Abstract
Natural selection drives populations of individuals towards local peaks in a fitness landscape. These peaks are created by the interactions between individual mutations. Fitness landscapes may change as an environment changes. In a previous contribution, we discovered a variant of the Azoarcus group I ribozyme that represents a local peak in the RNA fitness landscape. The genotype at this peak is distinguished from the wild-type by four point mutations. We here report ribozyme fitness data derived from constructing all possible combinations of these point mutations. We find that these mutations interact epistatically. Importantly, we show that these epistatic interactions change qualitatively in the three different environments that we studied. We find examples where the relative fitness of a ribozyme can change from neutral or negative in one environment, to positive in another. We also show that the fitness effect of a specific GC–AU base pair switch is dependent on both the environment and the genetic context. Moreover, the mutations that we study improve activity at the cost of decreased structural stability. Environmental change is ubiquitous in nature. Our results suggest that such change can facilitate adaptive evolution by exposing new peaks of a fitness landscape. They highlight a prominent role for genotype–environment interactions in doing so.
Keywords: epistasis, ribozymes, fitness landscape, environmental change
1. Introduction
Evolution is often seen as a process of biological optimization, where natural selection drives populations of individuals towards peaks in a fitness landscape [1–3]. During this process, individuals move incrementally ‘uphill’ in such a landscape by acquiring beneficial mutations. For the optimization of an individual enzyme, moving ‘uphill’ can mean increasing enzyme activity, specificity or stability. Even within individual enzymes, mutations do not operate in isolation, but they interact in their effects on fitness, a phenomenon known as epistasis [4]. A particularly severe form of epistasis, also called sign epistasis, occurs when individual mutations are deleterious in some genetic backgrounds, but beneficial in others. Sign epistasis creates peaks and valleys in the fitness landscape. It has received much attention in evolutionary biology, because fitness valleys can be sufficient to impede the biological optimization process [5–7]. The crossing of these valleys requires a temporary decrease in fitness that can be difficult to achieve through the forces of evolution [8–10].
Ultimately, the effect of mutations depends on interactions between the genotype and the environment. In the study of many common forms of human disease, for example, both genetic and environmental factors are required to predict the disposition of individuals to disease [11]. However, the genetic basis of complex traits, including disease traits, is often incompletely understood. This is evident in the fact that genetic polymorphisms associated with common (disease) traits usually explain only a small fraction of the variation in the trait [12,13]. This phenomenon has been referred to as the ‘missing heritability’ in complex traits. It may be explained by epistatic interactions among genotypes, unknown environmental factors, or both [14,15]. Until very recently, the complicated nature of gene–gene interactions (epistasis) affecting complex traits has prevented a systematic analysis of how epistatic interactions are affected by the environment [16]. Nevertheless, such analysis could improve our understanding of the tempo and mode of evolution in changing environments, and could ultimately help us to direct our efforts to dissect the components of heritability in complex traits.
An experimental approach to understanding the relationships between epistasis, fitness landscapes and evolution involves reconstructing all molecular intermediates between a reference genotype and a genotype at a local peak in the fitness landscape [5,17–21]. On the basis of this information, the fitness landscape can be reconstructed by characterizing the phenotype of each intermediate. All the possible pathways to higher fitness through these intermediates can be compared side by side. Previous experiments of this type have revealed that interactions between multiple mutations can differ depending on whether the mutations occur in different genes or within a single gene. For example, mutations occurring in different genes in Escherichia coli show very little sign epistasis [21]. Instead, the interactions between these different-gene mutations seem to be dominated by negative or ‘diminishing returns’ epistasis. That is, when several individually beneficial mutations from different genes are combined, the total fitness advantage is less than expected from each individual benefit, and this discrepancy increases with the number of mutations considered [20,21]. In contrast, when multiple mutations occur within individual enzyme-coding genes, extensive sign epistasis occurs, which can render most pathways to higher fitness inaccessible [8–10]. However, all of these experiments consider only epistatic interactions in a single environment (e.g. fitness in the presence of a single antibiotic). In nature, environments change constantly, and a more complete characterization of epistatic fitness landscapes requires experiments that consider environmental change. Here, we reconstruct the fitness landscape of a catalytic RNA molecule (ribozyme), including all possible intermediates that lead to a local fitness peak, and determine the ‘fitness’ (relative activity) of each intermediate in three different environments.
