Abstract
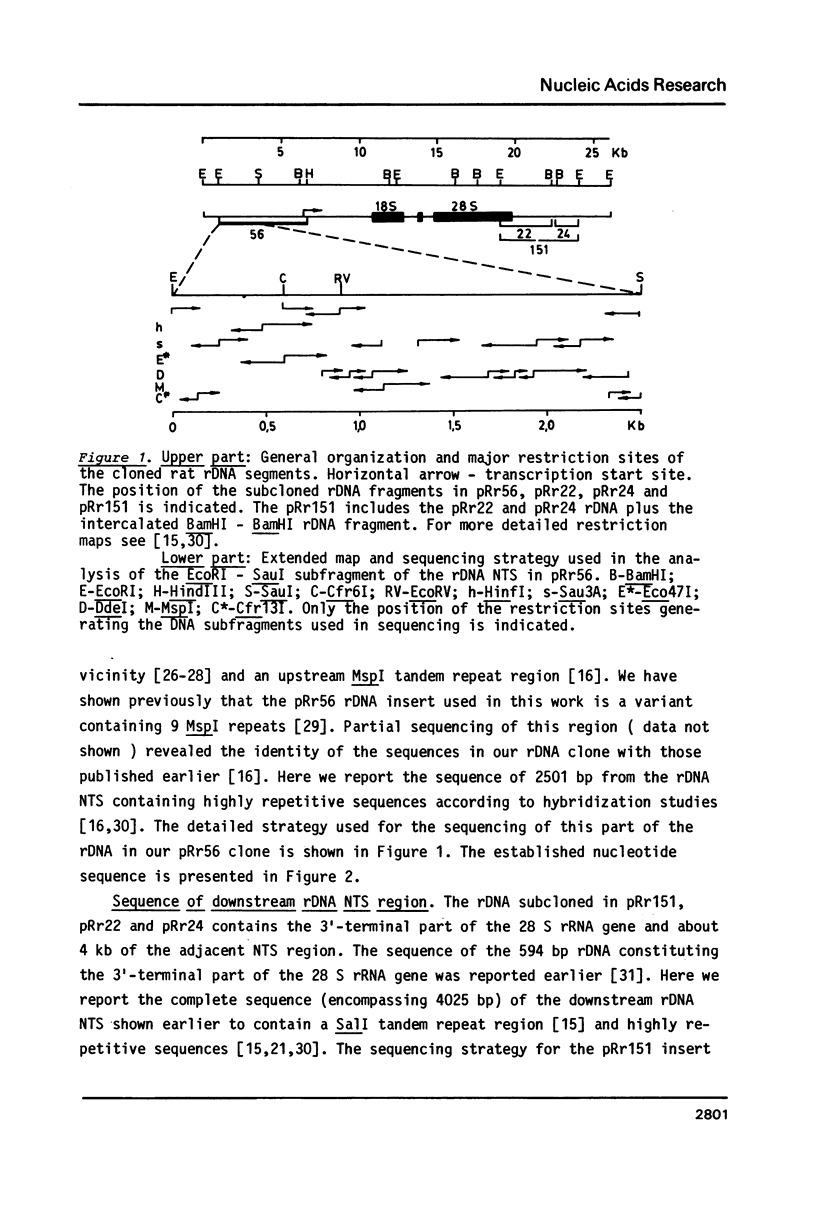
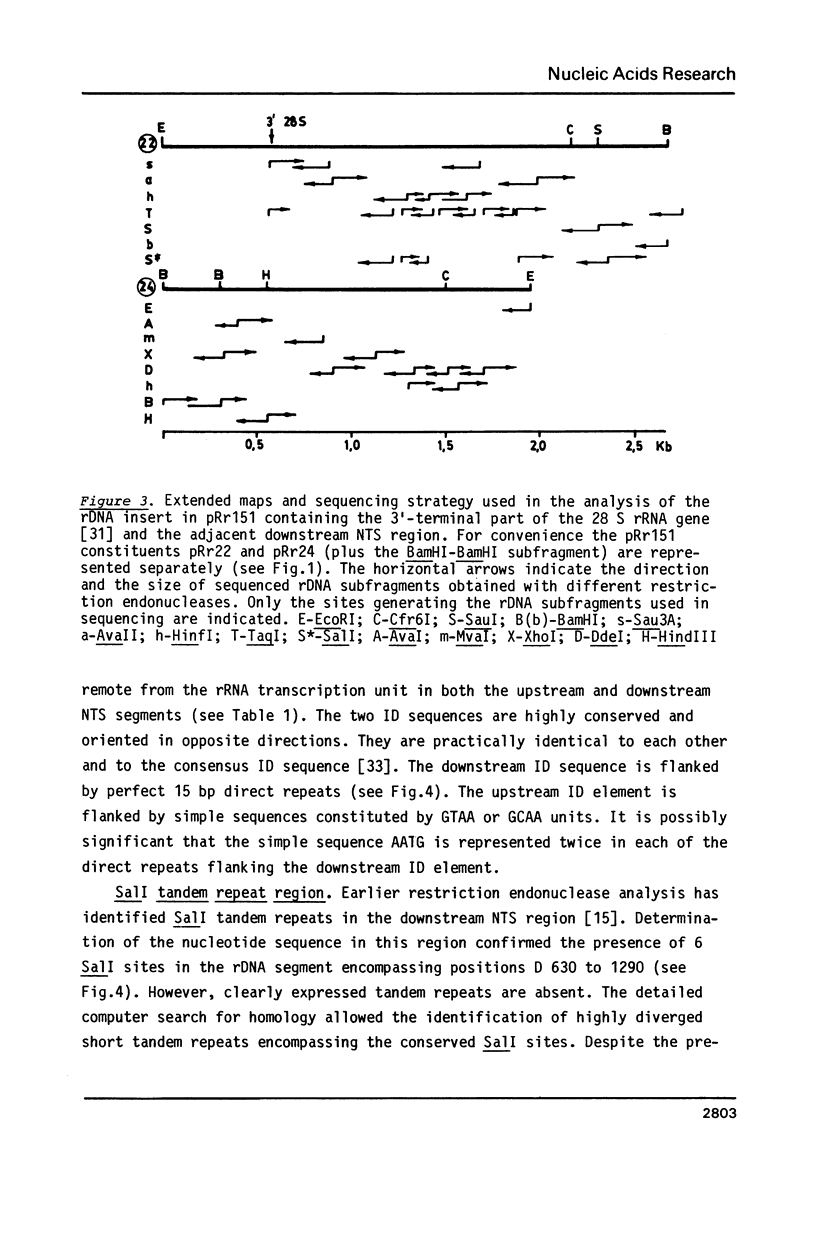
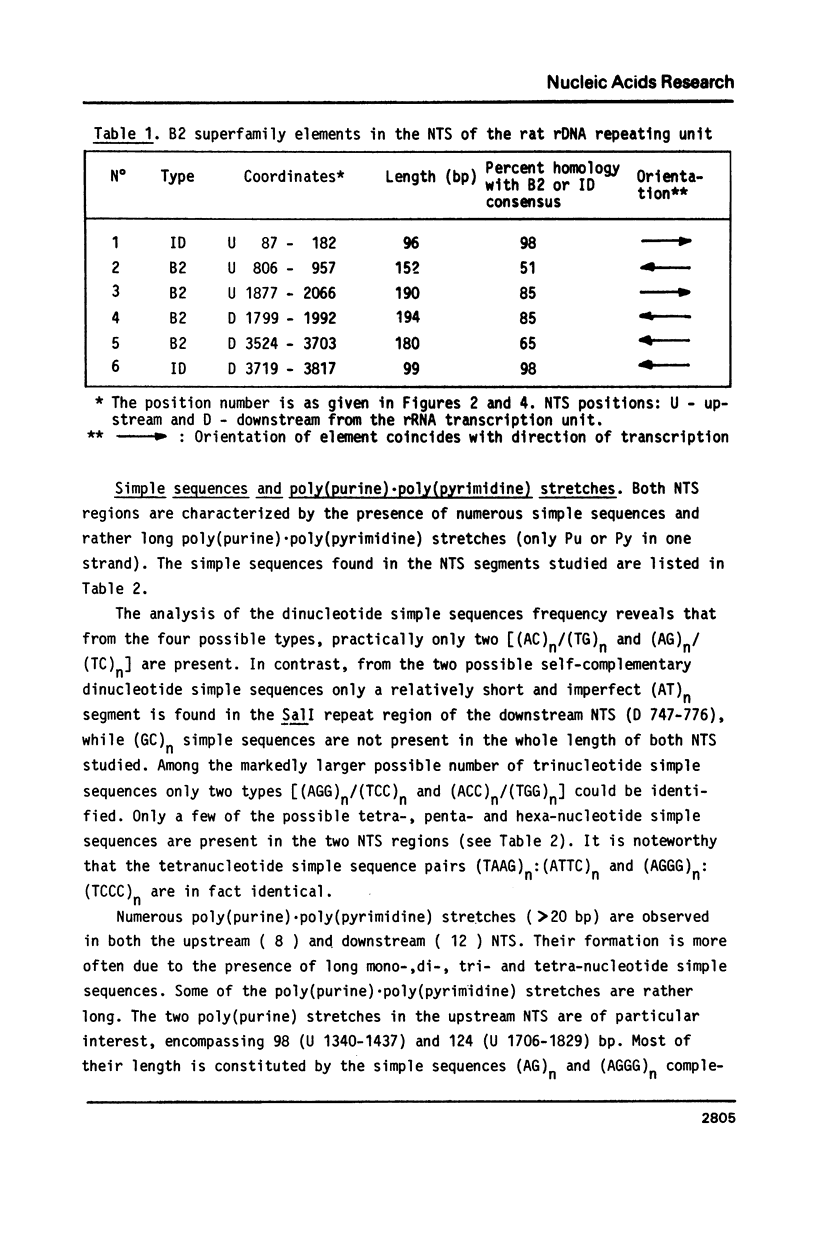
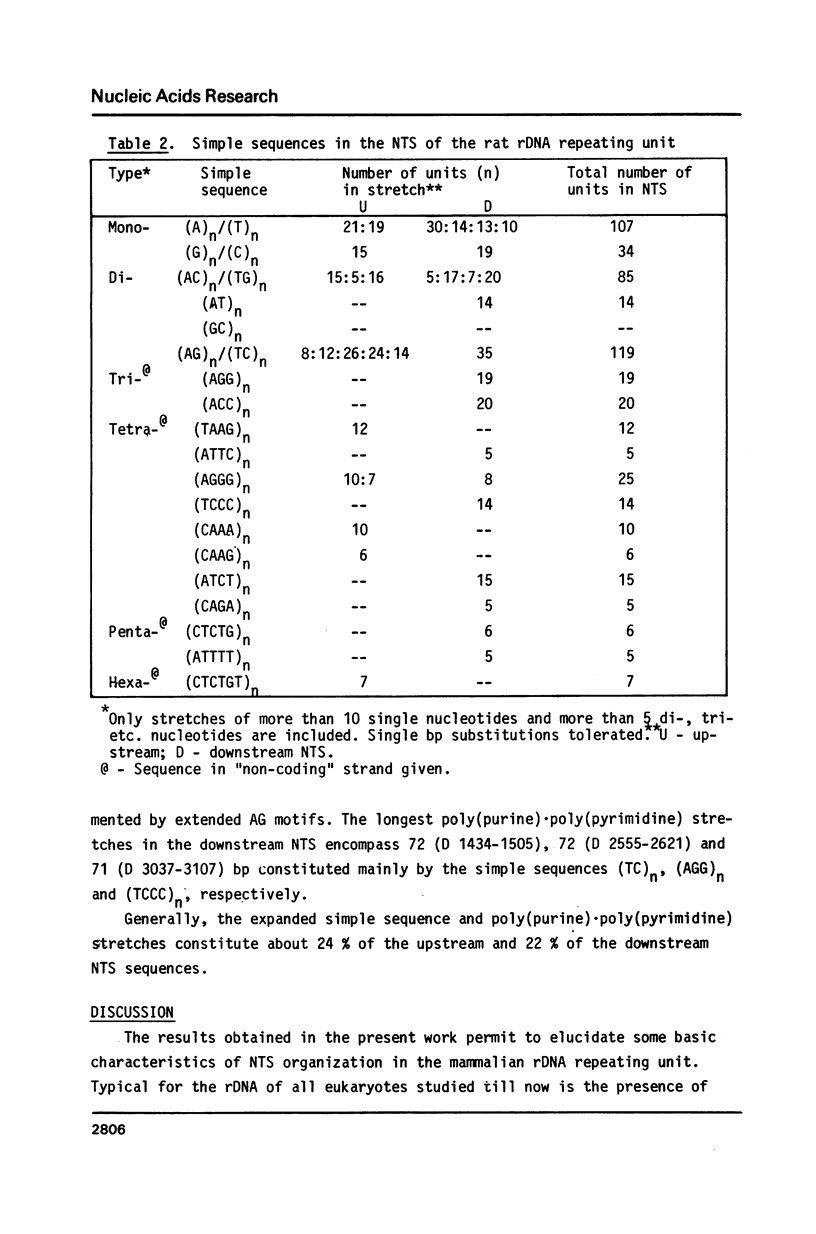
We investigated the organization of the rat rDNA non-transcribed spacer (NTS) by determining the sequence of large NTS segments located upstream (2501 bp) and downstream (4025 bp) from the rRNA transcription unit. We identified four B2-like and two ID mobile elements. They may be grouped in three pairs with the members of each pair located in the upstream and downstream NTS. The ID sequences are identical to the consensus sequence, while the pairs of B2-like elements show 85% and 50/65% homology to the consensus B2 sequence. The proximal part of the downstream NTS contains a region of widely diverged SalI tandem repeats. A considerable part of the analyzed upstream and downstream NTS sequences is constituted by different types of simple sequences and long poly(purine) X poly(pyrimidine) tracts. These data show that the rat rDNA NTS regions flanking the rRNA transcription unit are characterized by a combination of short interspersed (B2-superfamily) and various simple sequences.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnheim N., Seperack P., Banerji J., Lang R. B., Miesfeld R., Marcu K. B. Mouse rDNA nontranscribed spacer sequences are found flanking immunoglobulin CH genes and elsewhere throughout the genome. Cell. 1980 Nov;22(1 Pt 1):179–185. doi: 10.1016/0092-8674(80)90166-x. