Abstract
Eight intestinal cell lines, established from different animal species were submitted to DSMZ (German Collection of Microorganisms and Cell Cultures) in order to analyze their species of origin and their microbial contamination. Species identity was determined by PCR targeting mitochondrial genes and hence confirmed by sequencing the amplified PCR products. For three cell lines (CIEB, CLAB, PSI-1) we confirmed the species identity, whereas the species of origin of the three other cell lines (B6, B10XI and IPEC) was not the expected one: B6 and B10XI cells, which were supposed to be of chicken origin were identified as porcine cells. IPEC, allegedly a sub clone of the well-known porcine intestinal cell line IPEC-J2, was of bovine instead of porcine origin. However, two further IPEC-clones, namely IPEC-1 and IPEC-J2, provided by another source were shown to be derived from the correct species (i.e. pig). Furthermore, six out of these eight cell lines turned out to be highly contaminated with mycoplasma. Alerted by this high incidence of infected and false specified cell lines, we feel obliged to inform all those working with animal intestinal cell lines and we strongly recommend verifying the species identity before using them. Also, the presence of mycoplasma should be tested when taking the cells in culture for the first time, and this mycoplasma control should be repeated at regular time intervals (e.g. every 4 weeks).
Keywords: Cell Lines, Identity control of cell lines, Quality control of cell lines, Species-PCR, Speciation, Mycoplasma Infection, Intestinal cell lines, IPEC-cells
Introduction
Continuous cell lines represent widely used models for a large number of life science research fields. Two basic requirements for the use of continuous cell lines in fundamental and applied research are their unequivocal identity and absence of contamination with microorganisms such as mycoplasma. Therefore, special emphasis must be taken (1) to detect and to prevent a microbial contamination and (2) to regularly monitor the identity of cell line(s) in use. The importance of quality control as an essential part of good laboratory practice has been continuously addressed (Nelson-Rees et al. 1981; Stevenson 1987; Zoon 1993; MacLeod et al. 2002; Stacey et al. 2000; Masters 2002). Moreover, the recurrent reports of cross-contaminated and misidentified cell lines, mostly of human origin, underline the importance and necessity of identity testing (MacLeod et al., 1999; Drexler et al. 2003; Nardonne 2007; Lacroix 2008; Capes-Davis et al. 2010; Allston-Roberts et al. 2010).
In principle, cell lines can either be transformed or non-transformed, whereby the number of non-transformed, non-carcinogenic cell lines is widely underrepresented compared to tumor-derived ones. However, in the last few years a number of non-transformed human and animal cell lines have been proposed as model systems to study diverse aspects of intestinal physiology, toxicology and microbiology (Chantret et al. 1988).
Several animal intestinal cell lines are available from the investigating researchers and/or from public or commercial cell banks. Among these, especially two non-transformed animal cell lines, IPEC-1 and IPEC-J2, have been proven as valuable models studying gastrointestinal morphology and function (Gonzalez-Vallina et al. 1996; Schierack et al. 2006; Diesing et al. 2011; Nossol et al. 2011). Only very recently a functional and ultra structural characterization of IPEC-J2 was performed by Geens and Niewold (2011, this journal) emphasizing IPEC-J2 as a valuable in vitro model system for the intestinal epithelium.
In the present study the species of origin as well as the mycoplasma status of several animal intestinal cell lines from different laboratories were analyzed. The results show that it is imperative to determine the identity and the quality of intestinal cell lines of animal origin before the planned experiments are performed.
Materials and methods
Cell lines and culture conditions
CIEB, CLAB, IPEC, PSI-1, B6 and B10XI cells were first submitted from a third party to the Institute for Food Toxicology and Analytical Chemistry of the University of Veterinary Medicine (Hannover, Germany) and subsequently cultivated there, whereas IPEC-1 and IPEC-J2 were deposited at the DSMZ cell line bank by Prof. Dr. Rothkötter, University of Magdeburg, (Germany). All cell lines were propagated in DMEM medium (high glucose) supplemented with 10–20% FBS, non-essential amino acids and in case of B6 and B10XI additionally with 140 μg/mL pituitary extract (Sigma-Aldrich, Steinheim, Germany).
Other cell lines, serving as standards (Table 1), were taken from the stock of DSMZ. Growth conditions and cell line characteristics of accessioned cell lines are described in the DSMZ Catalogue of Human and Animal Cell Lines (http://www.dsmz.de). Two samples of purified reference DNA from horse and donkey were obtained from Cibus Biotech (Rheda-Wiedenbrück, Germany).