The fitness peak used in this study is occupied by a variant of the Azoarcus group I ribozyme that we call Azo*. Group I ribozymes can be studied experimentally through an assay that requires the cleavage of a synthetic RNA oligonucleotide, because this activity is a component of the ribozyme's natural self-splicing activity [22]. Four mutations in the Azo* variant significantly increase the activity of this ribozyme relative to the wild-type ribozyme (Azowt; figure 1). The Azo* variant was discovered during a previous set of directed evolution experiments that evolved populations of ribozymes in three different environments, which we call native, thermal stress and new (for a description of the evolution procedure, see Supporting Information, electronic supplementary material). These three environments were chosen as part of a two-step experimental design that tested the effect of cryptic variation on adapting populations [23]. In these experiments, we first subjected populations of ribozymes to a period of mutagenesis and purifying selection for the native ribozyme activity (RNA oligonucleotide cleavage). One population was selected under typical ribozyme reaction conditions (native environment), whereas the other was selected in the presence of formamide, which is a denaturant that lowers the stability of RNA structures (thermal stress environment). Second, we selected a new chemical environment, in which the standard substrate was replaced with an RNA oligonucleotide containing a phosphorothioate bond at the cleavage site (new environment). High-throughput sequencing analysis revealed that the populations from the first part of the experiment moved close in genotype space to the Azo* sequence, resulting in its rapid increase in frequency during the second step [23].
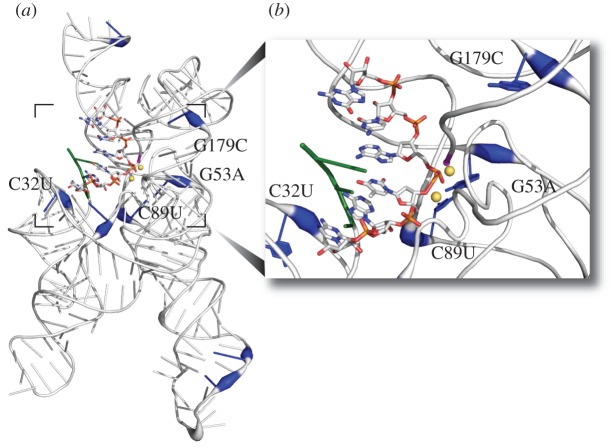
Figure 1.
The Azo* mutations in the context of the ribozyme structure. The four mutations of our study are labelled in the format XNY, where X is the wild-type base at position N and Y is the mutant base at this position. (a) The crystal structure of the Azoarcus ribozyme (1ZZN) is shown using the ‘PUTTY’ function in Pymol (Schrödinger). Positions where mutations rose to high frequency during our previous evolution in the new environment are shown in blue. The thickness of the tube representing the RNA backbone is scaled to the rate at which the mutation at that position increased in frequency [23]. The Azo* mutations were a unique set of these mutations that showed high co-occurrence and the most rapid increase in frequency. The active site of the ribozyme is indicated by the substrate (stick representation, coloured by chemical element). (b) A close-up view of the Azo* mutations in relationship to the active site.
2. Results and discussion
The four mutations that distinguish the superior Azo* activity can arise in any order, creating 4! = 24 mutational trajectories between Azowt and Azo*. These mutational trajectories consist of 16 riboyzme variants that comprise all the possible combinations of the four mutations, including Azowt (no mutations) and Azo* (all four mutations). For this work, we synthesized all 16 ribozymes and characterize their relative fitness (enzymatic activity) in each of the three environments (native, thermal stress and new) in order to study the environmental impact on the epistatic interactions (for full dataset, see Spreadsheets, electronic supplementary material). From the data, we can reconstruct the fitness landscapes composed of all the possible mutational trajectories that could be navigated to reach the Azo* landscape peak.
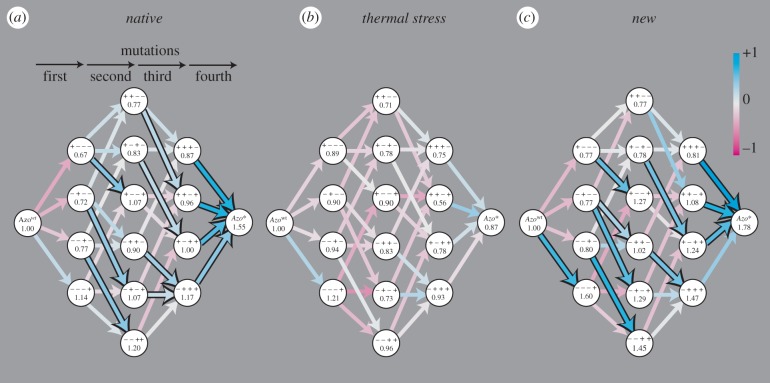
The landscapes are shown in figure 2. The trajectories between Azowt and Azo* constitute a discrete four-dimensional space (a hypercube) because each variant has four ‘neighbours’ that differ by exactly one mutation. We present a projection of this space onto two dimensions. Moving from left to right on a landscape in figure 2 takes one from Azowt to Azo* one mutation at a time. Each combination of mutations (genotype) is presented as a circle. The acquisition of a mutation (step to the right) can cause a change in activity from the previous variant, creating ups and downs in the landscape that are represented as arrows. We use colour to depict this ‘ruggedness’ (see key), where cyan indicates an increase in activity (uphill step) and magenta represents a decrease in activity (downhill step). The colour saturation of each arrow is scaled to the magnitude of change, so that mutational steps that result in no activity change are colourless (light grey arrows). Arrows with a black border reflect mutations that entail a significant increase in fitness (two tailed t-test, p < 0.05, n ≥ 3, allowing a multiple testing false discovery rate of 10% [24]; see §3). We highlight these transitions because they are highly favoured directions of change relative to neutral or deleterious transitions under the conditions of our previous evolution experiments (for discussion, see ‘Population genetics’ in Supporting Information, electronic supplementary material). The figure suggests the existence of sign epistasis in these landscapes, because significant uphill steps are separated by neutral or downhill steps.
