-Pacific disjunct taxa are known among animals. These include the kagu of New Caledonia (Rhynochetos jubatus) and its putative sister lineage, the Neotropical sunbittern Eurypyga helias [15]. The charismatic Fijian and Tongan iguanas (Brachylophus [16]) are postulated to have colonized Melanesia by transoceanic rafting from the Neotropics, but neither the mechanism nor the timing of the colonization is well understood. Similarly, the fossorial lizard family Dibamidae is represented by 22 species in southeast Asia and a single species that is endemic to Mexico, likely the result of transoceanic or overland dispersal across Beringia in the Eocene [17].
Among arthropods, lineages that colonize oceanic islands typically originate in the proximate continental regions, perhaps best exemplified by multiple, unrelated lineages of the spider family Thomisidae [8,18]. Thus, Melanesian and Micronesian islands are typically colonized by Australian or southeast Asian lineages, whereas the Galapagos and Polynesian islands are largely colonized by Neotropical lineages [4]. True trans-Pacific disjunct distributions are known (e.g. the ant genera Adelomyrmex, Gnamptogenys and Rogeria [19]), but infrequently examined in a phylogenetic framework that establishes the monophyly of the group and the direction of colonization.
Zalmoxidae is a constituent lineage within the suborder Laniatores—the largest subgroup of Opiliones—which includes much of the morphological, behavioural and ecological diversity within the order [20]. Several lineages of Laniatores are characterized by ornate, sexually dimorphic and striking armature. Multiple forms of paternal care, an unusual behaviour often associated with seahorses and sea spiders (pycnogonids), have evolved repeatedly in Laniatores, manifested as egg guarding or egg carrying behaviour [21]. Although zalmoxids do bear sexually dimorphic fourth legs, which are swollen and armed with spines, they are not particularly remarkable or superlative in most respects, save for one. Unlike the majority of arthropods, Zalmoxidae has a typical trans-Pacific disjunct distribution.
The ca 200 described species of Zalmoxidae are found throughout tropical leaf-litter habitats in Amazonia, Meso-America, parts of Melanesia and Micronesia, northern Australia, New Guinea, the Philippine Islands, Java and Borneo [22–25]. Two additional species are known from Mauritius and the Seychelle Islands [23]. Nearly all species exhibit micro-endemicity and are known from a single locality. Zalmoxids do not occur in southern South America, southern Australia, New Zealand, Tasmania or Africa; species from some of these regions once postulated to belong to Zalmoxidae have since been transferred to distantly related lineages, based on evidence from morphology and molecular sequence data [20,23].
Intercontinental distributions do occur among harvestmen, but these have been attributed to ancient vicariant events—particularly the break-up of the supercontinent Gondwana—based on tree topology, analysis of ancestral areas and/or molecular dating [26–28]. The predominance of vicariance scenarios in the study of harvestman biogeography stems from the ancient age of the group, which is evidenced by Upper Devonian crown-group fossils [29]. However, recent phylogenetic analyses have demonstrated that Zalmoxidae is derived among harvestmen [20,30], potentially challenging an explanation based on vicariance. Moreover, Zalmoxidae has only been represented in such studies by two or three terminals, which is inadequate for ruling out taxonomic oversights, e.g. the placement of unrelated Indo-Pacific and Neotropical lineages in the same family. Consequently, we assessed the phylogeny of Zalmoxidae in order to test the monophyly of the group and evaluate biogeographic scenarios for the trans-Pacific disjunction.
2. Material and methods
Methods are described in greater detail with full references in the electronic supplementary material.
(a). Taxon and gene sampling
We selected 147 specimens representing all families of the sister superfamilies Zalmoxoidea and Samooidea [20] (figure 1). To test the monophyly of Zalmoxidae, we included 116 zalmoxids sampled from throughout their known range. We used routine DNA extraction, amplification and sequencing protocols for partial fragments of two mitochondrial protein-encoding (cytochrome b, cytochrome c oxidase subunit I) and two nuclear protein-encoding (histone H3, histone H4) loci; and complete or nearly complete nuclear ribosomal genes (18S rRNA, 28S rRNA), yielding approximately 6500 bp of data.
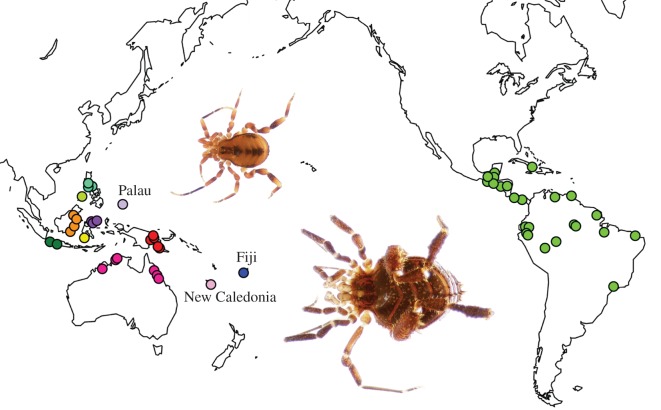
Figure 1.
Distribution map of sampled Zalmoxoidea. Coloured circles indicate cluster(s) of collection localities; colours correspond to areas designated for biogeographic analysis (indicated in figure 2). Smaller islands and archipelagos are as indicated. Insets: Undescribed male Zalmoxis; top: Zalmoxis sp. DNA102528 from Papua New Guinea, bottom: Zalmoxis sp. DNA102487 from North Sulawesi.
