Abstract
The effects of body mass and temperature on metabolic rate (MR) are among the most widely examined physiological relationships. Recently, these relationships have been incorporated into the metabolic theory of ecology (MTE) that links the ecology of populations, communities and ecosystems to the MR of individual organisms. The fundamental equation of MTE derives the relation between mass and MR using first principles and predicts the temperature dependence of MR based on biochemical kinetics. It is a deliberately simple, zeroth-order approximation that represents a baseline against which variation in real biological systems can be examined. In the present study, we evaluate the fundamental equation of MTE against other more parameter-rich models for MR using an information-theoretic approach to penalize the inclusion of additional parameters. Using a comparative database of MR measurements for 1359 species, from 11 groups ranging from prokaryotes to mammals, and spanning 16 orders of magnitude in mass and a 59°C range in body temperature, we show that differences between taxa in the mass and temperature dependence of MR are sufficiently large as to be retained in the best model for MR despite the requirement for estimation of 22 more parameters than the fundamental equation of MTE.
Keywords: scaling, allometry, metabolic theory, Q10, universal temperature dependence, metabolic rate
1. Introduction
The relationship between body mass and metabolic rate (MR) has been of interest since at least 1838 when Sarrus and Rameaux (cited in Brody [1]) hypothesized that MR should scale in proportion to body surface area, rather than to body mass. While some studies have reported that MR scales in proportion to body mass2/3 [2–4], as does body surface area [5], other studies reject this value in favour of exponents that are simple multiples of one quarter [6–10]. The metabolic theory of ecology (MTE) [7] combines a mechanistic explanation for quarter-power scaling and a description of the universal temperature dependence (UTD) of metabolic processes, and links individual organisms to the ecology of populations, communities and ecosystems. This is accomplished using a quantitative theory describing the relationship between body size, temperature and MR as described by the fundamental equation of MTE [6,7]:
where i0 is a normalization constant independent of body size and temperature, M is body mass, E is the activation energy, k is Boltzmann's constant and T is absolute temperature in Kelvin. Importantly, the fundamental equation contains only one parameter (i0) that must be determined empirically. The mass exponent is derived from first principles of chemistry and physics, and the temperature term is predicted based on the kinetics of biochemical reactions that contribute to MR [6,11,12]. The fractal geometry model for three-fourth-power scaling is a zeroth-order approximation of real biological systems [13], as is the MTE built it upon. These theories are not designed to capture all biological variation but are regarded as representing baselines or points of departure against which variation in real biological systems can be examined [7,13,14]. In such a role, the strength of MTE lies in its ability to predict a wide range of ecological patterns [7,15–19], its grounding in first principles (although some contention exists about the extent to which this holds for the temperature term: [20,21]), and its deliberate simplicity [14]. This contrasts with other theories also grounded in first principles (e.g. dynamic energy budget theory; [22]) that include many more variables than MTE, and have been criticized on the grounds that they are therefore less parsimonious [23]. Implicit in such a criticism is the idea that a model should be evaluated not only on how well it fits available data, but that comparisons of alternative models should incorporate information about how many parameters are required to describe the data. Such ideas form the basis of information-theoretic approaches to model comparison [24–26], in which the best of a candidate set of models is not necessarily the one that provides the best absolute fit to the data, but the one that provides the most acceptable fit with the least parameters.
In an information-theoretic framework, and in contrast to most analyses of metabolic scaling [4,27–33], the appropriateness of the M3/4 term in the fundamental equation of MTE is not assessed by calculating a scaling exponent (b) for a dataset and determining if the 95% confidence interval of the exponent excludes or includes 0.75. Instead, multiple models are constructed to explain the data. One model, following MTE, fits the theoretically predicted parameter values to the data, whereas the parameters in the other models are allowed to vary in various ways. The fits of the different models are then compared with models in which parameters are estimated being penalized for the necessary estimation of additional parameters [24–26]. Such an approach has been advocated for examination of other ecological theories [34–39], and has recently been used to evaluate the mass-dependent term in the fundamental equation of MTE for a limited group of metazoans [40]. The information-theoretic approach to model selection eliminates the risk that models can be accepted statistically on the basis of wide confidence intervals, a criticism levelled at some tests of metabolic theories in ecology [41]. Traditional statistical comparison of model predictions with empirical data invites a risk of failing to reject a model when it is false, because, for example, the weaker the relationship between MR and body mass, the wider the standard error of the slope and the more difficult it is to reject the model or falsify the null hypothesis.