Table 1.
Cell lines serving as controls
| Cell line | Origin | Speciesa |
|---|---|---|
| BHK-21 | Kidney | Syrian hamster |
| DT-40 | Blood | Chicken |
| EBL | Lung | Bovine |
| FLK-BLV-044 | Kidney | Ovine |
| GM-7373 | Aorta | Bovine |
| HD-11b | Blood | Chicken |
| HeLa | Cervix | Human |
| LAT | Aorta | Ovine |
| LLC-PK-1 | Kidney | Pig |
| LMHb | Liver | Chicken |
| MDBK | Kidney | Bovine |
| MDCKb | Kidney | Dog |
| NIH-3T3 | Embryo | Mouse |
| PK-15 | Kidney | Pig |
| PC-12 | Adrenal gland | Rat |
aIdentified by Species-PCR (animals) or DNA fingerprinting (human)
bNot available from the DSMZ cell bank; submitted to DSMZ only for internal use
PCR analysis and primer pairs
In order to minimize the risk of false PCR amplification, cell culture, DNA isolation, preparation of the reaction mix and final PCR were performed in different laboratories. PCR amplification and electrophoretic analysis of the products were described recently (Steube et al. 2003, 2008). Genomic sequencing of the PCR products was conducted by Eurofins MWG-Operon, Ebersberg, Germany. Oligonucleotide primers were obtained from Invitrogen, Darmstadt, Germany. Table 2 lists the applied primers for the identification of the animal species. Primers as well as method used for the mycoplasma detection were described earlier by Uphoff and Drexler (2011a).
Table 2.
Primer pairs used in the Species-PCR
| Primer name | 5′ to 3′ sequence | Species | Target gene | Gene bank no. | Temperature (°C) |
|---|---|---|---|---|---|
| Single and Duplex-PCR | |||||
| Mito-Bov F1 | gccatatactctccttggtgac | Bovine | ATPase 8/6 | NC_006853 | 56 |
| Mito Bov R1 | gtaggcttgggaatagtacgat | Bovine | ATPase 8/6 | NC_006853 | 56 |
| Mito Sus F | ctaaatctcccctcaatggtatg | Pig | ATPase 8/6 | AF034253 | 57 |
| Mito Sus R | gaatcctgtgaatacggttgc | Pig | ATPase 8/6 | AF034253 | 57 |
| Mito-Chk F2 | cgagtaatcatcaccgctgatga | Chicken | COII + ATPase 8 | AP003580 | 60 |
| Mito-Chk R2 | gcttaggttcatggtcaggttca | Chicken | COII + ATPase 8 | AP003580 | 60 |
| Mito-Doga | gaactaggtcagcccggtactt | Dog | COI | AY729880 | 60 |
| Mito-Dog | cggagcaccaattattaacggc | Dog | COI | AY729880 | 60 |
| Multiplex-PCR | |||||
| Sus ND F2 | ctgctaattggatgatgacacg | Pig | NADH5 | AF034253 | 58 |
| Sus ND R2 | ttcctgttaatgccaggcttc | Pig | NADH5 | AF034253 | 58 |
| Mito-Bov F2 | atcgtactattcccaagcctac | Bovine | ATPase 6 | NC_006853 | 58 |
| Mito Bov R3 | ctgttaaccgcacggcga | Bovine | ATPase 6 | NC_006853 | 58 |
| Mito OV F | cgatacacgggcttacttcacg | Sheep | COI | NC_001941 | 58 |
| Mito Ov R | aaatacagctcctattgataat | Sheep | COI | AF010406 | 58 |
| Mito-Hrs Fa | ctgccctaagcctcctaat | Horse | COI | HM102300 | 58 |
| Mito-Hrs Ra | agaagtaggaatgatggggg | Horse | COI | HM102300 | 58 |
Column “target genes”, are the various mitochondrial genes analyzed: ATPase 8/6 ATPase subunit 8 and ATPase subunit 6, COII cytochrome oxidase subunit II, COI cytochrome C oxidase subunit I, NADH5 NADH Dehydrogenase subunit 5; the respective gene bank reference numbers are indicated
Last column “temperature” gives the annealing temperatures used in the individual PCRs
Mito mitochondrial (all sequences are from mitochondrial DNA) of the respective species given in the third column
aPrimer sequences were adapted from Cooper et al. 2007
Elimination of mycoplasma
Mycoplasma eradication was carried out by treatment with Baytril, Plasmocin and BM-Cyclin (in three independent subcultures) as previously described in detail (Uphoff and Drexler 2011b).