(b). Phylogenetic analysis and molecular dating
We conducted analyses using four phylogenetic approaches: maximum likelihood (ML), Bayesian inference; weighted parsimony under dynamic homology; and simultaneous Bayesian inference of topology and divergence times. Divergence time estimation was conducted using relaxed clock methods [31]. We specified a unique general time-reversible (GTR) model of sequence evolution with corrections for a discrete gamma distribution and a proportion of invariant sites (GTR + Γ + I) for each partition, as recommended by Modeltest under the Akaike information criterion [32,33]. Calibration for the 147-taxon dataset was drawn from a larger phylogenetic study of the suborder Laniatores [20]. To test the validity of normal distribution priors as secondary calibrations, we also constructed a 228-taxon dataset for the same six genes, combining the 147 focal taxa with exemplars of all known families of Laniatores, as well as representatives of the other three suborders of Opiliones. Calibration of the 228-taxon dataset was conducted using Palaeozoic fossils of crown-group Opiliones.
(c). Ancestral areas and diversification through time
We inferred ancestral areas using a Bayesian approach as implemented in rasp [34], and using a dispersal–cladogenesis–extinction model, implemented in lagrange [35,36]. We used a series of dispersal constraint models in lagrange, including unconstrained, stepping-stone and a stratified model with seven matrices incorporating geological history (electronic supplementary material, table S3). Tests of diversification rate constancy were conducted using the R package laser [37].
3. Results
(a). Phylogenetic analysis
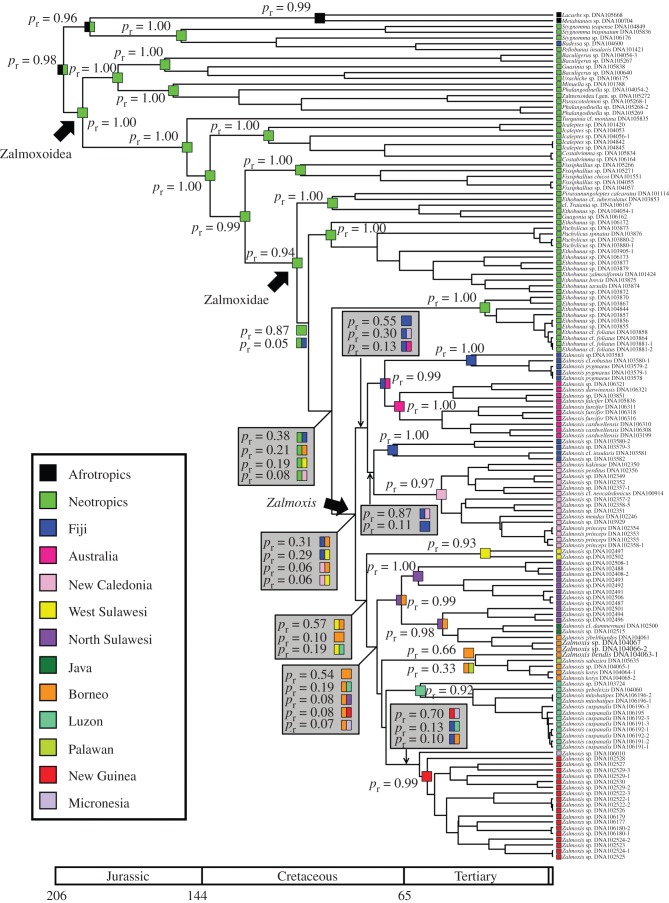
Analysis of the six-gene, 147-taxon dataset using beast v. 1.6.1 reached stationarity after ca 107 generations; 2 × 107 generations (20%) were discarded as burn-in. In the resulting tree topology (figure 2; electronic supplementary material, figure S1), the superfamilies Samooidea and Zalmoxoidea (the latter includes Zalmoxidae) form mutually monophyletic groups (both with posterior probabilities (PPbeast) of 1.00). Within Zalmoxoidea, early-diverging lineages are divided between two groups: the families Escadabiidae, Guasiniidae and Kimulidae form one group (PPbeast = 0.99); and Icaleptidae, Fissiphaliidae, Zalmoxidae and the Cuban genus Turquinia form another (PPbeast = 1.00). Zalmoxidae sensu stricto is monophyletic (PPbeast = 1.00), and early-diverging lineages within Zalmoxidae that are endemic to the Neotropics (e.g. Ethobunus, Pachylicus) form a grade with respect to Indo-Pacific zalmoxids. By contrast, the Indo-Pacific Zalmoxidae constitute a derived, monophyletic group (PPbeast = 1.00), i.e. the genus Zalmoxis. Two clades of Zalmoxis are recovered: a southwest Pacific clade, with constituent species endemic to Australia, Fiji and New Caledonia; and a southeast Asian clade, with constituent species endemic to Borneo, Java, New Guinea, the Philippines, Palau and Sulawesi. Relationships at the base of these clades are poorly supported.
Figure 2.
Ancestral area reconstruction based on Bayesian inference analysis (from beast ver. 1.6.1) of all molecular data using lagrange. Coloured squares at terminals indicate ranges occupied by sampled species. Coloured squares at nodes indicate ranges reconstructed for hypothetical ancestors. Numbers on nodes indicate relative probability of ranges reconstructed.
ML analysis using RAxML v. 7.2.7 resulted in a tree topology with ln L = −57789.507 (electronic supplementary material, figure S2) with major similarities to the beast tree topology, with respect to the monophyly of Zalmoxoidea (bootstrap resampling frequency (BS) of 76%) and the recovery of a basal grade that is endemic to the Neotropics. Early-diverging lineages of Zalmoxis are endemic to three terranes: the West Sulawesi plate, Fiji and New Caledonia. As with the beast topology, relationships among the constituent lineages of Zalmoxis are unstable and poorly supported.
Three of the four runs of MrBayes v. 3.1.2 reached stationarity in 5 × 106 generations, but the fourth did not converge with the other three runs until ca 1.75 × 107 generations; 2 × 107 generations (50%) were hence discarded as burn-in. The Bayesian inference (BI) analysis resulted in a topology largely similar to the ML tree (electronic supplementary material, figure S3). Both the monophyly of Zalmoxidae and Zalmoxis are supported in the BI analysis (posterior probability (PP) of 0.95 and 1.00, respectively). Some differences exist with respect to supported nodes within Zalmoxidae.