In the present study, we use an information-theoretic approach to evaluate the fundamental equation of MTE against a range of alternative statistical models that describe variation in MR, including the extreme alternative in which free parameters describe all aspects of the mass and temperature dependence of MR for each group considered. Our aim is to determine whether a robust MTE is best underpinned by a simple parameter sparse description of the associations between mass, temperature, and MR (the fundamental equation of MTE), or whether more complex models describe sufficient extra detail to justify the inclusion of extra parameters. The alternative models that we consider include a range of mass and temperature dependencies of MR as well as their interactions: (i) b is fitted statistically, but does not vary between groups; (ii) b is fitted statistically, but varies between groups (i.e. the model includes an interaction between lnM and group); (iii) the UTD term (e−E/kT) is replaced with a Q10 term that describes the factorial increase in MR associated with a 10° increase in temperature, to test the assumption that UTD is a more biologically realistic and accurate representation of temperature dependence [6]; (iv) the temperature dependence of MR (either UTD or Q10) varies between groups (i.e. an interaction between temperature and group); (v) both b and the temperature dependence of MR vary between groups (i.e. interactions between mass and group and between temperature and group); and (vi) both b and the temperature dependence of MR vary between groups, and the temperature dependence of MR depends on mass (i.e. interactions between mass and group, between temperature and group, and between mass and temperature). To test among the candidate set of models (table 1), data were compiled for 1359 species, including prokaryotes, protists, arachnids, insects, fishes, amphibians, reptiles, birds and mammals, spanning a mass range of 16 orders of magnitude. A total of 3622 measurements of MR were obtained at body temperatures ranging from 1°C to 60°C.
Table 1.
Akaike's information criterion (AIC) for a candidate set of models that explain variation in metabolic rate (MR) in terms of body mass (M) and temperature (T); where E is activation energy, b is the scaling exponent and Q10 (the factorial increase in MR associated with a 10°C increase in temperature) is calculated as e10c. (The best model is the one with the lowest AIC, and the probability that a given model provides the best fit of those tested is provided by its Akaike weight, wi.)
| model | scaling exponent, b | temperature dependence | interaction between mass and temperature | AIC | wi |
|---|---|---|---|---|---|
| MR = i0 Mb ecT | estimated, variesa | Q10, variesa | −1139 | 0.37 | |
| MR = i0 Mb e−E/kT | estimated, variesa | UTD, variesa | −1139 | 0.34 | |
| MR = i0 Mb ecT | estimated, variesa | Q10, variesa | lnM × Q10 | −1137 | 0.14 |
| MR = i0 Mb e−E/kT | estimated, variesa | UTD, variesa | lnM × UTD | −1137 | 0.13 |
| MR = i0 Mb ecT | estimated, variesa | Q10, constantb | −1133 | 0.02 | |
| MR = i0 Mb e−E/kT | estimated, variesa | UTD, constantb | −1127 | 0.001 | |
| MR = i0 Mb ecT | estimated, constantb | Q10, variesa | −1025 | <0.0001 | |
| MR = i0 Mb e−E/kT | estimated, constantb | UTD, variesa | −1024 | <0.0001 | |
| MR = i0 Mb ecT | estimated, constantb | Q10, constantb | −1015 | <0.0001 | |
| MR = i0 Mb e−E/kT | estimated, constantb | UTD, constantb | −1010 | <0.0001 | |
| MR = i0 Mb e−E/kT | fixedc: 0.75 | UTD, constantb | −982 | <0.0001 | |
| MR = i0 M3/4 e−7.4(1000/T) | fixedc: 0.75 | UTD, fixedd: −7.4 | −957 | <0.0001 |
aScaling exponent or temperature fitted separately for each group. Note that temperature is in Kelvin for UTD and degrees Celsius for Q10.
bScaling exponent or temperature dependence constant between groups.
2. Methods
The goodness of fit of the set of candidate models (table 1) to a database comprising 3622 measurements of MR for 1359 species of prokaryotes, protists, arachnids, insects, fishes, amphibians, reptiles, birds and mammals was assessed using Akaike's information criterion (AIC) as a measure of model fit [26]. The database was assembled using published compilations of data for MR [4,42–45], and data for MR were included only if they were accompanied by appropriate estimates of temperature (ambient or body temperature for ecotherms; body temperature for endotherms). We included all data available in the published compilations that satisfied these conditions. Multiple values were available for some species (particularly ectotherms) that were measured at multiple temperatures and/or stages of development; all of these values were included in the analysis. The dataset includes species spanning 16 orders of magnitude range in body mass, and measured at body temperatures ranging from 1°C to 60°C (see the electronic supplementary material). Mass and MR were loge transformed for analysis, and the relationship between loge MR, loge M and temperature (degree Celsius for calculation of Q10 values or 1000/kT with T in Kelvin for calculation of UTD values) was calculated using general linear modelling in JMP v. 8.0 or v. 9.0.2 (SAS Institute, Cary, NC, USA). Log-likelihoods were calculated according to Burnham and Anderson [26], and the best out of all of the models tested to explain MR was that with the lowest AIC. The probability that any given model is actually the best fit out of those tested was measured by its Akaike weight [26], the relative likelihood of the model compared with all others (the likelihood of the model divided by the sum of the likelihoods of all other models).