Results and discussion
The cell lines CLAB, PSI-1, IPEC (expected to be of porcine origin) and CIEB (expected to be of bovine origin) were sent from a third party to the University of Veterinary Medicine Hannover and subsequently cultivated there. All cultures exhibited a low viability and a very poor growth. Initial testing revealed that they were contaminated by mycoplasma and therefore were submitted to DSMZ for both elimination of the bacteria and evaluation of the species of origin. At the DSMZ mycoplasma infection was confirmed by PCR, speciation revealed Mycoplasma hyorhinis, and the eradication of the cultures was started.
In parallel several PCR analyses were performed to determine the species origin using the specific primers given in Table 2. As controls several well-characterized and speciated cell lines from the DSMZ cell bank were included in the assays (Table 1).
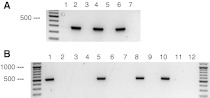
The species origin of cell lines CLAB, CIEB and PSI-1 turned out to be as expected (Fig. 1a, b). In contrast, IPEC, a cell line related to the porcine intestinal cell line IPEC-J2, (Berschneider 1989), revealed a strong amplification product when bovine primer pairs were used (Fig. 1a), but none when porcine primer pairs were applied (Fig. 1b), strongly suggesting a false-specified origin (bovine instead of porcine). These results were again verified, applying different sets of porcine and bovine primer pairs.
Fig. 1.
Analysis of genomic DNA from different cell lines using bovine (a) or porcine primer pairs (b). Amplified DNA fragments were detected after ethidium bromide staining of 1.2% agarose gels. DNA molecular weight markers of 500 bp and 1,000 bp are bold. a, primer pair: Mito-Bov. Lane 1: CLAB; lane 2: CIEB; lane 3: PSI-1; lane 4: IPEC; lane 5: PK-15 (pig, DSMZ ACC 640); lane 6: GM-7373 (bovine, DSMZ ACC 109); lane 7: water control (no DNA). Expected fragment size: 271 bp. b, primer pair: Mito-Sus. Lane 1: CLAB; lane 2: HELA (human, DSMZ ACC 57); lane 3: CIEB; lane 4: NIH-3T3 (mouse, DSMZ ACC 59); lane 5: PSI-1; lane 6: PC-12 (rat, DSMZ ACC 159); lane 7: IPEC; lane 8: PK-15 (pig, DSMZ ACC 640); lane 9: LAT (ovine, DSMZ ACC 349); lane 10: LLC-PK1 (pig, DSMZ ACC 637); lane 11: BHK-21 (hamster, DSMZ ACC 61); lane 12: water control (no DNA). Expected fragment size: 532 bp
To finally clarify whether these IPEC cells were indeed of porcine origin or an incorrect species at source, two additional IPEC-samples, obtained from another laboratory, were analyzed. These two samples, IPEC-1 and IPEC-J2, have been intensively characterized in various studies before (Mariani et al. 2009; Pinton et al. 2010; Nossol et al. 2011; Diesing et al. 2011).
As shown in Fig. 2, the two additional IPEC samples are indeed of porcine origin, this observation being in accordance with the results reported originally (Gonzalez-Vallina et al. 1996; Schierack et al. 2006) and supports the exclusion of a possible contamination during the course of the establishment of the original cell line IPEC-J2 (Berschneider 1989).
Fig. 2.
Analysis of genomic DNA from different cell lines using a duplex-PCR with dog and chicken primer pairs. Amplified DNA fragments were detected after ethidium bromide staining of 1.2% agarose gels. DNA molecular weight marker of 500 bp is bold. Lane 1: B6; lane 2: B10XI; lane 3: MDCK (dog); lane 4: HELA (human, DSMZ ACC 57); lane 5: MDCK (dog); lane 6: NIH-3T3 (mouse, DSMZ ACC 59); lane 7: DT-40 (chicken, DSMZ ACC 636); lane 8: MDBK (bovine, DSMZ ACC 174); lane 9: HD-11 (chicken); lane 10: EBL (bovine, DSMZ ACC 192); lane 11: LMH-2 (chicken); lane 12: water (no DNA). Expected fragment size for dog: 154 bp and chicken: 474 bp
Amplification products of the cell lines IPEC-J2 and IPEC-1, as well as IPEC and CIEB and from the control cell lines PK-15 (porcine) and EBL (bovine, both DSMZ cell bank) obtained with bovine and porcine primers were purified and submitted to sequencing. The provided sequences were blasted and found to be 99% identical to published sequences for bovine mitochondrion in the case of EBL, CIEB, and IPEC and for porcine mitochondrion in case of PK-15, IPEC-J2 and IPEC-1 cells, thereby again confirming that the species origin of IPEC was wrong.