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braga E. A., Avdonina T. A., Zhurkin V. B., Nosikov V. V. Structural organization of rat ribosomal RNA genes: interspersed sequences and their putative role in the alignment of nucleosomes. Gene. 1985;36(3):249–262. doi: 10.1016/0378-1119(85)90180-5. [DOI] [PubMed] [Google Scholar]
- Braga E. A., Yussifov T. N., Nosikov V. V. Structural organization of rat ribosomal genes restriction endonuclease analysis of genomic and cloned ribosomal DNAs. Gene. 1982 Dec;20(2):145–156. doi: 10.1016/0378-1119(82)90033-6. [DOI] [PubMed] [Google Scholar]
- Busby S. J., Reeder R. H. Spacer sequences regulate transcription of ribosomal gene plasmids injected into Xenopus embryos. Cell. 1983 Oct;34(3):989–996. doi: 10.1016/0092-8674(83)90556-1. [DOI] [PubMed] [Google Scholar]
- Chikaraishi D. M., Buchanan L., Danna K. J., Harrington C. A. Genomic organization of rat rDNA. Nucleic Acids Res. 1983 Sep 24;11(18):6437–6452. doi: 10.1093/nar/11.18.6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen E. S., Dover G. A. Multiple Pol I initiation sequences in rDNA spacers of Drosophila melanogaster. Nucleic Acids Res. 1982 Nov 11;10(21):7017–7026. doi: 10.1093/nar/10.21.7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion E. A., Warner J. R. The major promoter element of rRNA transcription in yeast lies 2 kb upstream. Cell. 1984 Dec;39(3 Pt 2):663–673. doi: 10.1016/0092-8674(84)90473-2. [DOI] [PubMed] [Google Scholar]