Iterative rounds of tree-fusing and driven searches using direct optimization in poy v. 4.1.2 resulted in two most parsimonious trees (MPTs) with a length of 18 137 weighted steps. A strict consensus of the MPTs (electronic supplementary material, figure S4) indicates that the difference between the two is restricted to internal relationships of a derived group of Ethobunus from southern Mexico. In general, the parsimony direct optimization (DO) tree topology is very similar to the topologies based on static alignments, particularly with respect to relationships among outgroups and the derived clades of Zalmoxidae. As with ML and BI analyses, the monophyly of Zalmoxidae and Zalmoxis are supported (jack-knife resampling frequencies (JF) of 85% and 97%, respectively), but internal relationships of Zalmoxis are unstable and poorly supported.
(b). Estimation of divergence times
Diversification of relevant lineages using beast for the 147-taxon dataset is estimated as follows: origin of Zalmoxidae sensu stricto, 127.4 Ma (95% HPD: 106.6–148.1 Ma); diversification of Zalmoxidae sensu stricto, 105.9 Ma (95% highest posterior density interval (HPD): 87.3–124.5 Ma); origin of Zalmoxis, 91.5 Ma (95% HPD: 75.0–107.8 Ma); diversification of Zalmoxis, 81.7 Ma (95% HPD: 67.8–94.8 Ma).
Subsequent to alignment and culling ambiguously aligned positions, the 228-taxon dataset constructed to test the validity of the secondary calibrations was smaller than the 147-taxon dataset (6172 versus 6563 nucleotide positions), owing to sequence variability in the nuclear ribosomal markers across Opiliones. Divergence time estimates were effectively identical to those obtained using the 147-taxon dataset (electronic supplementary material, figure S5). Median ages obtained for the three nodes used as secondary calibrations differed from the calibration medians by 7.7–12.7 Myr, and standard deviations by 2.7–3.93 Myr. Ages of relevant nodes in the 228-taxon dataset were as follows: origin of Zalmoxidae sensu stricto, 124.0 Ma (95% HPD: 104.1–143.0 Ma); diversification of Zalmoxidae sensu stricto, 101.9 Ma (95% HPD: 84.8–117.7 Ma); origin of Zalmoxis, 93.3 Ma (95% HPD: 75.4–108.2 Ma); diversification of Zalmoxis, 81.0 Ma (95% HPD: 65.7–94.4 Ma).
(c). Biogeographic analysis
To maintain comparability between the beast, ML, BI and DO topologies, and given the similar results of the two calibrations, we used the dated tree topology of the 147-taxon dataset for biogeographic analyses. All probabilistic approaches for inferring ancestral areas reconstruct the origin of Zalmoxidae (i.e. the split between Zalmoxidae and its sister family Fissiphalliidae) as unambiguously Neotropical (figure 2, table 1). The split between Neotropical and Indo-Pacific zalmoxids is reconstructed as the union of the Neotropics and the Fijian Islands under the unconstrained and stepping-stone models in lagrange analysis, or alternatively the union of the Neotropics and either Borneo, West Sulawesi or the Philippine Islands (excluding Palawan). Results from the Bayesian reconstruction of ancestral areas across all four topologies are largely congruent, favouring an ancestral distribution of Zalmoxis that united the Neotropics and either Fiji, Borneo or Sulawesi.
Table 1.
Biogeographic analysis of ancestral areas, with associated probabilities, across all topologies recovered using rasp or lagrange models. AUS, Australia; BRN, Borneo; FJI, Fiji; LZN, Philippines (excluding Palawan); NCA, New Caledonia; NGA, New Guinea; NSL, North Sulawesi; NTR, Neotropics; WSL, West Sulawesi.
|
rasp analyses |
lagrange analyses |
||||||
|---|---|---|---|---|---|---|---|
| ML-rasp | BI-rasp | DO-rasp | beast-rasp | unconstrained | stepping-stone | stratified | |
| origin of Zalmoxidae | NTR: 1.00 | NTR: 1.00 | NTR: 1.00 | NTR: 1.00 | NTR: 0.94 | NTR: 0.99 | NTR: 0.95 |
| origin of Zalmoxis | NTR: 0.98 | NTR: 0.99 | NTR: 0.99 | NTR: 0.99 | NTR + FJI: 0.38 | NTR + FJI: 0.34 | NTR + BRN: 0.32 |
| NTR + BRN: 0.21 | NTR + LZN: 0.24 | NTR: 0.30 | |||||
| NTR + WSL: 0.19 | NTR + WSL: 0.13 | NTR + AUS: 0.20 | |||||
| NTR + NCA: 0.08 | NTR + NGA: 0.13 | NTR + NGA: 0.07 | |||||
| NTR: 0.10 | |||||||
| diversification of Zalmoxis | NTR: 0.57 | NTR: 0.65 | NTR: 0.86 | NTR: 0.81 | FJI + BRN: 0.31 | FJI + LZN: 0.48 | BRN + AUS: 0.29 |
| FJI: 0.31 | FJI: 0.22 | FJI + WSL: 0.29 | FJI + NGA: 0.20 | NTR + BRN: 0.20 | |||
| NTR + FJI: 0.05 | NCA + BRN: 0.06 | WSL + AUS: 0.14 | BRN: 0.19 | ||||
| NCA + WSL: 0.06 | NTR + FJI: 0.07 | ||||||
As the geology of some candidate ancestral areas (e.g. Fiji) is inconsistent with the timing of the Zalmoxidae radiation, a stratified model replete with geological scenarios of landmass movements, fragmentations, appearance and subsidence was implemented to reconcile the biogeographic history of Zalmoxidae with geology and estimated divergence times. The stratified model reconstructs the split between Neotropical and Indo-Pacific zalmoxids as the union of the Neotropics and Borneo (pr = 0.32) or alternatively, the Neotropics alone (pr = 0.30). Diversification of Zalmoxis in the Indo-Pacific is reconstructed as the union of Borneo and Australia (pr = 0.29), the latter being the only terrane of the South Pacific clade (in the beast topology) that has consistently maintained suitable habitat for Opiliones since ca 81.7 Ma (while Gondwanan in origin, New Caledonia did not emerge in its present form until ca 37 Ma; [6,38]). Alternative reconstructions are a pan-Pacific diversification uniting Borneo and the Neotropics (pr = 0.20) or diversification out of Borneo alone (pr = 0.19).
