The discovery of the Archaea (1), the third domain of life, has reinvigorated efforts to understand the phylogenetic relationships among all forms of life. It is therefore not surprising that the recent completion of four archaeal genomes (2–5) created a flurry of excitement among biologists (Genetics, Vol. 152). A comparison of genome sequences reveals that the prokaryotes, Archaea and Bacteria, have closer similarities in their metabolic processes, but Archaea and Eukarya have more similarities in their information processing machineries (DNA replication, transcription, and translation) (6, 7). If this initial observation were supported by further biochemical analyses, then it would suggest that Archaea and Bacteria diverged before the evolution of mechanisms that substantially increased the fidelity of information transfer. That pressure for accuracy may have increased as genomes became more and more complex. Furthermore, it would suggest Archaea and Eukarya share a common ancestor more recently than either shares with Bacteria (Fig. 1). This observation has drawn intense interests from biochemists who have long been battling with the complexities of eukaryal biology. If Archaea embody a more primitive form of the highly evolved Eukarya, then simplified models for complex systems of Eukarya may be found in Archaea. Indeed, a composite of the putative replication proteins identified from four archaeal genomes supports this view (Table 1) (7). This rationale provided the motivation for the biochemical characterization of the Methanobacterium thermoautotrophicum MCM protein.
Figure 1.
The three domains of life.
Table 1.
Replication proteins of Eukarya, Bacteria, and Archaea
| Function | Archaeal | Eukaryal | Bacterial |
|---|---|---|---|
| Origin recognition | Orc1/Cdc6-like proteins | Origin recognition complex (ORC) six proteins | DnaA |
| Helicase | Dna2-like, MCM-like | Dna2, MCMs-six proteins | DnaB |
| Single-stranded DNA binding | RPA-like protein | Replication protein A (RPA, three subunits) | SSB |
| Primer synthesis | Eukaryotic-like primase | DNA Polα | DnaG |
| Clamp loader | RFC-like proteins (small, large) | RFC | γ-complex |
| Clamp (elongation factor) | PCNA-like proteins | PCNA | Pol III β subunit |
| DNA strand synthesis | Family B DNA Pol | DNA Pol α, δ, ɛ | Pol III |
| Family D DNA Pol | |||
| DNA strand ligation (on lagging strand) | DNA ligase (ATP-dependent) | DNA ligase (ATP-dependent) | DNA ligase (NAD-dependent) |
| Removal of primers | FEN1, RnaseH | FEN1, RnaseH | Pol I, Rnase H |
Adapted from ref. 7.
MthMCM shows similarities to a family of six related minichromosome maintenance proteins, Mcm2-Mcm7, that are conserved from yeast to human and are known for their central role in the highly regulated replication initiation process (8). These six proteins have been postulated to form a hexameric ring complex (MCM complex) that serves as the DNA helicase to unwind DNA at replication origins as well as at replication forks. However, although inferences for biochemical activities for the eukaryal MCMs are strong, evidence for such activities has been weak. In two recent issues of PNAS, two groups (9, 10) independently reported the exciting finding that purified recombinant MthMCM protein forms a double hexamer that acts as a 3′-5′ DNA helicase as postulated for the eukaryal MCM proteins.