Figure 2.
(a–c) Functional landscapes of combinations of the Azo* mutations. The landscapes are shown in three environments, all of which are at 37°C, 25 mM Mg2+ and pH 7.5 (native). Additionally, the thermal stress environment contained 5 M formamide. The new environment was identical to the native environment, but contained a substrate with a phosphorothioate at the scissile bond. Each circle corresponds to a genotype with a specific combination of the Azo* mutations shown as four ‘positive’ or ‘negative’ symbols that indicate, from left to right, the presence or absence of the mutations C32U, G53A, C89U and G179C. The value within each circle is the measured activity of that ribozyme relative to Azowt. The arrows between two circles are coloured to indicate the relative activity change in the direction of the arrow (acquired mutation). The colour saturation of each arrow is scaled to the activity effect: 100% saturation = fitness change of +1 (cyan), or −1 (magenta); 0% saturation = no effect (light grey). Arrows that correspond to a significant increase in activity have a black border.
In our ribozyme variants, each individual mutation can occur with any of eight different combinations of the other three mutations. We determined the effect of each mutation in each of the eight genetic contexts and classified the effect as positive or negative depending on a significant (see earlier text) increase or decrease in activity relative to the same genotype without the mutation. We classified effects as negligible if the change in activity was not significant. Table 1 lists the number of times each of the four mutations has a positive or non-positive (negative or negligible) effect (see also electronic supplementary material, figure S1 and ‘Fitness effects’ in Supporting Information). Sign epistasis exists in all cases where a mutation has both positive and non-positive effects. For example, in the native environment, the mutation C89U shows sign epistasis: in the background of three variants, it has a positive effect, whereas in the context of five others, it has a non-positive effect (table 1, row 3).
Table 1.
Summary of the effects of individual mutations.
| non-positive |
||||
|---|---|---|---|---|
| environment | mutation | positive | negative | negligible |
| native | C32U | 1 | 3 | 4 |
| G53A | 1 | — | 7 | |
| C89U | 3 | 1 | 4 | |
| G179C | 6 | — | 2 | |
| thermal stress | C32U | — | — | 8 |
| G53A | — | — | 8 | |
| C89U | — | — | 8 | |
| G179C | — | — | 8 | |
| new | C32U | — | — | 8 |
| G53A | 1 | — | 7 | |
| C89U | 2 | — | 6 | |
| G179C | 7 | — | 1 | |
The data in table 1 also indicate that epistasis changes with environments. Changes in sign epistasis in particular are revealed by the differing number of positive and non-positive effects of each mutation between the environments. For example, the mutations C32U and C89U lose instances of positive effects when moved from the native to the new environments, whereas G179C gains such effects. Although we group the negative and negligible effects together as ‘non-positive’ because they are both unlikely to reach fixation by natural selection [6], table 1 also shows that several instances of significantly negative effects in the native environment become negligible when the environment changes.
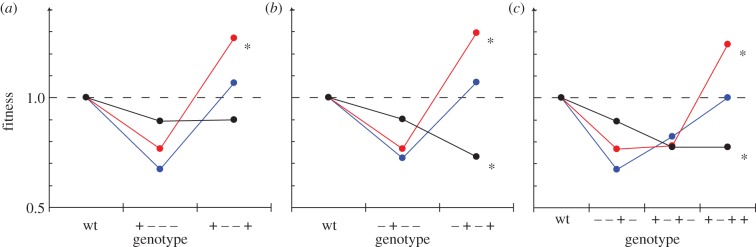
In some cases, the effect of the environments on a mutational interaction results in a relative fitness advantage only in the new environment (figure 3). This occurs in two examples of ribozyme variants that are two mutations away from the wild-type (figure 3a,b), and in one case where the variant is three mutations away (figure 3c). For example, in the trajectory shown in figure 3a, acquiring the mutation C32U (going from left to right) results in a decrease in fitness that is followed by a fitness increase if the additional mutation G179C occurs. In the native environment (blue), the increase in fitness caused by the second mutation is not enough to increase the fitness of the double mutant significantly above that of the wild-type. Thus, the net result of the two mutations is a neutral change. However, in the new environment (red), the same variant has a relative fitness that is significantly higher than the wild-type. Under thermal stress, the same double mutant has lower fitness than the wild-type. Qualitatively, identical patterns hold for the mutations in figure 3b,c.
Figure 3.