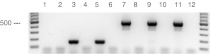
Additionally, two other cell lines, B6 and B10XI, supposed to be derived from chicken intestine were analyzed. These two cell lines were also found to be heavily contaminated with M. hyorhinis and again, the reputed species origin (chicken) could not be confirmed. No amplification products were detected after PCR analysis using chicken and dog (as a control) primer pairs (Fig. 2), whereas subsequent assays clearly revealed porcine signals for B6 and B10XI cells.
To summarize, a multiplex-PCR with bovine, ovine, porcine and horse primer pairs clearly confirmed once again that the species of origin of IPEC, B6 and B10XI cells (Fig. 3) was not the expected one (see also summary of the results in Table 3). Since the well-characterized IPEC-1 and IPEC-J2 cells, obtained from a different provider, were shown to be derived from the correct species, it can be deduced that the first examined IPEC sample, was mixed up or contaminated at a later stage of cultivation but not during original establishment of the IPEC-cell lines published by Berschneider (1989) and Gonzalez-Vallina et al. (1996).
Fig. 3.
Analysis of genomic DNA from different cell lines using a multiplex-PCR with bovine, horse, ovine and porcine primer pairs. Amplified DNA fragments were detected after ethidium bromide staining of 1.2% agarose gels. DNA molecular weight markers of 500 and 1,000 bp are bold. Lane 1: IPEC-J2; lane 2: IPEC; lane 3: LAT (ovine, DSMZ ACC 349); lane 4: reference DNA horse; lane 5: IPEC-1; lane 6: CIEB (bovine); lane 7: FLK-BLV-044 (ovine, DSMZ ACC 153); lane 8: reference DNA donkey; lane 9: DT-40 (chicken, DSMZ ACC 636); lane 10: B6; lane 11: B10XI; lane 12: water (no DNA). Expected fragment sizes: porcine, 691 bp; bovine 415 bp; ovine 300 bp; horsel/donkey, 244 bp
Table 3.
Summary of identity and quality testing results
| Cell linea | Species expected | Species identified | Mycoplasma infection |
|---|---|---|---|
| B6 | Chicken | Pig | Yes |
| B10XI | Chicken | Pig | Yes |
| CIEB | Bovine | Bovine | Yes |
| CLAB | Pig | Pig | Yes |
| IPEC | Pig | Bovine | Yes |
| IPEC-1a | Pig | Pig | No |
| IPEC-J2a | Pig | Pig | No |
| PSI-1 | Pig | Pig | Yes |
aCorrectly specified and mycoplama-free samples from the University of Magdeburg, Germany
Although it was not possible to obtain the cell lines B6 and B10XI from an alternative provider, a similar conclusion may be drawn for these two cell lines.
During the last years scientists at the DSMZ and other institutions have established easy-to-use methods for the analyses of animal cell lines (Stacey et al. 1997; Liu et al. 2003; Cooper et al. 2007; Ono et al. 2007; Steube et al. 2008). However, many cell lines, especially animal cell lines may be shared between laboratories without further testing and the laboratories in which cell lines were established are often reluctant to deposit the cell lines in international cell repositories for quality assessments and further distribution (MacLeod and Drexler 2006). Despite repeated warnings there is still a high number of false or contaminated cell lines in use (Capes-Davis et al. 2010; Allston-Roberts et al. 2010) and waste of time and money due to invalid data can only be estimated (Stacey et al. 2000; Chatterjee 2007).
One may speculate that our findings are just an exception. However, based on the experience of DMSZ and the survey of the literature the presented cases most probably are the tip of the iceberg.
Therefore, it is strongly recommended that all laboratories working with cell lines, especially with animal cell lines, should take special care: the identity and quality of all cell lines should be examined before using them and repeatedly in regular intervals.
Acknowledgments
We gratefully thank Prof. Dr. Hermann-Josef Rothkötter, University of Magdeburg for donation of the IPEC-1 and IPEC-J2 cells to DSMZ.