Following the analyses upon which MTE is based [6–8] as well as other comparative analyses of similarly diverse datasets [45], we do not incorporate phylogenetic information in the analysis. Meta-analyses of allometric scaling exponents have revealed little qualitative differences between phylogenetic and non-phylogenetic analyses [28], and the major conclusions of the present study are consistent with a number of recent phylogenetically informed studies of mammals [29–31]. Thus, while we consider that the inclusion of phylogenetic information would be unlikely to alter the conclusion, we nevertheless suggest that the analysis be repeated when appropriate trees become available.
3. Results and discussion
The fundamental equation of MTE is considerably less likely to provide the best fit to the data than the best model, which described MR in terms of a mass scaling exponent that varied between groups (figure 1) and a Q10 value that also varied between groups (figure 2). This is despite the best model being penalized for the additional 22 parameters required to describe the overall mass and temperature dependence of MR, as well as to describe the unique mass and temperature dependences of each group (figures 1 and 2). Overall, models that described the temperature dependence of MR using Q10 values provided a better fit to the data than models describing the relationship according to UTD, although the difference was small (table 1).
Figure 1.
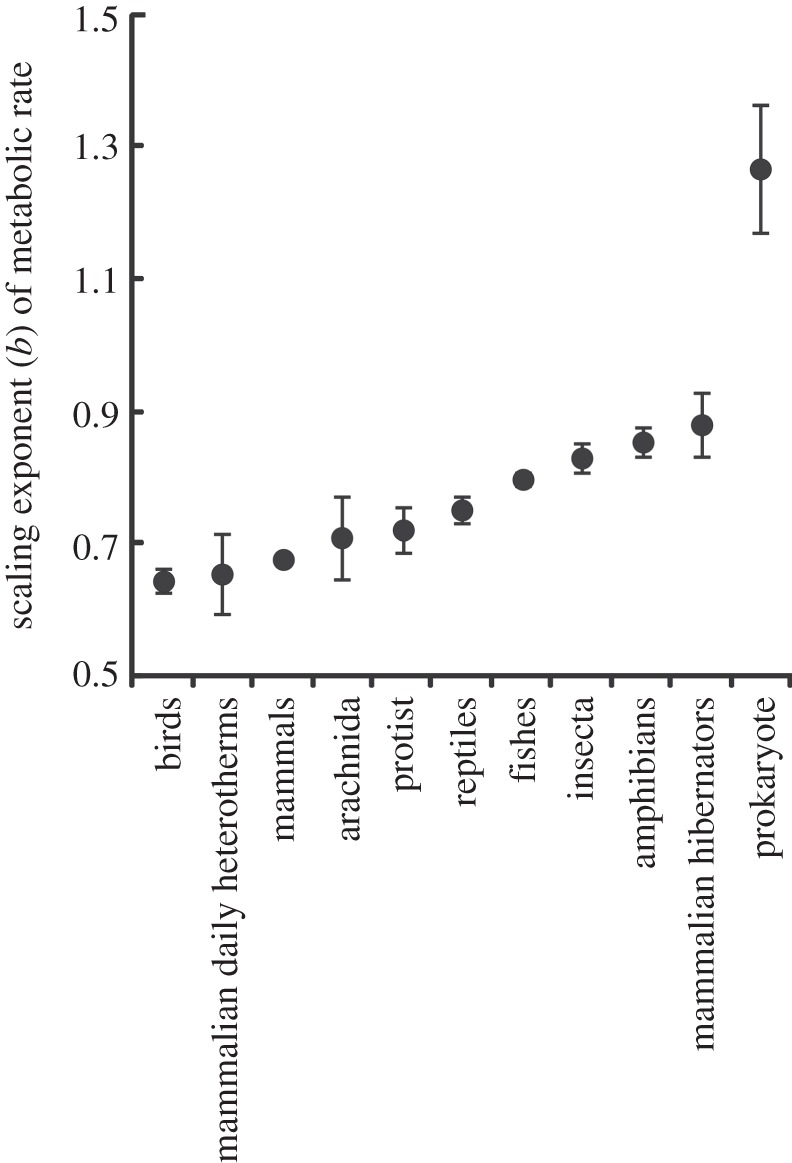
Scaling exponents (b) relating metabolic rate (MR) to body mass (M, where MR ∝ Mb) for a range of taxa. Scaling exponents are shown ± s.e.
Figure 2.