4. Discussion
Like many arthropod lineages, nearly all families of Opiliones are restricted to specific biogeographic provinces, such as the Neotropics or southeast Asia (Wallacea/Sundaland), or to constituents of ancient supercontinents, such as Gondwana and Laurasia [20,28]. The fidelity of opilionid distributions to particular regions, in conjunction with the ancient age of the group, has engendered the prevalence of vicariance scenarios to account for the evolution of most divisions of Opiliones—especially within the suborder Cyphophthalmi—and the concomitant conformity of systematics with putative vicariance scenarios [28,39]. Trans-Pacific disjunct distributions strongly discord with this pattern, and could potentially indicate either the remnant of a formerly pantropical clade or a dispersing taxon. We investigated the biogeography of an exceptional tropical opilionid family presenting a classic trans-Pacific disjunction, testing three hypotheses for its present distribution: taxonomic oversight, ancient vicariance followed by range extinction and colonization by dispersal. Each hypothesis engenders a set of phylogenetic and/or temporal predictions, which we evaluated in turn.
The recovery of zalmoxid monophyly across all topologies obtained (figure 2; electronic supplementary material, figures S1–S5), with nodal support (BS = 76%; PP = 0.95; JF = 85%; PPbeast = 1.00), rules out dismissal of zalmoxid distribution based on taxonomic error. Although we included genera of uncertain monophyly and lineages of unknown familial placement, these did not contradict the monophyly of Zalmoxidae, nor did they confound biogeographic inference, insofar as all these lineages are endemic to the Neotropics and form a paraphyletic group with respect to the Indo-Pacific zalmoxids (the genus Zalmoxis; figure 2; electronic supplementary material, figures S1–S4). Zalmoxis constitutes a derived and monophyletic, yet comparatively diverse, group containing ca 33 per cent of described zalmoxid species.
A hypothesis of ancient Gondwanan diversification engenders two predictions. First, the origin of Zalmoxis has to coincide with the age of the geological event that precipitated the vicariance between the Neotropics and the Indo-Pacific landmasses. Second, as a corollary, Zalmoxidae must be sufficiently old to predate the vicariance, i.e. the family must have been present before the fragmentation of Tropical Gondwana. Both predictions are falsified by the divergence time estimates. The molecular dating of the radiation indicates that the split between the Indo-Pacific and the Neotropical Zalmoxidae occurred ca 92 Ma (95% HPD: 75.0–107.8 Ma), or approximately 80 Myr after the fragmentation of Tropical Gondwana (figure 2; electronic supplementary material figure S1 and S5). Even if the upper bound of the 95% HPD interval is taken as the estimate of this node's age, the origin of the Indo-Pacific lineage occurred over 60 million years after the geological event that precipitated the separation of the Neotropics from the Palaeotropics. In fact, the age of this node postdates the timing of the isolation of the Neotropics from other tropical landmasses (110 Ma). Secondly, both the paraphyletic group of Neotropical lineages at the base of Zalmoxidae and their age estimates corroborate diversification in the Neotropics after it became isolated from the rest of Tropical Gondwana. Diversification of the family is estimated to have occurred ca 106 Ma (95% HPD: 87.3–124.5 Ma). Again taking the estimate's upper limit, Tropical Gondwana had fragmented long before the family began to diversify in the Neotropics—too young for the disjunction to be characterized as the remnant of a former pantropical lineage (figure 2).
The establishment of zalmoxid diversity in the Indo-Pacific is therefore consistent with a scenario of colonization by a New World lineage, given an unambiguous origin in the Neotropics (figure 2; table 1). As with other taxa with trans-Pacific disjunct distributions, colonization of the Indo-Pacific could have followed multiple routes, such as (i) overland dispersal via Beringia; (ii) overland dispersal via trans-Antarctic connections between the Australian plate and South America; and (iii) transoceanic dispersal. However, overland dispersal hypotheses are inconsistent a priori with zalmoxid distribution and tree topology. Distributions for which Beringian migration has been invoked include Nearctic and Palaearctic elements, such as parts of North America, Japan and/or southeast Eurasia [40–42], none of which is inhabited by Zalmoxidae [23,25]. Similarly, no Zalmoxidae occur in the former constituents of the trans-Antarctic connection, such as southern Australia, Tasmania, New Zealand or southern South America.
Reconciling overland dispersal scenarios with the present restriction of zalmoxid range to the tropical belt on both sides of the Pacific requires invocation of significant extinction events, due to the micro-endemicity exhibited by almost all known Zalmoxoidea. To investigate the possibility of cryptic extinction, we investigated temporal shifts in diversification rates within the dated zalmoxid phylogeny. We postulated that cryptic extinction in the evolutionary history of Zalmoxidae would engender a log-lineage through time plot with an anti-sigmoidal shape, characteristic of cryptic extinction events [43]. If a hypothetical extinction event is not sufficiently drastic, the curve may not be anti-sigmoidal, but could still exhibit a recent increase in diversification rate—a documented manifestation of a birth–death process [44].