References
- Allston-Roberts C, Barallon R, Bauer SR, Butler J, Capes-Davis A, Dirks WG, Elmore E, Furtado M, Kerrigan L, Kline MC, Kohara A, Los GV, MacLeod RAF, Masters JR, Nardone M, Nims RW, Price PJ, Reid YA, Shewale J, Steuer AF, Storts DR, Sykes G, Taraporewala Z, Thomson J. Cell line misidentification: the beginning of the end. Nat Rev Cancer. 2010;10:441–448. doi: 10.1038/nrc2852. [DOI] [PubMed] [Google Scholar]
- Berschneider HM. Development of normal cultured small intestinal epithelial cell lines which transport Na and Cl. Gastroenterol. 1989;96:A41. [Google Scholar]
- Capes-Davis A, Theodosopoulos G, Atkin I, Drexler HG, Kohara A, MacLeod RAF, Masters JR, Nakamura Y, Reid YA, Reddel RR, Freshney RA. Check your cultures! A list of cross-contaminated or misidentified cell lines. Int J Cancer. 2010;127:1–8. doi: 10.1002/ijc.25242. [DOI] [PubMed] [Google Scholar]
- Chantret I, Barbat A, Dussaulx E, Brattain MG, Zweibaum A. Epithelial polarity, villin expression, and enterocytic differentiation of cultured human colon carcinoma cells: a survey of twenty cell lines. Cancer Res. 1988;48:1936–1942. [PubMed] [Google Scholar]
- Chatterjee R. Cell biology: cases of mistaken. Science. 2007;315:928–931. doi: 10.1126/science.315.5814.928. [DOI] [PubMed] [Google Scholar]
- Cooper JK, Sykes G, King S, Cottrill K, Ivanova NV, Hanner R, Ikonomi P. Species identification in cell culture: a prolonged molecular approach. In Vitro Cell Dev Biol Anim. 2007;43:344–351. doi: 10.1007/s11626-007-9060-2. [DOI] [PubMed] [Google Scholar]
- Diesing A-K, Nossol C, Panther P, Walk N, Post A, Kluess J, Kreutzmann P, Dänicke S, Rothkötter H-J, Kahlert S. Mycotoxin deoxynivalenol (DON) mediates biphasic cellular response in intestinal porcine epithelial cell lines IPEC-1 and IPEC-J2. Toxicol Lett. 2011;200:8–18. doi: 10.1016/j.toxlet.2010.10.006. [DOI] [PubMed] [Google Scholar]
- Drexler HG, Dirks WG, Matsuo Y, MacLeod RAF. False leukemia- lymphoma cell lines: an update on over 500 cell lines. Leukemia. 2003;17:416–426. doi: 10.1038/sj.leu.2402799. [DOI] [PubMed] [Google Scholar]
- Geens MM, Niewold TA. Optimizing culture conditions of a porcine epithelial cell line IPEC-J2 through a histological and physiological characterization. Cytotechnolgy. 2011;63:415–423. doi: 10.1007/s10616-011-9362-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Vallina R, Wang H, Zhan R, Berschneider HM, Lee RM, Davidson NO, Black DD. Lipoprotein and apolipoprotein secretion by a newborn piglet intestinal cell line (IPEC-1) Am J Physiol. 1996;271:G249–G259. doi: 10.1152/ajpgi.1996.271.2.G249. [DOI] [PubMed] [Google Scholar]
- Lacroix M. Persistent use of "false” cell lines. Int J Cancer. 2008;122:1–4. doi: 10.1002/ijc.23233. [DOI] [PubMed] [Google Scholar]
- Liu MY, Lin SC, Liu H, Candal F, Vafai A. Identification and authentication of animal cell culture by polymerase chain reaction amplification and DNA sequencing. In Vitro Cell Dev Biol Anim. 2003;39:424–427. doi: 10.1290/1543-706X(2003)039<0424:IAAOAC>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- MacLeod RAF, Drexler HG. Public repositories: users reluctant to give materials. Nature. 2006;439:912. doi: 10.1038/439912b. [DOI] [PubMed] [Google Scholar]
- MacLeod RAF, Dirks WG, Matsuo Y, Kaufmann M, Milch H, Drexler HG. Widespread intraspecies cross-contamination of human tumor cell lines arising at source. Int J Cancer. 1999;83:555–563. doi: 10.1002/(SICI)1097-0215(19991112)83:4<555::AID-IJC19>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- MacLeod RAF, Dirks WG, Dexler HG. Persistent use of misidentified cell lines and its prevention. Genes Chromosom Cancer. 2002;33:103–105. doi: 10.1002/gcc.1217. [DOI] [PubMed] [Google Scholar]