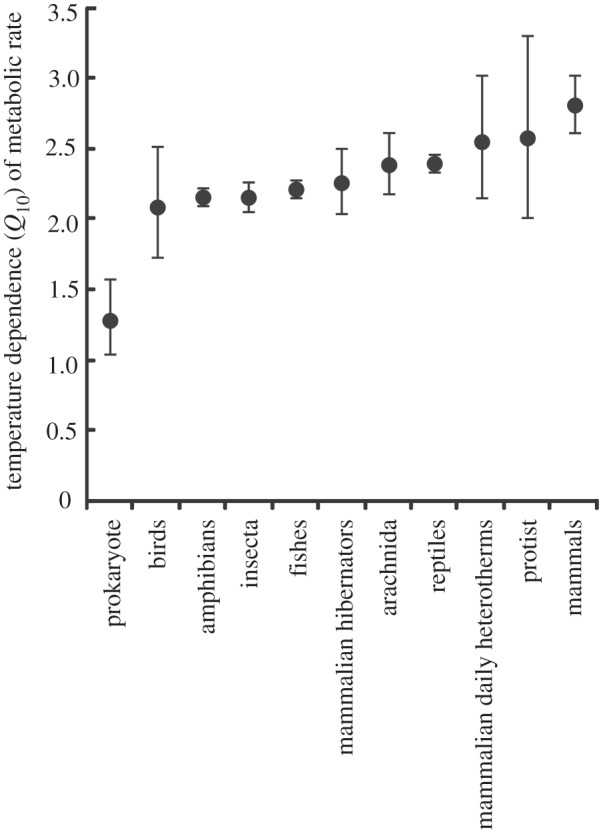
Temperature dependence of metabolic rate (MR) for a range of taxa. Q10 is the factorial increase in MR associated with a 10°C increase in body temperature. Values of Q10 are shown ± s.e.
With data for prokaryotes, which show the most extreme values of b (figure 1) and Q10 (figure 2) excluded, the best overall model (wi = 0.69) described MR in terms of a mass scaling exponent that varied between groups, a Q10 value that varied between groups, and an interaction between mass and temperature. The second best model (wi = 0.17) included a mass scaling exponent that varied between groups and a Q10 value that varied between groups, but no interaction between mass and temperature, and the third best model (wi = 0.12) included a mass scaling exponent that varied between groups, a UTD value that differed between groups, and an interaction between temperature and mass. The probability that one of these three models best described the data with prokaryotes excluded is 0.97.
The appropriate method for describing the temperature dependence of MR has been the subject of lively debate [6,20,46–48]. It has been argued that the Boltzmann–Arrhenius relationship is to be preferred over the Van't Hoff (Q10) equation on the grounds that the former incorporates both the general theory for the kinetics of chemical reactions and the empirically determined activation energies for the critical reactions of cellular respiration [6,47]. Q10, on the other hand, is by definition an approximation of the Boltzmann–Arrhenius relationship. The functions differ by up to 15 per cent over the ‘biologically relevant’ temperature range [49,50], suggesting a clear need to chose one over the other. Such a choice is not simple, however, because the Boltzmann–Arrhenius relationship is also regarded as a phenomenological approximation [50,51] suggesting that neither can be chosen simply on the basis of being grounded in first principles. In practice, there is also little statistical support for choosing one over the other when all data are considered, since Q10 provides only a 1.1-fold better fit to the data than UTD. With data for prokaryotes excluded, however, Q10 provides a 5.8-fold better fit to the data than UTD.
Importantly, irrespective of the method of temperature dependence preferred, models that incorporated variation in both the scaling exponent and temperature dependence between groups provided a better fit to the data than those that did not (table 1). Thus, the fundamental equation of MTE is unable to predict variation in the effects of temperature and mass on MR associated with major evolutionary transitions and life-history differences among groups, as has been reported previously for smaller sets of taxa [40,45,52]. The present analysis shows that these effects predominate across the spectrum of living organisms, as exemplified by a recent study demonstrating variation in the temperature dependence of a broad set of 112 physiological and ecological traits for a sample of 309 species [53].
In summary, the present study shows that the relationship between body mass, temperature and MR cannot be adequately described by a single equation, and identifies differences in values of the scaling exponent and Q10 between taxa and metabolic states (i.e. between hibernating and normothermic mammals; figures 1 and 2). Such differences are not captured by the fundamental equation of MTE, and the consequences of this variation can therefore not be understood by strictly adopting the fundamental equation of MTE. While the fundamental equation has been successful in explaining some ecological patterns [7,15–19], its failure in other cases [41,54–58] may stem, at least in part, from an imprecise description of the effects of temperature and mass on MR. Recent work has demonstrated that variation in the mass-dependent term of MTE can be achieved [59–62]. The best model from table 1 requires estimation of 34 parameters, so it seems likely that complex models with many terms [e.g. dynamic energy budget theory: 22] will outperform simple ones. Further statistical assessments of the reasons for variation in the scaling and temperature dependence of metabolism are therefore unlikely to improve understanding without reference to a wider set of alternative mechanistic models [63,64], including those which explicitly incorporate phylogenetic, ecological and spatial effects, which are significant contributors to variation in scaling and temperature dependence of MR [21,53,65,66] and which may be difficult to distinguish [67]. The ability of more parameter-rich theories for the mass and temperature dependence of MR to explain ecological patterns should also be explored.