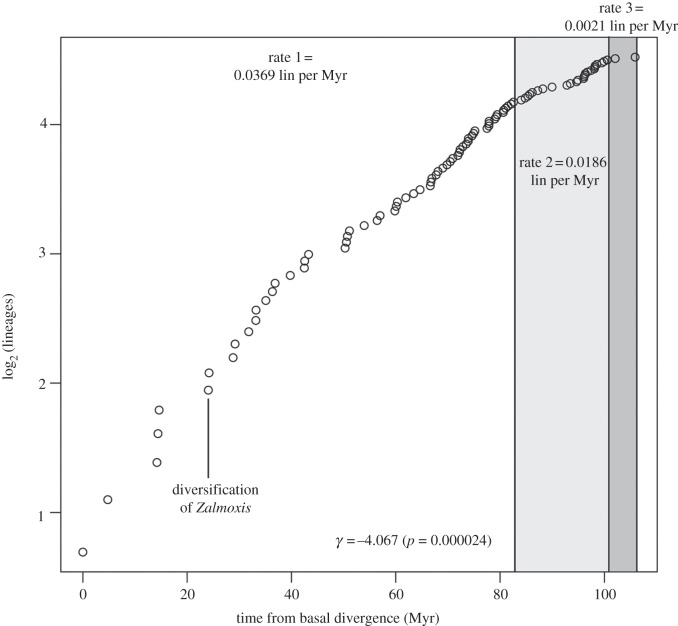
Of the eight competing models we examined, the optimal model was a Yule-three-rate model, which indicates that the diversification rate of Zalmoxidae has decreased steadily over time (figure 3 and table 2). Other suboptimal models examined similarly indicate diversification rate slowdowns. These data are inconsistent with the extinction episodes required to engender the trans-Pacific disjunct distribution of Zalmoxidae, assuming a scenario of overland dispersal. Diversification rate slowdowns must be interpreted cautiously, as they may be artefactual manifestations of inadequate lineage sampling [45], or a result of variable rates of cladogenesis and extinction over time [46]. Nevertheless, that neither an anti-sigmoidal curve nor recent recovery of diversification rate is observed for Zalmoxidae disfavours scenarios requiring lineage extinctions. This also applies, and in greater measure, to the hypothesis of ancient Gondwanan diversification followed by drastic range extinction.
Figure 3.
Log-lineage through time plot of Zalmoxidae, indicating diversification rates and timing of rate shifts inferred by the optimal, Yule-three-rate model. Note that the second best model, a Yule-two-rate, infers the same timing of the first rate shift (ca 82.6 Myr from basal divergence) as the optimal model.
Table 2.
Fit of models to the zalmoxid log-lineage through time curve. DDL and DDX refer to density-dependent logistic and exponential models, respectively. Boldface text indicates optimal model.
| model | no. parameters | ln L | AIC | ΔAIC |
|---|---|---|---|---|
| pure birth | 1 | −98.558 | 199.117 | 19.983 |
| birth–death | 2 | −98.558 | 201.117 | 21.983 |
| DDL | 2 | −92.282 | 188.565 | 9.431 |
| DDX | 2 | −91.748 | 187.495 | 8.361 |
| Yule-two-rate | 3 | −89.341 | 184.683 | 5.549 |
| Yule-three-rate | 5 | −84.567 | 179.134 | 0 |
| variable speciation | 3 | −91.711 | 189.421 | 10.287 |
| variable extinction | 3 | −98.942 | 203.884 | 24.75 |
The biogeographic history of Zalmoxidae is in accordance with a rare phenomenon: an amphitropical distribution achieved by trans-Pacific dispersal. Ancestral area analyses across all topologies corroborate the Neotropical origin of the family and an initial colonization of the Indo-Pacific that began in the Fijian Islands, Borneo, West Sulawesi (a terrane that was connected to Borneo ca 55 Myr [47]) or some combination of these (table 1). The reconstruction of any of these areas at the base of Zalmoxis is consistent with the hypothesis of transoceanic dispersal. In the case of the Fijian Archipelago, these islands are oceanic in origin [6]. In the case of Borneo and Sulawesi, as mentioned previously, the last recent connection of the southeast Asian tropics to the Neotropics predates the origin of Zalmoxidae by at least 60 million years, based on the upper limit of the age estimate [47,48]. Although the ages of the Fijian Islands postdate the diversification of Zalmoxis, the stratified model incorporating the geological histories of all 14 areas circumscribed for analysis reconstructs Borneo as the most likely target of colonization by the hypothetical ancestor of Zalmoxis (table 1; electronic supplementary material, table S3). The size, age and geological stability of Borneo are consistent with the availability of this landmass for colonization during the Late Cretaceous [47].
Consistent with this scenario, Zalmoxidae have demonstrable transoceanic dispersal capability, with locality records including remote islands, such as the Moluccas, Palau, Pohnpei and the Jaluit Atoll [23]. Furthermore, the ancient continental fragments of the Indo-Pacific—areas that are traditionally held to be the source regions of Australasian diversity—are either not inhabited by Zalmoxidae (e.g. southern Australia, Tasmania and New Zealand) or are inhabited by a single, derived clade (e.g. New Guinea, New Caledonia, northern Australia). New Guinea is an especially diverse island with complex geological history, and the putative source region of many diverse metazoan radiations, some of which extend worldwide [5,49,50]. The recovery of all New Guinean species, which constitute the most morphologically diverse Zalmoxis, within a single derived clade (that may include a Palauan species) is therefore an unexpected result. The biogeography of this region is already famous for its complexity, owing to Wallace's and Huxley's Lines and the interface of southeast Asian and Australian biotas; to our knowledge, Neotropical origins have never been demonstrated for diverse, endemic lineages in this biogeographic theatre.
5. Conclusion
The family Zalmoxidae, similar to the Pacific iguanas, constitutes the unusual case of a lineage of Neotropical origin that colonized the Indo-Pacific, likely by ancient transoceanic dispersal during the Late Cretaceous. While the mechanism of ancient dispersals is always a matter of speculation in the absence of a detailed fossil record, it is possible that Indo-Pacific colonization was achieved by rafting on floating vegetation propelled westwards by the Tethyan Seaway and/or southern equatorial currents, the directions of which are consistent with the direction of colonization. The timing here is well estimated and consistent with a Late Cretaceous dispersal event, and unlike other trans-Pacific taxa (e.g. iguanas, amphi-Pacific plant lineages) considerable diversity is retained on both sides of the Pacific Ocean. Zalmoxidae therefore presents a biogeographic conundrum: a radiation that has colonized the Indo-Pacific by ancient dispersal, engendering potentially hundreds of lineages endemic to Australasian terranes, but of Neotropical origins.