The genomes of eukaryotes and prokaryotes differ in size and organization. The generally larger genomes of eukaryotes are organized into multiple chromosomes, and initiation of DNA synthesis starts from not one but numerous sites on each chromosome. Compounding the complexity of this replication process is the necessity to coordinate these multiple initiation events such that on completion, the entire genome is duplicated exactly once every cell cycle. The strategy for this regulated process is beginning to emerge from the collective works of many laboratories. Key to this strategy is the periodic recruitment and discharge of the MCM complex at replication origins to forge a cycle of activity and inactivity at replication origins (8). Although this concept of the temporal separation of an active and inactive chromatin state at replication origins involving the MCM complex is relatively simple, the number of proteins involved to achieve this goal suggests a complex and intricate scheme. Chief among these proteins is the origin recognition complex (ORC), a complex of six nonidentical subunits, Orc1-Orc6, which binds an essential element at replication origins to act as a “bookmark” for these sites (11). During the G1 phase, the MCM complex is delivered to replication origins with the aid of Cdc6 (12, 13), a short-lived protein that also has some sequence similarities to Orc1. Initiation of DNA synthesis occurs after a rapid succession of events involving additional proteins (including Cdc45) cued by the cell cycle-dependent protein kinases (Cdc7-Dbf4 and Cdc28-Clb) resulting in the melting of DNA and the recruitment of elongation machineries to replication origins (14, 15). As replication transitions from initiation to elongation, the MCM complex is believed to change its alliance with the initiation complex to the elongation complex, where it acts as the processive helicase of the growing fork (16), much like the large T antigen of simian virus SV40 (17). The MCM-vacated replication origins now assume an inactive state that no longer supports the initiation of DNA synthesis. This sequence of events is not to repeat again until the next G1 phase.
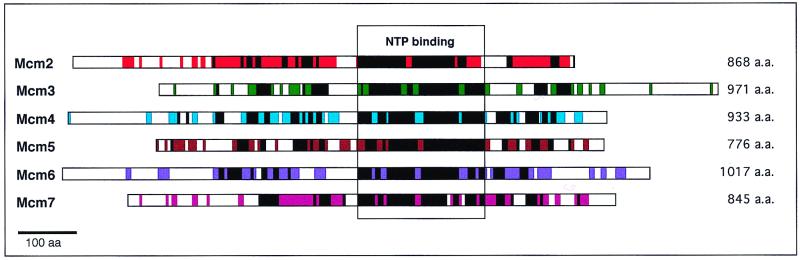
For the purpose of this commentary, the strategy of the eukaryal cell used to restrict DNA replication to one round per cell cycle can be simply viewed as a concerted effort to regulate the many activities of the MCM complex. Although this concept has gained popularity as a working hypothesis for those who study the MCM proteins, much of the information that generated this picture was derived from genetic or in vivo experiments without the support of biochemical evidence. Because of the number of protein factors involved, the prospect to study the activities of the MCM proteins in the context of the many accessory proteins in an in vitro assay for replication initiation is daunting. The existence of a single MCM homolog in M. thermoautotrophicum in contrast to the six MCM proteins found in all eukaryotes is welcome news. The six eukaryal MCM proteins, ranging in size from 776 to 1,017 residues, contain three highly conserved regions, including a NTP binding motif (Fig. 2). The MthMCM protein, although slightly smaller (667 amino acids), is also conserved in those regions (Fig. 2). All six MCM proteins are known to be essential for the initiation of DNA synthesis and can be purified in a hexameric complex that has a globular structure (18). However, to date, biochemical activities associated with this hexameric complex or any one of the six proteins have yet to be reported. The only report of enzymatic activities associated with the MCM proteins is a complex that contains three of the six MCMs (Mcm4, Mcm6, and Mcm7) formed during the course of purification of the larger complex in HeLa cells (19, 20). Weak helicase, ATPase, and single-strand DNA binding activities were found associated with this complex. These results suggest that the MCM proteins have an intrinsic helicase activity that can only be activated when assembled in a specific conformation. The search for the physiologically active form of the MCM complex becomes the key to understanding the regulation of eukaryote DNA replication (8).
Figure 2.
Sequence conservation among Saccharomyces cerevisiae MCM proteins. Black bars represent regions conserved between S. cerevisiae MCMs and the single MCM protein of M. thermoautotrophicum, and colored bars represent regions conserved between yeast and mammalian MCMs of the same class. The largest conserved domain contains the nucleotide binding motif. [Reproduced with permission from Kearsey and Labib (27) (Copyright 1998, Elsevier).]