Fitness advantages revealed after environmental change. Examples of combinations of (a,b) two or (c) three mutations that show a fitness advantage only in the new environment. Intermediate interactions encountered in each environment during a mutational trajectory that leads away from Azowt (wt) are also shown (for all trajectories, see electronic supplementary material, figure S1). The fitness (y-axis) of each variant is shown as a function of additional mutations (x-axis) introduced into Azowt (left to right on each graph correspond to increasing numbers of mutations). The specific mutation that is added is indicated on the x-axis using ‘positive’ and ‘negative’ symbols as in figure 2. Fitness values are measured relative to Azowt in each of the three environments: native (blue), thermal stress (black) and new (red). Fitness values that are significantly different than Azowt in the given environment are indicated by *p < 0.05 (t-test).
The data illustrate that environmental change can alter a fitness landscape and even create a fitness advantage where there was not one before. The examples in figure 3 are extreme examples of a more general pattern (i.e. an amplification of fitness advantages in the new environment). Comparing the fitness value of each variant in the new and native environment reveals that, on average, the fitness values are 21 per cent higher in the new environment, ignoring variants with a fitness decrease in both environments (see electronic supplementary material, figure S2). These same variants show an even greater advantage (49%) if the new fitness values are compared with those under thermal stress conditions.
In terms of the ribozyme's structural stability, the thermal stress conditions can be considered analogous to a temperature increase of 13.5°C [25]. We chose to introduce a denaturant (formamide) instead of an actual temperature change, because the latter can alter many other parameters, such as pH and kcat. In our experiments, the only change between the native and thermal stress environment is the presence of the denaturant, and we can therefore interpret the difference in fitness between these environments as an effect of structural stability. All fitness values we report for this environment are relative to the activity of Azowt in that environment. Therefore, a variant's decreased fitness in the thermal stress environment indicates that the variant has a decreased structural stability relative to Azowt.
Overall, we observe that the thermal stress environment flattens the fitness landscape, and sign epistasis is lost (table 1 and figure 2). In fact, not even the Azo* genotype itself is advantageous in this environment. This observation indicates that the four Azo* mutations cause increased activity at the expense of structural stability (function–stability trade-off). Under permissive conditions (native environment), the four mutations together are advantageous, but they compromise the stability of the enzyme, which is revealed under thermal stress. Interestingly, the single mutations alone do not reveal this trade-off because their fitness effects are similar in both environments (see electronic supplementary material, figure S2c). The effect is revealed only by combinations of two or more mutations that unanimously show decreased fitness effects in the thermal stress environment. The most striking example is Azo* itself. When all four mutations are present, they cause a 55 per cent fitness increase in the native environment, but a 13 per cent decrease under thermal stress. This is consistent with data from mutagenesis experiments on protein enzymes that have shown that most mutations, even beneficial ones, are destabilizing [26–28]. Our results indicate that this is also the case for our RNA enzyme. This trade-off amounts to antagonistic pleiotropy at the single enzyme level, and emphasizes that an enzyme cannot always optimize activity and structural stability simultaneously [27,28].
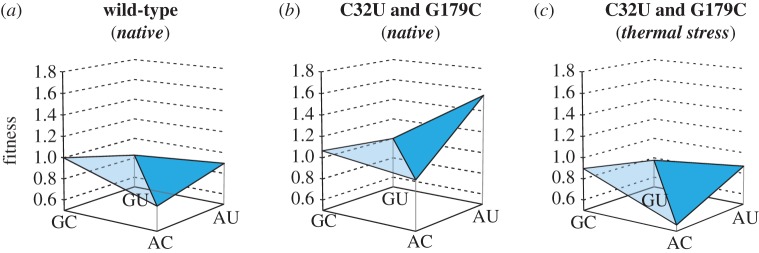
We next turn to an especially instructive pair of mutations in our data. These mutations cause a GC–AU base pair switch. A base pair switch of this type is a classic example of a compensatory mutation. Such mutations are frequent in naturally occurring RNA molecules and have important implications for phylogenetic analysis [29,30]. The two mutations occur at positions 53 and 89 in the Azoarcus ribozyme that form a base pair in the middle of the P4 stem of the conserved group I ribozyme structure [31]. Together, the two mutations preserve the secondary structure, and one might expect that they have little or no effect on function. Surprisingly, our results show otherwise. The change in activity resulting from this combination of mutations depends on both the genetic context and the environment (figure 4). In the native environment, the presence of two other mutations (C32U and G179C) can change the effect of the GC–AU switch from slightly negative to positive (compare figure 3a,b). Specifically, in the wild-type background, the AU base pair causes a 10 per cent decrease in activity relative to the GC base pair (figure 3b). However, when the mutations C32U and G179C are already present, the AU base pair causes a 48 per cent increase in activity. Furthermore, in this advantageous genetic context (C32U and G179C already present), a change to the thermal stress environment alters the effect of the base pair switch from positive (48% increase) to negligible (2.9% decrease; figure 3b,c). This change in fitness indicates that this base pair switch is an important component of the decreased stability of the Azo* genotype. In other genetic contexts where this double mutation can occur, a changing environment can also cause a negligible effect to become significantly negative (see electronic supplementary material, figure S3).