References
- 1.Brody S. 1945. Bioenergetics and growth. New York, NY: Reinhold Publishing Corporation [Google Scholar]
- 2.Rubner M. 1883. Über den Einfluss der Körpergrösse auf Stoff- und Kraftwechsel. Zeischrift für Biologie 19, 536–562 [Google Scholar]
- 3.Heusner A. A. 1991. Size and power in mammals. J. Exp. Biol. 160, 25–54 [DOI] [PubMed] [Google Scholar]
- 4.White C. R., Seymour R. S. 2003. Mammalian basal metabolic rate is proportional to body mass2/3. Proc. Natl Acad. Sci. USA 100, 4046–4049 10.1073/pnas.0436428100 (doi:10.1073/pnas.0436428100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reynolds P. S. 1997. Phylogenetic analysis of surface areas of mammals. J. Mammal. 78, 859–868 10.2307/1382944 (doi:10.2307/1382944) [DOI] [Google Scholar]
- 6.Gillooly J. F., Brown J. H., West G. B., Savage V. M., Charnov E. L. 2001. Effects of size and temperature on metabolic rate. Science 293, 2248–2251 10.1126/science.1061967 (doi:10.1126/science.1061967) [DOI] [PubMed] [Google Scholar]
- 7.Brown J. H., Gillooly J. F., Allen A. P., Savage V. M., West G. B. 2004. Toward a metabolic theory of ecology. Ecology 85, 1771–1789 10.1890/03-9000 (doi:10.1890/03-9000) [DOI] [Google Scholar]
- 8.Savage V. M., Gillooly J. F., Woodruff W. H., West G. B., Allen A. P., Enquist B. J., Brown J. H. 2004. The predominance of quarter-power scaling in biology. Funct. Ecol. 18, 257–282 10.1111/j.0269-8463.2004.00856.x (doi:10.1111/j.0269-8463.2004.00856.x) [DOI] [Google Scholar]
- 9.Moses M. E., Hou C., Woodruff W. H., West G. B., Nekola J. C., Zuo W., Brown J. H. 2008. Revisiting a model of ontogenetic growth: estimating model parameters from theory and data. Am. Nat. 171, 632–645 10.1086/587073 (doi:10.1086/587073) [DOI] [PubMed] [Google Scholar]
- 10.Kleiber M. 1932. Body size and metabolism. Hilgardia 6, 315–353 [Google Scholar]
- 11.West G. B., Brown J. H., Enquist B. J. 1997. A general model for the origin of allometric scaling laws in biology. Science 276, 122–126 10.1126/science.276.5309.122 (doi:10.1126/science.276.5309.122) [DOI] [PubMed] [Google Scholar]
- 12.West G. B., Brown J. H., Enquist B. J. 1999. The fourth dimension of life: fractal geometry and allometric scaling of organisms. Science 284, 1677–1679 10.1126/science.284.5420.1677 (doi:10.1126/science.284.5420.1677) [DOI] [PubMed] [Google Scholar]
- 13.West G. B., Brown J. H. 2005. The origin of allometric scaling laws in biology from genomes to ecosystems: towards a quantitative unifying theory of biological structure and organization. J. Exp. Biol. 208, 1575–1592 10.1242/jeb.01589 (doi:10.1242/jeb.01589) [DOI] [PubMed] [Google Scholar]
- 14.Brown J. H., Gillooly J. F., Allen A. P., Savage V. M., West G. B. 2004. Response to forum commentary on ‘toward a metabolic theory of ecology’. Ecology 85, 1818–1821 10.1890/03-0800 (doi:10.1890/03-0800) [DOI] [Google Scholar]
- 15.Allen A. P., Brown J. H., Gillooly J. F. 2002. Global biodiversity, biochemical kinetics, and the energetic-equivalence rule. Science 297, 1545–1548 10.1126/science.1072380 (doi:10.1126/science.1072380) [DOI] [PubMed] [Google Scholar]
- 16.Meehan T. D., Jetz W., Brown J. H. 2004. Energetic determinants of abundance in winter landbird communities. Ecol. Lett. 7, 532–537 10.1111/j.1461-0248.2004.00611.x (doi:10.1111/j.1461-0248.2004.00611.x) [DOI] [Google Scholar]
- 17.Meehan T. D. 2006. Energy use and animal abundance in litter and soil communities. Ecology 87, 1650–1658 10.1890/0012-9658(2006)87[1650:EUAAAI]2.0.CO;2 (doi:10.1890/0012-9658(2006)87[1650:EUAAAI]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 18.Buckley L. B., Rodda G. H., Jetz W. 2008. Thermal and energetic constraints on ectotherm abundance: a global test using lizards. Ecology 89, 48–55 10.1890/07-0845.1 (doi:10.1890/07-0845.1) [DOI] [PubMed] [Google Scholar]