- Hadjiolov A. A., Georgiev O. I., Nosikov V. V., Yavachev L. P. Primary and secondary structure of rat 28 S ribosomal RNA. Nucleic Acids Res. 1984 Apr 25;12(8):3677–3693. doi: 10.1093/nar/12.8.3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington C. A., Chikaraishi D. M. Identification and sequence of the initiation site for rat 45S ribosomal RNA synthesis. Nucleic Acids Res. 1983 May 25;11(10):3317–3332. doi: 10.1093/nar/11.10.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi R., Stang H. D., Browne J. K., Martin M. O., Huot M., Lipeles J., Salser W. Human ribosomal RNA gene spacer sequences are found interspersed elsewhere in the genome. Gene. 1981 Nov;15(2-3):177–186. doi: 10.1016/0378-1119(81)90127-x. [DOI] [PubMed] [Google Scholar]
- Kohorn B. D., Rae P. M. Nontranscribed spacer sequences promote in vitro transcription of Drosophila ribosomal DNA. Nucleic Acids Res. 1982 Nov 11;10(21):6879–6886. doi: 10.1093/nar/10.21.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kominami R., Urano Y., Mishima Y., Muramatsu M., Moriwaki K., Yoshikura H. Novel repetitive sequence families showing size and frequency polymorphism in the genomes of mice. J Mol Biol. 1983 Apr 5;165(2):209–228. doi: 10.1016/s0022-2836(83)80254-x. [DOI] [PubMed] [Google Scholar]
- Kominami R., Urano Y., Mishima Y., Muramatsu M. Organization of ribosomal RNA gene repeats of the mouse. Nucleic Acids Res. 1981 Jul 24;9(14):3219–3233. doi: 10.1093/nar/9.14.