Abstract
Human embryonic stem cells (hESCs) require specific niches for adhesion, expansion, and lineage-specific differentiation. In this study, we showed that a membrane substrate offers better tissue niches for hESC attachment, spreading, proliferation, and differentiation. The cell doubling time was shortened from 46.3±5.7 h for hESCs grown on solid substrates to 25.6±2.6 h for those on polyester (PE) membrane substrates with pore size of 0.4 μm. In addition, we observed an increase of approximately five- to ninefold of definitive endoderm marker gene expression in hESCs differentiated on PE or polyethylene terephthalate membrane substrates. Global gene expression analysis revealed upregulated expressions of a number of extracellular matrix and cell adhesion molecules in hESCs grown on membrane substrates. Further, an enhanced nuclear translocation of β-catenin was detected in these cells. These observations suggested the augmentation of Wnt signaling in hESCs grown on membrane substrates. These results also demonstrated that a membrane substrate can offer better physicochemical cues for enhancing in vitro hESC attachment, proliferation, and differentiation.
Introduction
Human embryonic stem cells (hESCs) are extremely valuable for many clinical applications owing to their capability of self-renewal and multilineage differentiation. However, the use of hESCs for cell replacement therapy has been very challenging so far partly due to the scarcity of knowledge about in vitro lineage-specific differentiation of hESCs. In vivo cells rely upon at least three interactions: cell–cell, cell–extracellular matrix (ECM), and cell–growth factors/signaling molecules to function within tissues. Soluble and insoluble signaling molecules combined with physiochemical factors constitute a tissue niche that offers optimal control and regulation of biological stimulations to instruct cell differentiation toward specific lineages. Ideally, an in vitro hESC differentiation system should mimic these in vivo environments that allow the orchestration of multiple signaling pathways for directing hESC lineage specification.
Several molecular mechanisms underlying cell response to surrounding environments have been identified to date. It has been demonstrated that cells sense environmental factors and signals mainly through cell–matrix and cell–cell interactions. For instance, cells grown on a substrate detect a mechanical signal using a set of molecules at their subcellular sites, such as focal adhesions. In these regions, a super family of transmembrane proteins, that is, integrins, plays a central role in transducing a mechanical signal into a biochemical signal, leading to the alternation of cell fates.1 Integrins act as mechanosensors to detect a variety of mechanical signals.2,3 Their cytoplasmic domains interact with talin, α-actinin, filamin, tesin, and other focal adhesion proteins to stabilize or destabilize the focal adhesions,1,4–6 resulting in the remodeling of microfilament and microtubule networks, and subsequently altering the gene expression. The signal transduction events involve several types of enzymes, including protein tyrosine kinase, protein tyrosine phosphatase, and serine-theronine kinase.1,7–9 A body of evidence suggests that mechanical stimulations occur at either adhesion sites or the extracellular subunits of integrins.10,11 Thus, the adhesion sites, that is, the interface between cell and substrates, are critical to the regulation and control of physiochemical signals. Clearly, the adhesion sites are influenced by the topographical structure of a substrate, including elements such as porosity and network structure of membrane substrates. There is ample evidence that the contact between cells and a substrate provides guidance for cells to attach, spread, proliferate, migrate, and differentiate on the surface. The contact-mediated guidance can alter cell focal adhesions, cytoskeletal architecture, nuclear shape, and nuclear orientation, thereby regulating cell signaling pathways and affecting cell morphology,12,13 adhesion,14 proliferation,15 motility,16 and differentiation.17
Extensive studies have been conducted to characterize the effect of topographical cues on cell behaviors. These studies have revealed many details on how surface-induced topographical stimulations affect stem cell behaviors. For instance, a nanostructured surface has been found to be able to significantly enhance osteogenesis of human mesenchymal stem cells (hMSCs).18 These nanostructured surfaces are fabricated by dispensing prepolymer polyurethane acrylate on a supporting polyethylene terephthalate (PET) film, forming surface conformal contacts with cells. Studies on other stem cells/progenitors, including retinal progenitors,19 human umbilical cord blood hematopoietic progenitors,20 osteoblasts,21–24 neural cells,25,26 and hESCs,27,28 all showed similar results. Further studies indicate that surface-induced topographical stimulation influences not only the differentiation efficiency, but also the lineage specification.29 For example, the study of hMSC differentiation on microcontact-printed surface revealed that cells on the periphery of the pattern sense the edge, affecting the net differentiation of cells on the interior.30,31 Another study on testing hMSC differentiation on stress gradients suggested that cells that attach to a high-stress region differentiate into osteoblasts, whereas those grown in a low-stress region differentiate into adipocytes.32 All these observations strongly suggest that the topographical structure of a substrate can remarkably influence cell behaviors, including directed differentiation of hESCs. During embryo development, cells are exposed to various topological and biochemical cues in their surrounding environments, including basement membranes.33 Basement membranes usually have a mixture of pores (∼72 nm), ridges, and fibers (∼77 nm in diameter).34,35 They separate tissues from each other and allow cells to develop their polarized morphology. Thus, we assumed that not only the porous structure but also the roughness of a substrate is essential for restoring the in vivo niche equilibrium crucial to hESC growth and lineage specification.