- 19.Munch S. B., Salinas S. 2009. Latitudinal variation in lifespan within species is explained by the metabolic theory of ecology. Proc. Natl Acad. Sci. USA 106, 13 860–13 864 10.1073/pnas.0900300106 (doi:10.1073/pnas.0900300106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clarke A. 2006. Temperature and the metabolic theory of ecology. Funct. Ecol. 20, 405–412 10.1111/j.1365-2435.2006.01109.x (doi:10.1111/j.1365-2435.2006.01109.x) [DOI] [Google Scholar]
- 21.Irlich U. M., Terblanche J. S., Blackburn T. M., Chown S. L. 2009. Insect rate-temperature relationships: environmental variation and the metabolic theory of ecology. Am. Nat. 174, 819–835 10.1086/647904 (doi:10.1086/647904) [DOI] [PubMed] [Google Scholar]
- 22.Kooijman S. A. L. M. 2010. Dynamic energy budget theory for metabolic organisation, 3rd edn Cambridge, UK: Cambridge University Press [Google Scholar]
- 23.Zuo W., Moses M. E., Hou C., Woodruff W. H., West G. B., Brown J. H. 2009. Response to comments on ‘Energy uptake and allocation during ontogeny’. Science 325, 1206. 10.1126/science.1171949 (doi:10.1126/science.1171949) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson J. B., Omland K. S. 2004. Model selection in ecology and evolution. Trends Ecol. Evol. 19, 101–108 10.1016/j.tree.2003.10.013 (doi:10.1016/j.tree.2003.10.013) [DOI] [PubMed] [Google Scholar]
- 25.Hobbs N. T., Hilborn R. 2006. Alternatives to statistical hypothesis testing in ecology: a guide to self teaching. Ecol. Appl. 16, 5–19 10.1890/04-0645 (doi:10.1890/04-0645) [DOI] [PubMed] [Google Scholar]
- 26.Burnham K. P., Anderson D. R. 2010. Model selection and multi-model inference: a practical information-theoretic approach, 2nd edn New York, NY: Springer [Google Scholar]
- 27.White C. R., Seymour R. S. 2005. Allometric scaling of mammalian metabolism. J. Exp. Biol. 208, 1611–1619 10.1242/jeb.01501 (doi:10.1242/jeb.01501) [DOI] [PubMed] [Google Scholar]
- 28.White C. R., Cassey P., Blackburn T. M. 2007. Allometric exponents do not support a universal metabolic allometry. Ecology 88, 315–323 10.1890/05-1883 (doi:10.1890/05-1883) [DOI] [PubMed] [Google Scholar]
- 29.White C. R., Blackburn T. M., Seymour R. S. 2009. Phylogenetically informed analysis of the allometry of mammalian basal metabolic rate supports neither geometric nor quarter-power scaling. Evolution 63, 2658–2667 10.1111/j.1558-5646.2009.00747.x (doi:10.1111/j.1558-5646.2009.00747.x) [DOI] [PubMed] [Google Scholar]
- 30.Sieg A. E., O'Conner M. P., McNair J. N., Grant B. W., Agosta S. J., Dunham A. E. 2009. Mammalian metabolic allometry: do intraspecific variation, phylogeny, and regression models matter? Am. Nat. 174, 720–733 10.1086/606023 (doi:10.1086/606023) [DOI] [PubMed] [Google Scholar]
- 31.Duncan R. P., Forsythe D. M., Hone J. 2007. Testing the metabolic theory of ecology: allometric scaling exponents in mammals. Ecology 88, 324–333 10.1890/0012-9658(2007)88[324:TTMTOE]2.0.CO;2 (doi:10.1890/0012-9658(2007)88[324:TTMTOE]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 32.McKechnie A. E., Wolf B. O. 2004. The allometry of avian basal metabolic rate: good predictions need good data. Physiol. Biochem. Zool. 77, 502–521 10.1086/383511 (doi:10.1086/383511) [DOI] [PubMed] [Google Scholar]
- 33.McKechnie A. E., Freckleton R. P., Jetz W. 2006. Phenotypic plasticity in the scaling of avian basal metabolic rate. Proc. R. Soc. B 273, 931–937 10.1098/rspb.2005.3415 (doi:10.1098/rspb.2005.3415) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gotelli N. J., McGill B. J. 2006. Null versus neutral models: what's the difference? Ecography 29, 793–800 10.1111/j.2006.0906-7590.04714.x (doi:10.1111/j.2006.0906-7590.04714.x) [DOI] [Google Scholar]
- 35.Thompson R., Townsend C. 2006. A truce with neutral theory: local deterministic factors, species traits and dispersal limitation together determine patterns of diversity in stream invertebrates. J. Anim. Ecol. 75, 476–484 10.1111/j.1365-2656.2006.01068.x (doi:10.1111/j.1365-2656.2006.01068.x) [DOI] [PubMed] [Google Scholar]
- 36.McGill B. J., et al. 2007. Species abundance distributions: moving beyond single prediction theories to integration within an ecological framework. Ecol. Lett. 10, 995–1015 10.1111/j.1461-0248.2007.01094.x (doi:10.1111/j.1461-0248.2007.01094.x) [DOI] [PubMed] [Google Scholar]