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krayev A. S., Markusheva T. V., Kramerov D. A., Ryskov A. P., Skryabin K. G., Bayev A. A., Georgiev G. P. Ubiquitous transposon-like repeats B1 and B2 of the mouse genome: B2 sequencing. Nucleic Acids Res. 1982 Dec 11;10(23):7461–7475. doi: 10.1093/nar/10.23.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn M., Arnheim N. Nucleotide sequence of the genetically labile repeated elements 5' to the origin of mouse rRNA transcription. Nucleic Acids Res. 1983 Jan 11;11(1):211–224. doi: 10.1093/nar/11.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Volpe A., Simeone A., D'Esposito M., Scotto L., Fidanza V., de Falco A., Boncinelli E. Molecular analysis of the heterogeneity region of the human ribosomal spacer. J Mol Biol. 1985 May 25;183(2):213–223. doi: 10.1016/0022-2836(85)90214-1. [DOI] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. Enhancer-like properties of the 60/81 bp elements in the ribosomal gene spacer of Xenopus laevis. Cell. 1984 May;37(1):285–289. doi: 10.1016/0092-8674(84)90324-6. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Milner R. J., Bloom F. E., Lai C., Lerner R. A., Sutcliffe J. G. Brain-specific genes have identifier sequences in their introns. Proc Natl Acad Sci U S A. 1984 Feb;81(3):713–717. doi: 10.1073/pnas.81.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T. A transcriptional function for the repetitive ribosomal spacer in Xenopus laevis. Nature. 1983 Mar 17;302(5905):223–228. doi: 10.1038/302223a0. [DOI] [PubMed] [Google Scholar]