It has long been known that cell fate can be manipulated using growth factors. This strategy has been very successful for most stem cells. However, it has shown limited success in directing hESC differentiation into clinically relevant cell lineages. In many cases, cells differentiated using growth factors alone are immature, suggesting that the use of soluble signaling molecules alone is insufficient to resemble in vivo tissue niches. Recently, it has become clear that not only growth factors but also insoluble factors, such as mechanical stress, mechanical, chemical properties, and topography of substrates, and so on, play roles in controlling and regulating hESC fate.36 hESCs secrete ECM proteins and various cytokines to coordinate cell–cell and cell–ECM interactions, which in turn regulate cell attachment, proliferation, and differentiation. In general, cells grown on a flat, solid, and impermeable substrate will be forced to grow into a nonpolarized status due to their limitation of expansion at the z-direction. The difficulty in maintaining optimal morphology can impair the cells' differentiation and maturation, as morphology is one of the key factors affecting cell fate.37,38 In addition, the impermeability of the substrate prevents cells from taking or secreting signaling molecules and nutrients from both basal and apical surfaces. This causes cells to deviate from their natural environment, in which they are always exposed to porous basement membranes and interact with hundreds of signals from basal and apical surfaces. Membrane substrates have been employed for growing various types of cells in the past.39 However, the use of membrane substrates for hESC proliferation and differentiation has not yet been explored. In this study, we intend to determine whether a porous and permeable membrane substrate provides better tissue niches for hESC proliferation and differentiation. In particular, we are interested in understanding how membrane substrates enhance hESC growth and lineage-specific differentiation.
Materials and Methods
hESC culture
The hESC line H9 (WiCell Institute) was maintained in undifferentiated state on growth-factor-reduced Matrigel-coated (BD Biosciences) dishes in a defined mTeSR1 (Stem Cell Technologies) medium at 37°C and 5% CO2. The medium was replaced daily. For passage, hESCs were treated with 1 mg/mL dispase (Stem Cell Technologies) for 7 min at 37°C, and then washed three times with Dulbecco's Modified Eagle Medium/F12 to completely remove the dispase. hESC colonies were gently scraped from the dishes and broken up into small colonies before seeding. Cells were split at a ratio of 1:3–1:5 every 3–4 days. PET and polyester (PE) membranes were obtained from Millipore and Corning. They were coated with Matrigel before cell seeding. The membrane substrates were placed inside a six-well plate and seeded with 50,000 cells/cm2 hESCs in the mTeSR1 medium. The viability of hESCs was determined through trypan blue staining assay after detaching the cells from culture plates using trypsin-ethylenediaminetetraacetic acid (EDTA).
Definitive endoderm differentiation
Undifferentiated H9 cells were seeded onto Matrigel-coated membrane substrates and cultured overnight in the mTeSR1 medium. Cells were then fed with a definitive endoderm (DE) differentiation medium containing RPMI1640, B27 (Invitrogen), 1 mM sodium-butyrate, and 4 nM activin A next day. The sodium butyrate concentration was reduced to 0.5 mM after 24 h of differentiation. The differentiation medium was exchanged every other day until day 7 postdifferentiation.