Figure 4.
(a–c) Context-dependent effect of a GC–AU base pair switch. The fitness effect of a switch from a GC to an AU base pair resulting from the two mutations: G53A and C89U. The fitness (relative activity) of the original base pair (GC), the two possible intermediates (GU or AC) and the final base pair (AU) are shown relative to the Azowt activity in the given environment. The environment is shown in italics above the graph (see figure 2 and §3 for environmental conditions). The other mutations that are present during the switch are shown in bold type above each graph.
Two trajectories are possible in converting a GC base pair to an AU base pair. One of them passes through a GU base pair intermediate, the other through an AC mismatch intermediate (see figure 4; electronic supplementary material, figure S3). In our data, a ribozyme with the GU intermediate is always more active than that with the AC intermediate (from +1.4 to +22.7%; average = +10.3%). This confirms predictions of the relative effects of these two intermediates based on the geometry of base pairs [32], experimental measurements [33] and bioinformatic analysis of tRNA evolution [34]. Our data support a ‘continuous ridge’ or ‘neutral network’ model (see ‘Landscapes caused by a base pair switch’ in Supporting Information, electronic supplementary material) of compensatory evolution in RNA secondary structure, because in our experiments, the GU intermediate shows a significant decrease in fitness only in a single instance, and in this instance the base pair switch is not compensatory but deleterious (see electronic supplementary material, figure S3d). In all other cases, no significant decrease in fitness occurs for any given genotype when the GC base pair is replaced by the GU intermediate (see electronic supplementary material, figure S3).
A further intriguing observation is that the fitness values in the new and native environments are correlated (Pearson's r = 0.95; electronic supplementary material, figure S2a). The correlation probably arises from the fact that the new substrate is a promiscuous substrate of the wild-type ribozyme, and that this substrate is structurally highly similar to the native substrate. It has the same sequence as the ‘native’ substrate, and differs only at the scissile phosphate, where a non-bridging oxygen is replaced by a sulphur. The correlation is intriguing because it might help explain why in nature, where environments constantly change, promiscuous activities can be preserved even in enzymes highly optimized for a primary substrate [35,36]. The correlation we observe suggests that pathways to improving promiscuous functions may stay open even while optimizing an enzyme's native activity, and vice versa. Further experimental evidence is needed to test the generality of these findings.
Our previous evolution experiments provide a unique opportunity to ask how the three epistatic fitness landscapes we study here altered the course of evolution under defined conditions. For this analysis, we turn to deep sequencing data of populations that resulted from our previous evolution experiments. Each population began from the same ‘wild-type’ population and was evolved for several (8 or 10) generations in each of the three environments (see electronic supplementary material, Supporting Information for a description of selection experiments). We find that the evolving populations contained variants that are several mutations away from Azowt, but that do not have increased fitness. In fact, some sequenced variants have no intermediates that show an increased activity relative to Azowt (see electronic supplementary material, figure S4). For example, the variant ‘+++–’ has a fitness of 0.87, 0.75 and 0.81 in the native, thermal stress and new environments, respectively (figure 2). All intermediates between Azowt and ‘+++–’ have fitness values of less than 1. Nevertheless, we find this genotype in the individuals sampled after 10 generations of selection in the thermal stress (0.3% of sample), and after eight generations in the new environment (0.4% of sample). Other similar examples exist.
The earlier-mentioned sequencing results are surprising because strong selection was acting in the evolution experiments that produced this sequence data (see ‘Population genetics’ in Supporting Information, electronic supplementary material). In these experiments, selection should have favoured trajectories that do not contain intermediates with negative or no fitness change. However, our evolution experiments also used large populations and high mutation rates. Our sequence data are consistent with predictions of valley/plateau crossing scenarios that consider parameters (population size, mutation rate and selection pressure) in the range of our experiments [9,37,38]. Specifically, for our experiments, we estimate an effective population size of 107 molecules, a mutation rate of 10−4 (Taq polymerase), and relative fitness effects of approximately 0.1 (see ‘Effective population size’ in Supporting Information, electronic supplementary material). Under these parameters, populations are expected to cross each of our three landscapes by establishing lineages based on neutral or deleterious variants, because such variants can persist in a population at mutation selection balance. In fact, under these conditions, populations are predicted to take neutral and slightly deleterious steps even when continuously uphill paths are possible [9]. Our sequence data support this prediction. We note that the parameter values we just mentioned are not restricted to our experiments, but can also occur in large populations of organisms, making the kind of valley/plateau crossing we observe a biologically relevant scenario [39–42]. Several examples are discussed briefly in the Supporting Information in the electronic supplementary material (see ‘Natural valley/plateau crossing populations’).