- Moss T., Boseley P. G., Birnstiel M. L. More ribosomal spacer sequences from Xenopus laevis. Nucleic Acids Res. 1980 Feb 11;8(3):467–485. doi: 10.1093/nar/8.3.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mroczka D. L., Cassidy B., Busch H., Rothblum L. I. Characterization of rat ribosomal DNA. The highly repetitive sequences that flank the ribosomal RNA transcription unit are homologous and contain RNA polymerase III transcription initiation sites. J Mol Biol. 1984 Mar 25;174(1):141–162. doi: 10.1016/0022-2836(84)90369-3. [DOI] [PubMed] [Google Scholar]
- Nordheim A., Rich A. Negatively supercoiled simian virus 40 DNA contains Z-DNA segments within transcriptional enhancer sequences. Nature. 1983 Jun 23;303(5919):674–679. doi: 10.1038/303674a0. [DOI] [PubMed] [Google Scholar]
- Page G. S., Smith S., Goodman H. M. DNA sequence of the rat growth hormone gene: location of the 5' terminus of the growth hormone mRNA and identification of an internal transposon-like element. Nucleic Acids Res. 1981 May 11;9(9):2087–2104. doi: 10.1093/nar/9.9.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy R., Busch H. Small nuclear RNAs and RNA processing. Prog Nucleic Acid Res Mol Biol. 1983;30:127–162. doi: 10.1016/s0079-6603(08)60685-6. [DOI] [PubMed] [Google Scholar]
- Reeder R. H. Enhancers and ribosomal gene spacers. Cell. 1984 Sep;38(2):349–351. doi: 10.1016/0092-8674(84)90489-6. [DOI] [PubMed] [Google Scholar]