- Mariani V, Palermo S, Fiorentini S, Lanubile A, Giuffra E. Gene expression study of two widely used pig intestinal epithelial cell lines: IPEC-J2 and IPI-2I. Vet Immunol Immunopathol. 2009;131:278–284. doi: 10.1016/j.vetimm.2009.04.006. [DOI] [PubMed] [Google Scholar]
- Masters JR. False cell lines: the problem and the solution. Cytotechnology. 2002;39:17–22. doi: 10.1023/A:1022908930937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardonne RM. Eradication of cross-contaminated cell lines. A call for action. Cell Biol Toxicol. 2007;23:367–372. doi: 10.1007/s10565-007-9019-9. [DOI] [PubMed] [Google Scholar]
- Nelson-Rees WA, Daniels DW, Flandermeyer RR. Cell cross-contamination in cell cultures. Science. 1981;212:446–452. doi: 10.1126/science.6451928. [DOI] [PubMed] [Google Scholar]
- Nossol C, Diesing AK, Walk N, Faber-Zuschratter H, Hertig R, Post A, Kluess J, Rothkötter HJ, Kahlert S. Air-liquid interface cultures enhance the oxygen supply and trigger the structural and functional differentiation of intestinal porcine epithelial cells (IPEC) Histochem Cell Biol. 2011;36:103–115. doi: 10.1007/s00418-011-0826-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K, Motonobu S, Yoshida T, Ozawa Y, Kohara A, Takeuchi M, Mizusawa H, Sawada H. Species identification of animal cells by nested PCR targeted to mitochondrial DNA. In Vitro Cell Dev Biol Anim. 2007;43:168–175. doi: 10.1007/s11626-007-9033-5. [DOI] [PubMed] [Google Scholar]
- Pinton P, Braicu C, Nougayrede JP, Laffitte J, Taranu I, Oswald IP. Deoxynivalenol impairs porcine intestinal barrier function and decreases the protein expression of claudin-4 through a mitogen-activated protein kinase-dependent mechanism. J Nutr. 2010;140:1956–1962. doi: 10.3945/jn.110.123919. [DOI] [PubMed] [Google Scholar]
- Schierack P, Nordhoff M, Pollmann M, Weyrauch KD, Amasheh S, Lodemann U, Jores J, Tachu B, Kleta S, Blikslager A, Tedin K, Wieler LH. Characterization of a porcine intestinal epithelial cell line for in vitro studies of microbial pathogenesis in swine. Histochem Cell Biol. 2006;125:293–305. doi: 10.1007/s00418-005-0067-z. [DOI] [PubMed] [Google Scholar]
- Stacey GN, Hoelzl H, Stephenson JR, Doyle A. Authentication of animal cell cultures by direct visualization of repetitive DNA, aldolase gene PCR and isoenzyme analysis. Biologicals. 1997;25:75–85. doi: 10.1006/biol.1996.0062. [DOI] [PubMed] [Google Scholar]
- Stacey GN, Masters JR, Hay RJ, Drexler HG, MacLeod RAF, Freshney RI. Cell contamination leads to inaccurate data: we must take action now. Nature. 2000;403:356. doi: 10.1038/35000394. [DOI] [PubMed] [Google Scholar]
- Steube KG, Meyer C, Uphoff CC, Drexler HG. A simple method using β-globin PCR for the species identification of animal cell lines. In Vitro Cell Dev Biol Anim. 2003;39:468–475. doi: 10.1290/1543-706X(2003)039<0468:ASMUGP>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Steube KG, Koelz AL, Drexler HG. Identification and verification of rodent cell lines by polymerase chain reaction. Cytotechnology. 2008;56:49–65. doi: 10.1007/s10616-007-9106-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson R. Development of cell banking in the US 1960–1985: a strategic approach to quality control. Adv Cell Cult. 1987;51:267–288. [Google Scholar]
- Uphoff CC, Drexler HG. Detecting Mycoplasma contamination in cell cultures by polymerase chain reaction. In: Cree I, editor. Cancer cell culture: methods and protocols. 2. Berlin: Springer; 2011. pp. 93–103. [DOI] [PubMed] [Google Scholar]
- Uphoff CC, Drexler HG. Elimination of mycoplasmas from infected cell lines using antibiotics. In: Cree I, editor. Cancer cell culture: methods and protocols. 2. Berlin: Springer; 2011. pp. 105–114. [DOI] [PubMed] [Google Scholar]
- Zoon KC. Points to consider in the characterization of cell lines used to produce biologicals. Rockville, MD: Center for Biological Evaluation and Research. Food and Drug Administration; 1993. pp. 7–8. [Google Scholar]