In the papers reported recently in PNAS, two groups purified and characterized the recombinant MthMCM protein and obtained similar results (9, 10). In a purified preparation of MthMCM protein, ≈80% of the protein assembled in an oligomeric complex of about 850–950 kDa, corresponding to a dodecamer, and ≈20% existed as a monomer of about 75 kDa. The dodecameric complex examined by scanning transmission electron microscopy appears to form a double hexamer with a ring structure (10) characteristic of proteins or enzymes that travel large distances along DNA such as the processivity factors (21) and helicases (22). When assayed for helicase and helicase associated activities, both forms exhibited single-strand DNA binding, ssDNA-stimulated ATPase, and ATP-dependent 3′-5′ helicase activities. The double hexamer was further tested for processivity of its helicase activity and was shown to be able to unwind DNA of up to 500 base pairs (10). Mutational analysis indicated that the N-terminal 110 amino acids as well as the putative Walker A motif of the conserved NTP binding site were dispensable for the ssDNA binding activity but essential for the ATPase as well as the helicase activities. The robust helicase activity is consistent with a role for the MCM complex in replication elongation as well as initiation as postulated for the eukaryal MCM complex.
Studying DNA replication in Archaea, the third domain of life, is important and interesting both in its own right and from an evolutionary standpoint. A major difference between Bacteria and Eukarya in replicating their genomes is the use of a single versus multiple replication origins. Where is Archaea in this evolutionary process? Is the evolution of a more complex eukaryal helicase containing six different MCMs a consequence of the expansion of genome sizes as well as the number of replication origins? The revelation that Archaea may have many of the basic components (Table 1) of the replication initiation machinery of Eukarya offers new opportunities for unraveling the complexities of eukaryal systems not offered by viral models such as the SV40 (17). The viral models provide their own initiator proteins that usurp the host replication elongation machineries and therefore are useful for studying replication elongation as well as understanding the principles, although not the mechanistics, of eukaryal replication initiation.
The study of the MthMCM suggests that Achaea as a model may provide useful information at two levels. The self-assembly of a single MthMCM protein into an active double hexamer is a dramatic simplification of the Eukaryal MCM studies. It allows one to explore the complete repertoire of enzymatic activities associated with the MCM complex in isolation. Furthermore, setting aside the intricacies of assembling six MCMs into an active complex, it allows one to focus on the interaction of this complex with other components of the replication initiation complex, such as ORC, Cdc6, and RPA. Indeed, the existence of two Cdc6/Orc1-like proteins in M. thermoautotrophicus instead of the six ORC subunits suggests significant simplification in the origin recognition function and bodes well for an in vitro system that is likely to have some resemblance to the eukaryal system.
There are also some important omissions in the Mt genome that suggest major differences between Eukarya and Archaea. Cdc45, an important factor in the transition of the initiation complex to elongation complex, and Cdc7-Dbf4 and Cdc28-Clb, the cell cycle-dependent protein kinases, all appear to be absent in the Mt genome. These omissions suggest a less elaborate regulatory mechanism for replication initiation in Archaea, although the existence of functional Mt homologs cannot be ruled out. Clearly, much work is needed for studies in both the archaeal and the eukaryal systems to begin to unravel the complexities of the regulation of DNA replication of eukaryotes that is fundamental to our understanding of the control of cell proliferation. It is worth mentioning here that because expression of the MCMs is tightly coupled to the regulation of cell proliferation (23–25), this property of the MCMs is currently being exploited as a diagnostic marker for neoplasticity in precancerous cells (26). It is therefore not a stretch of the imagination to look for answers in the ancient microbe for cures of human diseases. The reports on the MthMCM underscore the value of studying fundamental biological processes in diverse organisms and the prospect that these studies will reveal the underlying principles of life.
Acknowledgments
I thank Chip Aquadro, Jeff Roberts, and Sara Sawyer for critical reading of this commentary. I thank Tom Fox for discussions. Research from this laboratory is supported by a grant (GM34190) from the National Institutes of Health.