- Reeder R. H., Roan J. G., Dunaway M. Spacer regulation of Xenopus ribosomal gene transcription: competition in oocytes. Cell. 1983 Dec;35(2 Pt 1):449–456. doi: 10.1016/0092-8674(83)90178-2. [DOI] [PubMed] [Google Scholar]
- Rothblum L. I., Parker D. L., Cassidy B. Isolation and characterization of rat ribosomal DNA clones. Gene. 1982 Jan;17(1):75–77. doi: 10.1016/0378-1119(82)90102-0. [DOI] [PubMed] [Google Scholar]
- Rothblum L. I., Reddy R., Cassidy B. Transcription initiation site of rat ribosomal DNA. Nucleic Acids Res. 1982 Nov 25;10(22):7345–7362. doi: 10.1093/nar/10.22.7345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skryabin K. G., Eldarov M. A., Larionov V. L., Bayev A. A., Klootwijk J., de Regt V. C., Veldman G. M., Planta R. J., Georgiev O. I., Hadjiolov A. A. Structure and function of the nontranscribed spacer regions of yeast rDNA. Nucleic Acids Res. 1984 Mar 26;12(6):2955–2968. doi: 10.1093/nar/12.6.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. R., Bolla R. I., Rumbyrt J. S., Schlessinger D. DNase I-resistant nontranscribed spacer segments of mouse ribosomal DNA contain poly(dG-dT).poly(dA-dC). Proc Natl Acad Sci U S A. 1985 Nov;82(22):7595–7598. doi: 10.1073/pnas.82.22.7595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walseth T. F., Johnson R. A. The enzymatic preparation of [alpha-(32)P]nucleoside triphosphates, cyclic [32P] AMP, and cyclic [32P] GMP. Biochim Biophys Acta. 1979 Mar 28;562(1):11–31. doi: 10.1016/0005-2787(79)90122-9. [DOI] [PubMed] [Google Scholar]