The phenotype of our study system (ribozyme activity) is much simpler than that of complex organismal traits. However, our experimental system enables us to alter systematically genotypes and environments to reveal important interactions. Such manipulation is very difficult or impossible at the organismal level, where the genetic factors leading to complex traits are incompletely understood. It has been argued that some of the ‘missing heritability’ in genome-wide association studies may be due to the averaging of the effects of specific alleles over different environments [14,15]. In fact, for our data, if we average the fitness of each ribozyme variant over all three environments, then we find that no variant has a significant increase in fitness relative to Azowt (see electronic supplementary material, figure S5). This shows that even in our simple system the relationship between genotype and phenotype can be correctly characterized only if gene–environment interactions are considered. These results support the hypothesis that gene–environment interactions can be an important source of ‘missing heritability’.
Our experimental results highlight a positive role for environmental change in adaptive evolution. The Azo* genotype was actually discovered by first evolving populations in the native or thermal stress environments, then switching to the new environment. The fitness landscapes whose structure we determined reveal that in the first environments populations were required to explore genotype space through the preservation of neutral or even slightly deleterious variants. These variants have no fitness advantage in this environment, and they did not rise to high frequency in our evolving populations. However, in the new environment, some of these same variants do have a fitness advantage. These variants increased in frequency in our populations after being exposed to the new environment. Gene–environment interactions are critical in this scenario because some combinations of mutations become adaptive only on environmental change. Especially in large populations, the shifting patterns of epistasis that are caused by environmental change may render adaptive peaks more accessible and fitness landscapes less rugged than is sometimes assumed.
3. Methods
(a). Ribozyme and substrate synthesis
The dsDNA templates containing variants of the Azoarcus ribozyme were constructed by a two-step PCR-based assembly from six synthetic DNA oligonucleotides [43]. The templates contain 197 nucleotides of the Azoarcus group I intron, excluding the first eight nucleotides, but including the nucleophilic terminal guanosine (G205), all preceded by the T7 promoter sequence to allow in vitro transcription. In vitro transcription reactions were performed as previously described [23]. Substrate oligonucleotides GGCAU(AAAU)4A and GGCAUs(AAAU)4A (where ‘s’ denotes phosphorothioate bond) were produced by solid phase synthesis and purified by denaturing PAGE (Microsynth). Concentrations were determined by UV-absorbance on an ND-1000 spectrophotometer (NanoDrop Technologies).
(b). Activity measurements
Activities of ribozyme variants were determined in 10 μl reactions containing 10 pmol ribozyme, 50 pmol substrate oligonucleotide, 25 mM MgCl2 and 30 mM EPPS (pH 7.5). The new reactions contained 50 pmol each Rp and Sp phosphorothioate diastereomers (100 pmole racemic mixture), of which only the Rp diastereomer is a viable substrate. The reaction mixtures were incubated at 37°C for 1 h. Reacted and unreacted enzyme was separated by denaturing PAGE (6% polyacrylamide, 8 M urea), and visualized by UV transillumination after fluorescent staining with GelRed (Biotum). Activities were determined as the fraction of fluorescence from the reacted ribozyme relative to the total ribozyme fluorescence (reacted plus unreacted). Fitness values were determined as the ratio of variant activity to Azowt activity under the given reaction conditions. Activities were based on the average of at least three measurements. Some of the activity measurements (12) from the native environment were reported as supplementary material in a previous publication [23].
(c). Controlling the rate of false discovery
Significant differences in the fitness of neighbours in our landscapes were corrected for multiple hypothesis testing. Raw p-values from t-tests were rank-ordered, and converted to false discovery rate p-values by Hochberg & Benjamini [24] procedure. These false discovery rate p-values were used to assign the significance of fitness effects tabulated in table 1 and indicated by black borders in figure 2 of the main text.
Acknowledgements
We acknowledge support from Swiss National Science Foundation grants nos 315200-116814, 315200-119697 and 315230-129708, from the YeastX program of SystemsX.ch, and from the University Research Priority Programme: Systems Biology/Functional Genomics of the University of Zurich and the ETH.