Acknowledgements
We are indebted to Alan Andersen, Ligia R. Benavides, Sarah L. Boyer, Graham Brown, Rafe Brown, Perry A. C. Buenavente, Tokasaya Cakacaka, Ronald M. Clouse, Jesse Czekanski-Moir, Arvin C. Diesmos, Dave General, Ben Hoffmann, Milan Janda, Corrie S. Moreau, Jerome Murienne, Abel Pérez González and Cahyo Rahmadi for providing collections and/or assistance during collecting efforts. Most of the Neotropical collections were provided by the Leaf Litter Arthropods of Meso-America (LLAMA) project to J. Longino (NSF award no. 0640015). Peter J. Schwendinger (MHNG) collected and provided most of the material for study from southeast Asia. Collecting in the South Pacific was greatly facilitated by Hervé Jourdan (IRD, Noumea, New Caledonia) and Marika Tuiwawa (USP, Suva, Fiji). Fieldwork in New Caledonia was supported by the Goelet Award and the Explorers Club award. Fieldwork in Indonesia, Fiji, Vanuatu and the Philippines was supported by three MCZ Putnam Expedition Grants. Fieldwork in Australia was supported by a Deakin-Royce Fellowship to P.P.S. Molecular study was supported by the BioNeoCal grant to P. Grandcolas. Maximilian Telford and two anonymous reviewers provided comments that helped to improve earlier versions of this article. P.P.S. collected samples, designed and performed research, analysed data, and wrote the paper; G.G. collected samples, and supervised and provided input through different stages of the research.
References
- 1.de Quieroz A. 2005. The resurrection of oceanic dispersal in historical biogeography. Trends. Ecol. Evol. 20, 68–73 10.1016/j.tree.2004.11.006 (doi:10.1016/j.tree.2004.11.006) [DOI] [PubMed] [Google Scholar]
- 2.Cowie R. H., Holland B. S. 2006. Dispersal is fundamental to biogeography and the evolution of biodiversity on oceanic islands. J. Biogeogr. 33, 193–198 10.1111/j.1365-2699.2005.01383.x (doi:10.1111/j.1365-2699.2005.01383.x) [DOI] [Google Scholar]
- 3.Bellemain E., Ricklefs R. E. 2008. Are islands the end of the colonization road? Trends. Ecol. Evol. 23, 461–468 10.1016/j.tree.2008.08.002 (doi:10.1016/j.tree.2008.08.002) [DOI] [PubMed] [Google Scholar]
- 4.Keppel G., Lowe A. J., Possingham H. P. 2009. Changing perspectives on the biogeography of the tropical South Pacific: influences of dispersal, vicariance and extinction. J. Biogeogr. 36, 1035–1054 10.1111/j.1365-2699.2009.02095.x (doi:10.1111/j.1365-2699.2009.02095.x) [DOI] [Google Scholar]
- 5.Gillespie R. G., Roderick G. K. 2002. Arthropods on islands: colonization, speciation, and conservation. Ann. Rev. Entomol. 47, 595–632 10.1146/annurev.ento.47.091201.145244 (doi:10.1146/annurev.ento.47.091201.145244) [DOI] [PubMed] [Google Scholar]
- 6.Neall V. E., Trewick S. A. 2008. The age and origin of the Pacific islands: a geological overview. Phil. Trans. R. Soc. B 363, 3293–3308 10.1098/rstb.2008.0119 (doi:10.1098/rstb.2008.0119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Myers N., Mittermeler R. A., Mittermeler C. G., da Fonseca G. A. B., Kent J. 2000. Biodiversity hotspots for conservation priorities. Nature 403, 853–858 10.1038/35002501 (doi:10.1038/35002501) [DOI] [PubMed] [Google Scholar]
- 8.Garb J. E., Gillespie R. G. 2006. Island hopping across the central Pacific: mitochondrial DNA detects sequential colonization of the Austral Islands by crab spiders (Araneae: Thomisidae). J. Biogeogr. 33, 201–220 10.1111/j.1365-2699.2005.01398.x (doi:10.1111/j.1365-2699.2005.01398.x) [DOI] [Google Scholar]
- 9.Harbaugh D. T., Wagner W. L., Allan G. J., Zimmer E. A. 2009. The Hawaiian Archipelago is a stepping stone for dispersal in the Pacific: an example from the plant genus Melicope (Rutaceae). J. Biogeogr. 36, 230–241 10.1111/j.1365-2699.2008.02008.x (doi:10.1111/j.1365-2699.2008.02008.x) [DOI] [Google Scholar]
- 10.Spalik K., Piwczynski M., Danderson C. A., Kurzyna-Mlynik R., Bone T. S., Downie S. R. 2010. Amphitropic amphiantarctic disjunctions in Apiaceae subfamily Apioideae. J. Biogeogr. 37, 1977–1994 10.1111/j.1365-2699.2010.02334.x (doi:10.1111/j.1365-2699.2010.02334.x) [DOI] [Google Scholar]
- 11.Olmstead R. G., Zjhra M. L., Lohmann L. G., Grose S. O., Eckert A. J. 2009. A molecular phylogeny and classification of Bignoniaceae. Am. J. Bot. 96, 1731–1743 10.3732/ajb.0900004 (doi:10.3732/ajb.0900004) [DOI] [PubMed] [Google Scholar]
- 12.Schaefer H., Heibl C., Renner S. S. 2009. Gourds afloat: a dated phylogeny reveals an Asian origin of the gourd family (Cucurbitaceae) and numerous overseas dispersal events. Proc. R. Soc. B 276, 843–851 10.1098/rspb.2008.1447 (doi:10.1098/rspb.2008.1447) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michalak I., Zhang L.-B., Renner S. S. 2010. Trans-Atlantic, trans-Pacific and trans-Indian Ocean dispersal in the small Gondwanan Laurales family Hernandiaceae. J. Biogeogr. 37, 1214–1226 10.1111/j.1365-2699.2010.02306.x (doi:10.1111/j.1365-2699.2010.02306.x) [DOI] [Google Scholar]