Gene expression assay
To detect gene expression in cells grown on membrane substrates, the total RNA was isolated from cells using an RNA extraction kit from Qiagen. Genomic DNA was eliminated during the RNA purification. RNA was synthesized to cDNA in a reverse transcription reaction using an RT kit from Applied Biosystems. Samples were then subjected to cell adhesion and ECM gene expression array plates (Invitrogen) using TaqMan RT-PCR Master Mix (Invitrogen) according to the manufacturer's instruction. GAPDH served as an endogenous control. To determine DE marker gene expression, the SOX17 and FOXA2 primer probe sets were used, as reported elsewhere.40 The cyclophilin (Applied Biosystems), a human endogenous gene, served as a housekeeping gene for normalization. Their relative gene expression was expressed as fold changes to the corresponding values detected from RNA prepared using adult human pancreata (Stratagene). A control assay was performed to ensure the absence of genomic DNA contamination in the quantitative real-time–polymerase chain reaction (qRT-PCR) assay.41 The following primer pairs were used for characterization of mRNA expression. OCT4 forward: 5′-TGGGCTCGAGAAGGATGTG-3′, OCT4 reverse: 5′-GCATAGTCGCTGCTTGATCG-3′; FLK1 forward: ACTTTGGAAGACAGAACCAAATTATCTC-3′, FLK1 reverse: 5′-TGGGCACCATTCCACCA-3′; FOXA2 forward: 5′-GGGAGCGGTGAAGATGGA-3′, FOXA2 reverse: 5′-TCATGTTGCTCACGGAGGAGTA-3′; SOX3 forward: 5′-GAGGGCTGAAAGTTTTGCTG-3′, SOX3 reverse: 5′-CCCAGCCTACAAAGGTGAAA-3′; and β-actin forward: 5′-CTGGAACGGTGAAGGTGACA-3′, β-actin reverse: 5′-AAGGGACTTCCTGTAACAATGCA-3′.
Scanning electron microscopy
Scanning electron microscopy (SEM) imaging was performed, as described in our previous work.42 In brief, membrane substrates were fixed in 2.5% (v/v) glutaraldehyde (EM Sciences) for 2 h, followed by washing three times with phosphate-buffered saline (PBS). Samples were fixed again with 2% osmium tetroxide (EM Sciences) for 2 h and dehydrated in a lyophilizer (Labconco). The lyophilized samples were gold-sputtered and examined using a JEOL Field Emission SEM JSM-6335F with acceleration voltage of 5 kV (JSM-6335F; JEOL USA).
Immunofluorescence microscopy
To detect the expression of two DE marker proteins, SOX17 and FOXA2, in differentiated hESCs, cells were fixed in 4% paraformaldehyde (Fisher Scientific) and permeabilized with 0.1% Triton X-100/PBS buffer. Cells were blocked with 5% goat serum and 0.1% Triton X-100/PBS followed by incubation with primary antibodies and fluorescence dye-conjugated secondary antibodies. The samples were cross-stained with diaminophenylindole (Invitrogen). Cells were visualized under an Olympus IX-71 fluorescence microscope. Mouse monoclonal anti-human SOX17 (1:50; R&D Systems), rabbit monoclonal anti-human FOXA2 (1:1000; Abcam), mouse anti-OCT4 (1:100; Millipore), and mouse anti-SSEA4 (1:100; Millipore) were used as primary antibodies. Goat anti-mouse IgG FITC conjugates (1:100; Sigma) and donkey anti-rabbit IgG TRITC (1:50) conjugates, goat anti-mouse Alexa Fluro 488 IgG1 (1:200) and goat anti-mouse Alexa Fluro 488 IgG3 (1:200) antibodies from Invitrogen were used as secondary antibodies for labeling cells.
Western blotting
Cytoplasmic and nuclear proteins were extracted using a kit from Thermo Scientific. Immunoblotting was performed, as described in our previous work.43 Rabbit anti-human β-catenin antibodies (1:2000) and anti-rabbit IgG HRP conjugates (1:1000; Sigma) were used as primary and secondary antibodies, respectively. HRP-conjugated β-actin antibody (1:35,000; Sigma) served as a loading control for the assay. Proteins were visualized with chemiluminescent substrate reagents (Thermo Scientific).