References
- 1.Wright S. 1932. The roles of mutation, inbreeding, crossbreeding and selection in evolution. In Proc. 6th Int. congress of genetics (ed. Jones D. F.), pp. 356–366 Menasha, WI: Brooklyn Botanic Garden [Google Scholar]
- 2.Kauffman S., Levin S. 1987. Towards a general theory of adaptive walks on rugged landscapes. J. Theor. Biol. 128, 11–45 10.1016/S0022-5193(87)80029-2 (doi:10.1016/S0022-5193(87)80029-2) [DOI] [PubMed] [Google Scholar]
- 3.Smith J. M. 1970. Natural selection and the concept of a protein space. Nature 225, 563–564 10.1038/225563a0 (doi:10.1038/225563a0) [DOI] [PubMed] [Google Scholar]
- 4.Phillips P. C. 2008. Epistasis: the essential role of gene interactions in the structure and evolution of genetic systems. Nat. Rev. Genet. 9, 855–867 10.1038/nrg2452 (doi:10.1038/nrg2452) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weinreich D. M., Delaney N. F., Depristo M. A., Hartl D. L. 2006. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 312, 111–114 10.1126/science.1123539 (doi:10.1126/science.1123539) [DOI] [PubMed] [Google Scholar]
- 6.Weinreich D. M., Watson R. A., Chao L. 2005. Perspective: sign epistasis and genetic constraint on evolutionary trajectories. Evolution 59, 1165–1174 [PubMed] [Google Scholar]
- 7.Poelwijk F. J., Kiviet D. J., Weinreich D. M., Tans S. J. 2007. Empirical fitness landscapes reveal accessible evolutionary paths. Nature 445, 383–386 10.1038/nature05451 (doi:10.1038/nature05451) [DOI] [PubMed] [Google Scholar]
- 8.Gillespie J. H. 1984. Molecular evolution over the mutational landscape. Evolution 38, 1116–1129 10.2307/2408444 (doi:10.2307/2408444) [DOI] [PubMed] [Google Scholar]
- 9.Weissman D. B., Desai M. M., Fisher D. S., Feldman M. W. 2009. The rate at which asexual populations cross fitness valleys. Theor. Popul. Biol. 75, 286–300 10.1016/j.tpb.2009.02.006 (doi:10.1016/j.tpb.2009.02.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weissman D. B., Feldman M. W., Fisher D. S. 2010. The rate of fitness-valley crossing in sexual populations. Genetics 186, 1389–1410 10.1534/genetics.110.123240 (doi:10.1534/genetics.110.123240) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manolio T. A., et al. 2009. Finding the missing heritability of complex diseases. Nature 461, 747–753 10.1038/nature08494 (doi:10.1038/nature08494) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCarthy M. I., Abecasis G. R., Cardon L. R., Goldstein D. B., Little J., Ioannidis J. P. A., Hirschhorn J. N. 2008. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 9, 356–369 10.1038/nrg2344 (doi:10.1038/nrg2344) [DOI] [PubMed] [Google Scholar]
- 13.Visscher P. M. 2008. Sizing up human height variation. Nat. Genet. 40, 489–490 10.1038/ng0508-489 (doi:10.1038/ng0508-489) [DOI] [PubMed] [Google Scholar]
- 14.Ober C., Vercelli D. 2011. Gene–environment interactions in human disease: nuisance or opportunity? Trends Genet. 27, 107–115 10.1016/j.tig.2010.12.004 (doi:10.1016/j.tig.2010.12.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas D. 2010. Gene–environment-wide association studies: emerging approaches. Nat. Rev. Genet. 11, 259–272 10.1038/nrg2764 (doi:10.1038/nrg2764) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hinkley T., Martins J., Chappey C., Haddad M., Stawiski E., Whitcomb J. M., Petropoulos C. J., Bonhoeffer S. 2011. A systems analysis of mutational effects in HIV-1 protease and reverse transcriptase. Nat. Genet. 43, 487–489 10.1038/ng.795 (doi:10.1038/ng.795) [DOI] [PubMed] [Google Scholar]
- 17.Lee Y. H., DSouza L. M., Fox G. E. 1997. Equally parsimonious pathways through an RNA sequence space are not equally likely. J. Mol. Evol. 45, 278–284 10.1007/PL00006231 (doi:10.1007/PL00006231) [DOI] [PubMed] [Google Scholar]
- 18.Reetz M. T., Sanchis J. 2008. Constructing and analyzing the fitness landscape of an experimental evolutionary process. ChemBioChem 9, 2260–2267 10.1002/cbic.200800371 (doi:10.1002/cbic.200800371) [DOI] [PubMed] [Google Scholar]
- 19.Lunzer M., Miller S. P., Felsheim R., Dean A. M. 2005. The biochemical architecture of an ancient adaptive landscape. Science 310, 499–501 10.1126/science.1115649 (doi:10.1126/science.1115649) [DOI] [PubMed] [Google Scholar]
- 20.Chou H.-H., Chiu H.-C., Delaney N. F., Segrè D., Marx C. J. 2011. Diminishing returns epistasis among beneficial mutations decelerates adaptation. Science 332, 1190–1192 10.1126/science.1203799 (doi:10.1126/science.1203799) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khan A. I., Dinh D. M., Schneider D., Lenski R. E., Cooper T. F. 2011. Negative epistasis between beneficial mutations in an evolving bacterial population. Science 332, 1193–1196 10.1126/science.1203801 (doi:10.1126/science.1203801) [DOI] [PubMed] [Google Scholar]
- 22.Zaug A. J., Cech T. R. 1986. The intervening sequence RNA of tetrahymena is an enzyme. Science 231, 470–475 10.1126/science.3941911 (doi:10.1126/science.3941911) [DOI] [PubMed] [Google Scholar]