- 14.Renner S. S., Strijk J. S., Strasberg D., Thèbaud C. 2010. Biogeography of the Monimiaceae (Laurales): a role for East Gondwana and long-distance dispersal, but not West Gondwana. J. Biogeogr. 37, 1227–1238 10.1111/j.1365-2699.2010.02319.x (doi:10.1111/j.1365-2699.2010.02319.x) [DOI] [Google Scholar]
- 15.Hackett S. J., et al. 2008. A phylogenomic study of birds reveals their evolutionary history. Science 320, 1763–1767 10.1126/science.1157704 (doi:10.1126/science.1157704) [DOI] [PubMed] [Google Scholar]
- 16.Keogh J. S., Edwards D. L., Fisher R. N., Harlow P. S. 2008. Molecular and morphological analysis of the critically endangered Fijian iguanas reveals cryptic diversity and a complex biogeographic history. Phil. Trans. R. Soc. B 363, 3413–3426 10.1098/rstb.2008.0120 (doi:10.1098/rstb.2008.0120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Townsend T. M., Leavitt D. H., Reeder T. W. 2011. Intercontinental dispersal by a microendemic burrowing reptile. Proc. R. Soc. B 278, 2568–2574 10.1098/rspb.2010.2598 (doi:10.1098/rspb.2010.2598) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garb J. E., Gillespie R. G. 2009. Diversity despite dispersal: colonization history and phylogeography of Hawaiian crab spiders inferred from multilocus genetic data. Mol. Ecol. 18, 1746–1764 10.1111/j.1365-294X.2009.04125.x (doi:10.1111/j.1365-294X.2009.04125.x) [DOI] [PubMed] [Google Scholar]
- 19.Bolton B., Alpert G., Ward P. S., Naskrecki P. 2005. Bolton's catalogue of ants of the world. Cambridge, MA: Harvard University Press [Google Scholar]
- 20.Sharma P. P., Giribet G. 2011. The evolutionary and biogeographic history of the armoured harvestmen: Laniatores phylogeny based on ten molecular markers, with the description of two new families of Opiliones (Arachnida). Invertebr. Syst. 25, 106–142 10.1071/IS11002 (doi:10.1071/IS11002) [DOI] [Google Scholar]
- 21.Machado G., Macías-Ordóñez R. 2007. Reproduction. In Harvestmen: the biology of Opiliones (eds Pinto-da-Rocha R., Machado G., Giribet G.), pp. 414–454 Cambridge, MA: Harvard University Press [Google Scholar]
- 22.Kury A. B. 2003. Annotated catalogue of the Laniatores of the New World (Arachnida, Opiliones). Revista Ibérica de Aracnología, Volúmen especial monográfico 1, 1–337 [Google Scholar]
- 23.Sharma P. P., Kury A. B., Giribet G. 2011. The Zalmoxidae (Arachnida: Opiliones: Laniatores) of the Paleotropics: a catalogue of Southeast Asian and Indo-Pacific species. Zootaxa 2972, 37–58 [Google Scholar]
- 24.Sharma P. P. 2012. New Australasian Zalmoxidae (Opiliones: Laniatores) and a new case of male polymorphism in Opiliones. Zootaxa 3236, 1–35 [Google Scholar]
- 25.Sharma P. P., Buenavente P. A. C., Clouse R. M., Diesmos A. C., Giribet G. 2012. Forgotten gods: Zalmoxidae of the Philippines and Borneo (Opiliones: Laniatores). Zootaxa. 3280, 29–55 [Google Scholar]
- 26.Boyer S. L., Clouse R. M., Benavides L. R., Sharma P., Schwendinger P. J., Karunarathna I., Giribet G. 2007. Biogeography of the world: a case study of globally distributed arachnids. J. Biogeogr. 34, 2070–2085 10.1111/j.1365-2699.2007.01755.x (doi:10.1111/j.1365-2699.2007.01755.x) [DOI] [Google Scholar]
- 27.Mendes A. C., Kury A. B. 2008. Intercontinental Triaenonychidae: the case of Ceratomontia (Opiliones, Insidiatores). J. Arachnol. 36, 273–279 10.1636/CH07-93.1 (doi:10.1636/CH07-93.1) [DOI] [Google Scholar]
- 28.Giribet G., et al. 2012. Evolutionary and biogeographical history of an ancient and global group of arachnids (Arachnida: Opiliones: Cyphophthalmi) with a new taxonomic arrangement. Biol. J. Linn. Soc. 105, 92–130 10.1111/j.1095-8312.2011.01774.x (doi:10.1111/j.1095-8312.2011.01774.x) [DOI] [Google Scholar]
- 29.Dunlop J. A. 2007. Paleontology. In Harvestmen: the biology of Opiliones (eds Pinto-da-Rocha R., Machado G., Giribet G.), pp. 247–265 Cambridge, MA: Harvard University Press [Google Scholar]
- 30.Giribet G., Vogt L., González A. P., Sharma P., Kury A. B. 2010. A multilocus approach to harvestmen (Arachnida: Opiliones) phylogeny with emphasis on biogeography and the systematics of Laniatores. Cladistics 26, 408–437 10.1111/j.1096-0031.2009.00296.x (doi:10.1111/j.1096-0031.2009.00296.x) [DOI] [PubMed] [Google Scholar]