Indentation testing
The mechanical properties of the membrane substrates were determined using an ultra nanoindentation tester by the CSM Instruments, Inc.
Statistical analyses
All data were presented as mean±SD. Student's t-test using a one-tailed algorithm was performed and p-values≤0.05 were considered statistically significant.
Results
Enhanced hESC attachment and proliferation by porous-structured substrates
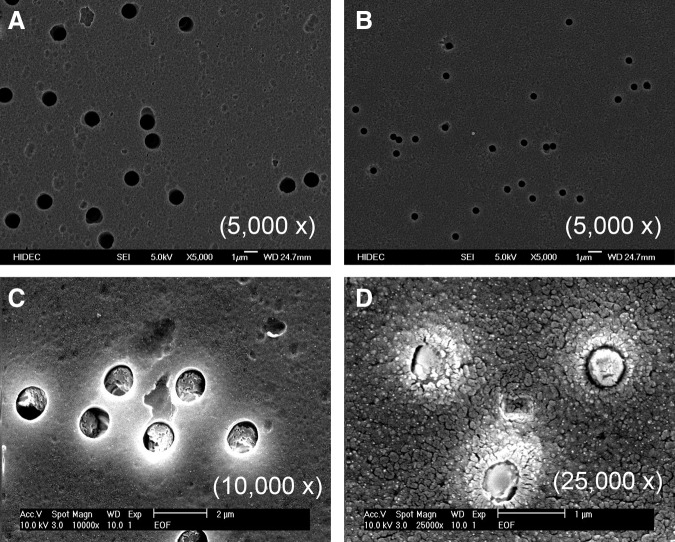
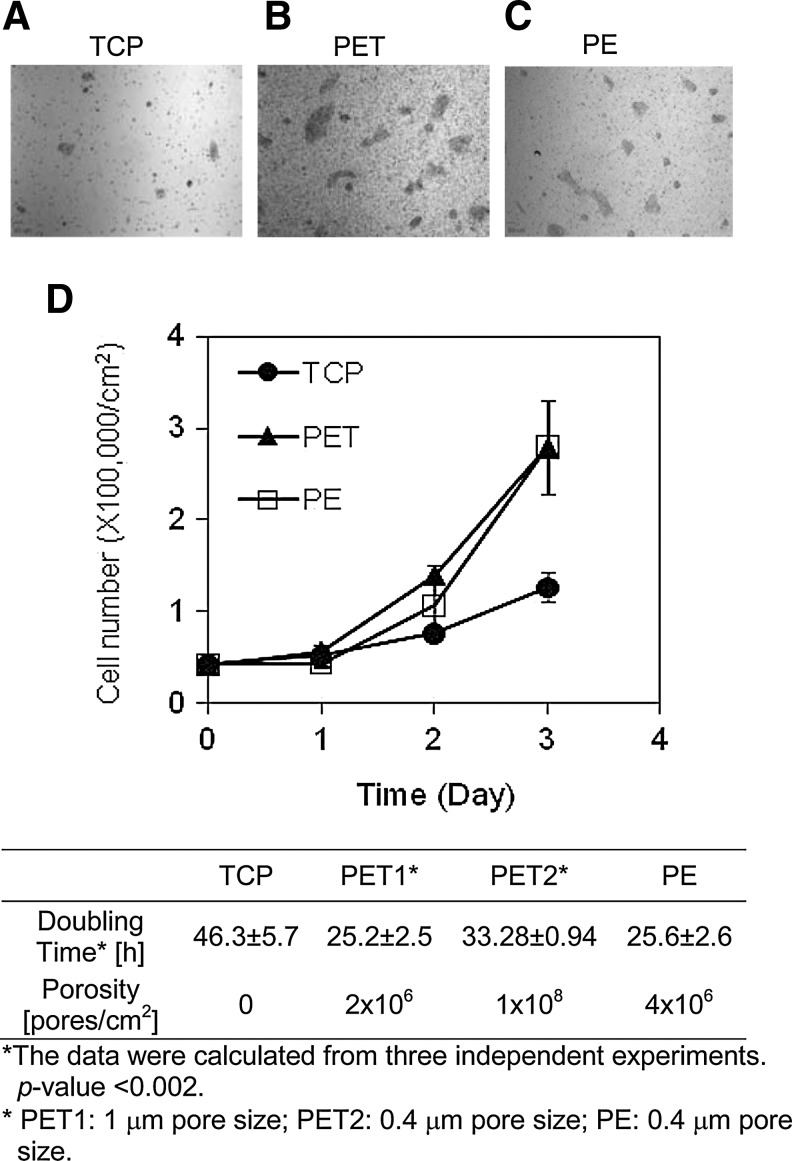
To determine whether hESC attachment and proliferation can be improved by growing them on a porous membrane substrate, we seeded hESCs on a porous PET or PE membrane substrate. Cells grown on a conventional tissue culture polystyrene (TCP) plate served as a control for comparison. We determined the structures of membrane substrates before and after Matrigel coating. As shown in Figure 1C and D, membrane substrates remained porous after Matrigel coating. The determination of cell growth rates suggested that hESCs grow much faster on a porous and permeable membrane substrate (Fig. 2D). The cell doubling time was 25.6±2.6 h when cultured on PE membrane substrates with pore size of 0.4 μm; it was 46.3±5.7 h when cells were cultured on TCP plates. Moreover, hESCs tend to form larger colonies on membrane substrates, as compared with those on TCP plates (Fig. 2A–C). Interestingly, we observed the shortening of cell doubling time on PET membrane substrates with 1 μm pores, as compared with those grown on membrane substrates with 0.4 pores (Fig. 2). However, the comparison of hESC growth on PET membrane substrates with 1 μm pores with those on PE membrane substrates with 0.4 μm pores revealed no significant differences in cell doubling time. Although they had two different pore sizes, they had similar porosity as indicated in Figure 2. These experimental results suggested that the topographical and chemical properties of a membrane substrate considerably influence hESC attachment and proliferation. Thus, we reasoned that hESC attachment and proliferation can benefit from physiochemical cues offered by a porous membrane substrate. The following experiments were designed to validate this hypothesis.