- 23.Hayden E. J., Ferrada E., Wagner A. 2011. Cryptic genetic variation promotes rapid evolutionary adaptation in an RNA enzyme. Nature 474, 92–95 10.1038/nature10083 (doi:10.1038/nature10083) [DOI] [PubMed] [Google Scholar]
- 24.Hochberg Y., Benjamini Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B Method. 57, 289–300 10.2307/2346101 (doi:10.2307/2346101) [DOI] [Google Scholar]
- 25.Blake R. D., Delcourt S. G. 1996. Thermodynamic effects of formamide on DNA stability. Nucleic Acids Res. 24, 2095–2103 10.1093/nar/24.11.2095 (doi:10.1093/nar/24.11.2095) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bloom J. D., Arnold F. H. 2009. Colloquium papers: in the light of directed evolution: pathways of adaptive protein evolution. Proc. Natl Acad. Sci. USA 106, 9995–10 000 10.1073/pnas.0901522106 (doi:10.1073/pnas.0901522106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bloom J. D., Wilke C. O., Arnold F. H., Adami C. 2004. Stability and the evolvability of function in a model protein. Biophys. J. 86, 2758–2764 10.1016/S0006-3495(04)74329-5 (doi:10.1016/S0006-3495(04)74329-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soskine M., Tawfik D. S. 2010. Mutational effects and the evolution of new protein functions. Nat. Rev. Genet. 11, 572–582 10.1038/nrg2808 (doi:10.1038/nrg2808) [DOI] [PubMed] [Google Scholar]
- 29.Kimura M. 1985. The role of compensatory neutral mutations in molecular evolution. J. Genet. 64, 7–19 10.1007/BF02923549 (doi:10.1007/BF02923549) [DOI] [Google Scholar]
- 30.Stephan W. 1996. The rate of compensatory evolution. Genetics 144, 419–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanner M., Cech T. 1996. Activity and thermostability of the small self-splicing group I intron in the pre-tRNA(lle) of the purple bacterium Azoarcus. RNA 2, 74–83 [PMC free article] [PubMed] [Google Scholar]
- 32.Leontis N. B., Stombaugh J., Westhof E. 2002. The non-Watson–Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 30, 3497–3531 10.1093/nar/gkf481 (doi:10.1093/nar/gkf481) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis A. R., Znosko B. M. 2007. Thermodynamic characterization of single mismatches found in naturally occurring RNA. Biochemistry 46, 13 425–13 436 10.1021/bi701311c (doi:10.1021/bi701311c) [DOI] [PubMed] [Google Scholar]
- 34.Meer M. V., Kondrashov A. S., Artzy-Randrup Y., Kondrashov F. A. 2010. Compensatory evolution in mitochondrial tRNAs navigates valleys of low fitness. Nature 464, 279–282 10.1038/nature08691 (doi:10.1038/nature08691) [DOI] [PubMed] [Google Scholar]
- 35.Soo V. W. C., Hanson-Manful P., Patrick W. M. 2011. Artificial gene amplification reveals an abundance of promiscuous resistance determinants in Escherichia coli. Proc. Natl Acad. Sci. USA 108, 1484–1489 10.1073/pnas.1012108108 (doi:10.1073/pnas.1012108108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aharoni A., Gaidukov L., Khersonsky O., Gould S. M., Roodveldt C., Tawfik D. S. 2005. The ‘evolvability’ of promiscuous protein functions. Nat. Genet. 37, 73–76 10.1038/ng1482 (doi:10.1038/ng1482) [DOI] [PubMed] [Google Scholar]
- 37.Desai M. M., Fisher D. S. 2007. Beneficial mutation–selection balance and the effect of linkage on positive selection. Genetics 176, 1759–1798 10.1534/genetics.106.067678 (doi:10.1534/genetics.106.067678) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Østman B., Hintze A., Adami C. 2011. Impact of epistasis and pleiotropy on evolutionary adaptation. Proc. R. Soc. B 279, 247–256 10.1098/rspb.2011.0870 (doi:10.1098/rspb.2011.0870) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weinreich D. M., Chao L. 2005. Rapid evolutionary escape by large populations from local fitness peaks is likely in nature. Evolution 59, 1175–1182 [PubMed] [Google Scholar]
- 40.Iwasa Y., Michor F., Nowak M. A. 2004. Stochastic tunnels in evolutionary dynamics. Genetics 166, 1571–1579 10.1534/genetics.166.3.1571 (doi:10.1534/genetics.166.3.1571) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Desai M. M., Fisher D. S., Murray A. W. 2007. The speed of evolution and maintenance of variation in asexual populations. Curr. Biol. 17, 385–394 10.1016/j.cub.2007.01.072 (doi:10.1016/j.cub.2007.01.072) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woods R. J., Barrick J. E., Cooper T. F., Shrestha U., Kauth M. R., Lenski R. E. 2011. Second-order selection for evolvability in a large Escherichia coli population. Science 331, 1433–1436 10.1126/science.1198914 (doi:10.1126/science.1198914) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rydzanicz R., Zhao X. S., Johnson P. E. 2005. Assembly PCR oligo maker: a tool for designing oligodeoxynucleotides for constructing long DNA molecules for RNA production. Nucleic Acids Res. 33, W521–W525 10.1093/nar/gki380 (doi:10.1093/nar/gki380) [DOI] [PMC free article] [PubMed] [Google Scholar]