- 31.Drummond A. J., Rambaut A. 2007. beast: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214. 10.1186/1471-2148-7-214 (doi:10.1186/1471-2148-7-214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Posada D., Buckley T. 2004. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst. Biol. 53, 793–808 10.1080/10635150490522304 (doi:10.1080/10635150490522304) [DOI] [PubMed] [Google Scholar]
- 33.Posada D., Crandall K. A. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14, 817–818 10.1093/bioinformatics/14.9.817 (doi:10.1093/bioinformatics/14.9.817) [DOI] [PubMed] [Google Scholar]
- 34.Yu Y., Harris A. J., He X.-J. 2011. rasp (reconstruct ancestral state in phylogenies) 2.0 beta. See http://mnh.scu.edu.cn/soft/blog/RASP [DOI] [PubMed]
- 35.Ree R. H., Moore B. R., Webb C. O., Donoghue M. J. 2005. A likelihood framework for inferring the evolution of geographic range on phylogenetic trees. Evolution 59, 2299–2311 10.1554/05-172.1 (doi:10.1554/05-172.1) [DOI] [PubMed] [Google Scholar]
- 36.Ree R. H., Smith S. A. 2008. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst. Biol. 57, 4–14 10.1080/10635150701883881 (doi:10.1080/10635150701883881) [DOI] [PubMed] [Google Scholar]
- 37.Rabosky D. L. 2006. LASER: a maximum likelihood toolkit for detecting temporal shifts in diversification rates from molecular phylogenies. Evol. Bioinform. 2, 247–250 [PMC free article] [PubMed] [Google Scholar]
- 38.Grandcolas P., Murienne J., Robillard T., Desutter-Grandcolas L., Jourdan H., Guilbert E., Deharveng L. 2008. New Caledonia: a very old Darwinian island? Phil. Trans. R. Soc. B 363, 3309–3317 10.1098/rstb.2008.0122 (doi:10.1098/rstb.2008.0122) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giribet G., Kury A. B. 2007. Phylogeny and biogeography. In Harvestmen: the biology of Opiliones (eds Pinto-da-Rocha R., Machado G., Giribet G.), pp. 62–87 Cambridge, MA: Harvard University Press [Google Scholar]
- 40.Hendrixon B. E., Bond J. E. 2007. Molecular phylogeny and biogeography of an ancient Holarctic lineage of mygalomorph spiders (Araneae: Antrodiaetidae: Antrodiaetus). Mol. Phylogenet. Evol. 42, 738–755 10.1016/j.ympev.2006.09.010 (doi:10.1016/j.ympev.2006.09.010) [DOI] [PubMed] [Google Scholar]
- 41.Nie Z.-L., Sun H., Beardsley P. M., Olmstead R. G., Wen J. 2006. Evolution of biogeographic disjunction between eastern Asia and eastern North America in Phryma (Phrymaceae). Am. J. Bot. 93, 1343–1356 10.3732/ajb.93.9.1343 (doi:10.3732/ajb.93.9.1343) [DOI] [PubMed] [Google Scholar]
- 42.Vila R., et al. 2011. Phylogeny and palaeoecology of Polyommatus blue butterflies show Beringia was a climate-regulated gateway to the New World. Proc. R. Soc. B 278, 2737–2744 10.1098/rspb.2010.2213 (doi:10.1098/rspb.2010.2213) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crisp M. D., Cook L. G. 2009. Explosive radiation or cryptic mass extinction? Interpreting signatures in molecular phylogenies. Evolution 63, 2257–2265 10.1111/j.1558-5646.2009.00728.x (doi:10.1111/j.1558-5646.2009.00728.x) [DOI] [PubMed] [Google Scholar]
- 44.Nee S., May R. M., Harvey P. H. 1994. The reconstructed evolutionary process. Phil. Trans. R. Soc. Lond. B 344, 305–311 10.1098/rstb.1994.0068 (doi:10.1098/rstb.1994.0068) [DOI] [PubMed] [Google Scholar]
- 45.Cusimano N., Renner S. S. 2011. Slowdowns in diversification rates from real phylogenies may not be real. Syst. Biol. 59, 458–464 10.1093/sysbio/syq032 (doi:10.1093/sysbio/syq032) [DOI] [PubMed] [Google Scholar]
- 46.Rabosky D. L., Lovette I. J. 2008. Explosive evolutionary radiations: decreasing speciation or increasing extinction through time? Evolution 62, 1866–1875 10.1111/j.1558-5646.2008.00409.x (doi:10.1111/j.1558-5646.2008.00409.x) [DOI] [PubMed] [Google Scholar]
- 47.Hall R. 2002. Cenozoic geological and plate tectonic evolution of SE Asia and the SW Pacific: computer-based reconstructions and animations. J. Asian Earth Sci. 20, 353–434 10.1016/S1367-9120(01)00069-4 (doi:10.1016/S1367-9120(01)00069-4) [DOI] [Google Scholar]
- 48.Sanmartín I., Ronquist F. 2004. Southern Hemisphere biogeography inferred by event-based models: plant versus animal patterns. Syst. Biol. 53, 216–243 10.1080/10635150490423430 (doi:10.1080/10635150490423430) [DOI] [PubMed] [Google Scholar]
- 49.Balke M., Pons J., Ribera I., Sagata K., Vogler A. P. 2007. Infrequent and unidirectional colonization of hyperdiverse Papuadytes diving beetles in New Caledonia and New Guinea. Mol. Phylogenet. Evol. 42, 505–516 10.1016/j.ympev.2006.07.019 (doi:10.1016/j.ympev.2006.07.019) [DOI] [PubMed] [Google Scholar]
- 50.Jønsson K. A., Fabre P.-H., Ricklefs R. E., Fjeldså J. 2011. Major radiation of corvoid birds originated in the proto-Papuan archipelago. Proc. Natl Acad. Sci. USA 108, 2328–2333 10.1073/pnas.1018956108 (doi:10.1073/pnas.1018956108) [DOI] [PMC free article] [PubMed] [Google Scholar]