Footnotes
References
- 1.Woese C R, Fox G E. Proc Natl Acad Sci USA. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bult C J, White O, Olsen G J, Zhou L X, Fleischmann R D, Sutton G G, Blake J A, Fitzgerald L M, Clayton R A, Gocayne J D, et al. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 3.Klenk H P, Clayton R A, Tomb J F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, et al. Nature (London) 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 4.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H M, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, et al. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawarabayasi Y, Sawada M, Horikawa H, Kaikawa Y, Hino Y. DNA Res. 1998;5:55–76. doi: 10.1093/dnares/5.2.55. [DOI] [PubMed] [Google Scholar]
- 6.Whitman W B, Pfeifer F, Blum P, Klein A. Genetics. 1999;152:1245–1248. doi: 10.1093/genetics/152.4.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caan I K O, Ishino Y. Genetics. 1999;152:1249–1267. doi: 10.1093/genetics/152.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tye B K. Annu Rev Biochem. 1999;68:649–686. doi: 10.1146/annurev.biochem.68.1.649. [DOI] [PubMed] [Google Scholar]
- 9.Kelman Z, Lee J-K, Hurwitz J. Proc Natl Acad Sci USA. 1999;96:14783–14788. doi: 10.1073/pnas.96.26.14783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chong J, Hayashi M K, Simon M N, Xu R-M, Stillman B. Proc Natl Acad Sci USA. 2000;97:1530–1535. doi: 10.1073/pnas.030539597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bell S, Stillman B. Nature (London) 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 12.Cocker J H, Piatti S, Santocanale C, Nasmyth K, Diffley J F X. Nature (London) 1996;379:180–182. doi: 10.1038/379180a0. [DOI] [PubMed] [Google Scholar]
- 13.Coleman T R, Carpenter P B, Dunphy W G. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- 14.Lei M, Kawasaki Y, Young M R, Kihara M, Sugino A, Tye B K. Genes Dev. 1997;11:3365–3374. doi: 10.1101/gad.11.24.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zou L, Stillman B. Science. 1998;280:593–596. doi: 10.1126/science.280.5363.593. [DOI] [PubMed] [Google Scholar]
- 16.Aparicio O M, Weinstein D M, Bell S P. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 17.Li J J, Kelly T J. Proc Natl Acad Sci USA. 1984;81:6973–6977. doi: 10.1073/pnas.81.22.6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adachi Y, Usukura J, Yanagida M. Genes Cells. 1997;2:467–479. doi: 10.1046/j.1365-2443.1997.1350333.x. [DOI] [PubMed] [Google Scholar]
- 19.Ishimi Y. J Biol Chem. 1997;272:24508–24513. doi: 10.1074/jbc.272.39.24508. [DOI] [PubMed] [Google Scholar]
- 20.You Z, Komamura Y, Ishimi Y. Mol Cell Biol. 1999;19:8003–8015. doi: 10.1128/mcb.19.12.8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong X-P, Onrust R, O'Donnell M, Kuriyan J. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- 22.Yu X, Egelman E H. Nat Struct Biol. 1997;4:101–104. doi: 10.1038/nsb0297-101. [DOI] [PubMed] [Google Scholar]
- 23.Springer P S, McCombe W R, Sundaresan V, Martienssen R A. Science. 1995;268:877–880. doi: 10.1126/science.7754372. [DOI] [PubMed] [Google Scholar]
- 24.Treisman J E, Follette P J, O'Farrell P H, Rubin G M. Genes Dev. 1995;9:1709–1715. doi: 10.1101/gad.9.14.1709. [DOI] [PubMed] [Google Scholar]
- 25.Young M, Tye B K. Mol Biol Cell. 1997;8:1587–1601. doi: 10.1091/mbc.8.8.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams G H, Romanowski P, Morris L, Madine M, Mills A D, Stoeber K, Marr J, Laskey R A, Coleman N. Proc Natl Acad Sci USA. 1999;95:14932–14937. doi: 10.1073/pnas.95.25.14932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kearsey S E, Labib K. Biochim Biophys Acta. 1998;1398:113–136. doi: 10.1016/s0167-4781(98)00033-5. [DOI] [PubMed] [Google Scholar]