- 37.Richards S. A. 2005. Testing ecological theory using the information-theoretic approach: examples and cautionary results. Ecology 86, 2805–2814 10.1890/05-0074 (doi:10.1890/05-0074) [DOI] [Google Scholar]
- 38.Stephens P. A., Buskirk S. W., del Rio C. M. 2007. Inference in ecology and evolution. Trends Ecol. Evol. 22, 192–197 10.1016/j.tree.2006.12.003 (doi:10.1016/j.tree.2006.12.003) [DOI] [PubMed] [Google Scholar]
- 39.Horne J. S., Garton E. O. 2006. Selecting the best home range model: an information-theoretic approach. Ecology 87, 1146–1152 10.1890/0012-9658(2006)87[1146:STBHRM]2.0.CO;2 (doi:10.1890/0012-9658(2006)87[1146:STBHRM]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 40.Isaac N. J. B., Carbone C. 2010. Why are metabolic scaling exponents so controversial? Quantifying variance and testing hypotheses. Ecol. Lett. 13, 728–735 (doi:10.1111/j.1461–0248.2010.01461.x) [DOI] [PubMed] [Google Scholar]
- 41.Hawkins B. A., et al. 2007. A global evaluation of metabolic theory as an explanation for terrestrial species richness gradients. Ecology 88, 1877–1888 10.1890/06-1444.1 (doi:10.1890/06-1444.1) [DOI] [PubMed] [Google Scholar]
- 42.White C. R., Seymour R. S. 2004. Does BMR contain a useful signal? Mammalian BMR allometry and correlations with a selection of physiological, ecological and life-history variables. Physiol. Biochem. Zool. 77, 929–941 10.1086/425186 (doi:10.1086/425186) [DOI] [PubMed] [Google Scholar]
- 43.White C. R., Phillips N. F., Seymour R. S. 2006. The scaling and temperature dependence of vertebrate metabolism. Biol. Lett. 2, 125–127 10.1098/rsbl.2005.0378 (doi:10.1098/rsbl.2005.0378) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.White C. R., Blackburn T. M., Martin G. R., Butler P. J. 2007. Basal metabolic rate of birds is associated with habitat temperature and precipitation, not primary productivity. Proc. R. Soc. B 274, 287–293 10.1098/rspb.2006.3727 (doi:10.1098/rspb.2006.3727) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Makarieva A. M., Gorshkov V. D., Li B.-L., Chown S. L., Reich P. B., Gavrilov V. M. 2008. Mean mass-specific metabolic rates are strikingly similar across life's major domains: evidence for life's metabolic optimum. Proc. Natl Acad. Sci. USA 105, 16 994–16 999 10.1073/pnas.0802148105 (doi:10.1073/pnas.0802148105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clarke A. 2004. Is there a universal temperature dependence of metabolism? Funct. Ecol. 18, 252–256 10.1111/j.0269-8463.2004.00842.x (doi:10.1111/j.0269-8463.2004.00842.x) [DOI] [Google Scholar]
- 47.Gillooly J. F., Allen A. P., Savage V. M., Charnov E. L., West G. B., Brown J. H. 2006. Response to Clarke and Fraser: effects of temperature on metabolic rate. Funct. Ecol. 20, 400–404 10.1111/j.1365-2435.2006.01110.x (doi:10.1111/j.1365-2435.2006.01110.x) [DOI] [Google Scholar]
- 48.Downs C. J., Hayes J. P., Tracy C. R. 2008. Scaling metabolic rate with body mass and inverse body temperature: a test of the Arrhenius fractal supply model. Funct. Ecol. 22, 239–244 10.1111/j.1365-2435.2007.01371.x (doi:10.1111/j.1365-2435.2007.01371.x) [DOI] [Google Scholar]
- 49.Allen A. P., Gillooly J. F. 2007. The mechanistic basis of the metabolic theory of ecology. Oikos 116, 1073–1077 10.1111/j.0030-1299.2007.16079.x (doi:10.1111/j.0030-1299.2007.16079.x) [DOI] [Google Scholar]
- 50.Gutman E. M. 2007. Empiricism or self-consistent theory in chemical kinetics? J. Alloys Compd 434–435, 779–782 10.1016/j.jallcom.2006.08.175 (doi:10.1016/j.jallcom.2006.08.175) [DOI] [Google Scholar]
- 51.del Rio C. M. 2008. Metabolic theory or metabolic models? Trends Ecol. Evol. 23, 256–260 10.1016/j.tree.2008.01.010 (doi:10.1016/j.tree.2008.01.010) [DOI] [PubMed] [Google Scholar]
- 52.DeLong J. P., Okie J. G., Moses M. E., Sibly R. M., Brown J. H. 2010. Shifts in metabolic scaling, production, and efficiency across major evolutionary transitions of life. Proc. Natl Acad. Sci. USA 107, 12 941–12 945 10.1073/pnas.1007783107 (doi:10.1073/pnas.1007783107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dell A. I., Pawar S., Savage V. M. 2011. Systematic variation in the temperature dependence of physiological and ecological traits. Proc. Natl Acad. Sci. USA 108, 10 591–10 596 10.1073/pnas.1015178108 (doi:10.1073/pnas.1015178108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Terribile L. C., Diniz-Filho J. A. F. 2009. Spatial patterns of species richness in New World coral snakes and the metabolic theory of ecology. Acta Oecol. 35, 163–173 10.1016/j.actao.2008.09.006 (doi:10.1016/j.actao.2008.09.006) [DOI] [Google Scholar]
- 55.Muller-Landau H. C., et al. 2006. Testing metabolic ecology theory for allometric scaling of tree size, growth and mortality in tropical forests. Ecol. Lett. 9, 575–588 10.1111/j.1461-0248.2006.00904.x (doi:10.1111/j.1461-0248.2006.00904.x) [DOI] [PubMed] [Google Scholar]
- 56.Rahbek C., Gotelli N. J., Colwell R. K., Entsminger G. L., Rangel T. F. L. V. B., Graves G. R. 2007. Predicting continental-scale patterns of bird species richness with spatially explicit models. Proc. R. Soc. B 274, 165–174 10.1098/rspb.2006.3700 (doi:10.1098/rspb.2006.3700) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Castro F., Gaedke U. 2008. The metabolism of lake plankton does not support the metabolic theory of ecology. Oikos 117, 1218–1226 10.1111/j.0030-1299.2008.16547.x (doi:10.1111/j.0030-1299.2008.16547.x) [DOI] [Google Scholar]
- 58.Algar A. C., Kerr J. T., Currie D. J. 2007. A test of metabolic theory as the mechanism underlying broad-scale species-richness gradients. Glob. Ecol. Biogeogr. 16, 170–178 10.1111/j.1466-8238.2006.00275.x (doi:10.1111/j.1466-8238.2006.00275.x) [DOI] [Google Scholar]
- 59.Price C. A., Enquist B. J., Savage V. M. 2007. A general model for allometric covariation in botanical form and function. Proc. Natl Acad. Sci. USA 104, 13 204–13 209 10.1073/pnas.0702242104 (doi:10.1073/pnas.0702242104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Savage V. M., Deeds E. J., Fontana W. 2008. Sizing up allometric scaling theory. PLoS Comput. Biol. 4, e1000171. 10.1371/journal.pcbi.1000171 (doi:10.1371/journal.pcbi.1000171) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Banavar J. R., Moses M. E., Brown J. H., Damuth J., Rinaldo A., Sibly R. M., Maritan A. 2010. A general basis for quarter-power scaling in animals. Proc. Natl Acad. Sci. USA 107, 15 816–15 820 10.1073/pnas.1009974107 (doi:10.1073/pnas.1009974107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kolokotrones T., Savage V. M., Deeds E. J., Fontana W. 2010. Curvature in metabolic scaling. Nature 464, 753–756 10.1038/nature08920 (doi:10.1038/nature08920) [DOI] [PubMed] [Google Scholar]
- 63.Kozłowski J., Konarzewski M., Gawelczyk A. T. 2003. Cell size as a link between noncoding DNA and metabolic rate scaling. Proc. Natl Acad. Sci. USA 100, 14 080–14 085 10.1073/pnas.2334605100 (doi:10.1073/pnas.2334605100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Glazier D. S. 2010. A unifying explanation for diverse metabolic scaling in animals and plants. Biol. Rev. 85, 111–138 10.1111/j.1469-185X.2009.00095.x (doi:10.1111/j.1469-185X.2009.00095.x) [DOI] [PubMed] [Google Scholar]
- 65.Wang Z., Brown J. H., Tang Z., Fang J. 2009. Temperature dependence, spatial scale, and tree species diversity in eastern Asia and North America. Proc. Natl Acad. Sci. USA 106, 13 388–13 392 10.1073/pnas.0905030106 (doi:10.1073/pnas.0905030106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stegen J. C., Enquist B. J., Ferriere R. 2009. Advancing the metabolic theory of biodiversity. Ecol. Lett. 12, 1001–1015 10.1111/j.1461-0248.2009.01358.x (doi:10.1111/j.1461-0248.2009.01358.x) [DOI] [PubMed] [Google Scholar]
- 67.Freckleton R. P., Jetz W. 2009. Space versus phylogeny: disentangling phylogenetic and spatial signals in comparative data. Proc. R. Soc. B 276, 21–30 10.1098/rspb.2008.0905 (doi:10.1098/rspb.2008.0905) [DOI] [PMC free article] [PubMed] [Google Scholar]