FIG. 1.
Scanning electron microscopy micrograph of porous membrane substrates used for supporting human embryonic stem cell (hESC) proliferation and differentiation. Polyethylene terephthalate (PET) (A) and polyester (PE) (B) porous membrane substrates were tested for hESC growth and differentiation. (C) and (D) are Matrigel-coated PET and PE membrane substrates. The pore sizes of the PE and PET membrane are 0.4 and 1 μm, respectively. Scale bars: 1 μm in (A) and (B), 2 μm in (C), and 1 μm in (D).
FIG. 2.
hESC attachment and proliferation on various substrates. hESCs were seeded onto tissue culture polystyrene (TCP) (A), 1 μm PET (B), and 0.4 μm PE (C) membrane substrates in mTeSR1 medium. The micrographic images of cell attachment were taken at day 2 after seeding. Scale bar: 50 μm. (D) Time courses of hESC proliferation on various substrates. Data were presented as mean±SD. All experiments were carried out at least three times.
First, we determined whether hESCs remain undifferentiated when being grown on membrane substrates. We analyzed pluripotency marker protein expression in hESCs after five consecutive passages on PET or PE membrane substrate. As shown in Figure 3A, we detected a high-level expression of both OCT4 and SSEA4 in hESCs after five consecutive passages on membrane substrates, suggesting the undifferentiated state of the cells during passages. To rule out any possible spontaneous differentiation of hESCs during passage on the membrane substrates, we further characterized gene expression levels of ESC marker OCT4 and three germ layer, mesoderm, endoderm, and ectoderm, markers in these cells after their five passages on membrane substrates. As shown in Figure 3B, we observed no considerable difference in the expression level of these marker genes as compared with those detected from hESCs grown on Matrigel-coated TCPs (control in Fig. 3B). These data demonstrated that it is possible to maintain hESCs on membrane substrates without compromising their pluripotency.
FIG. 3.
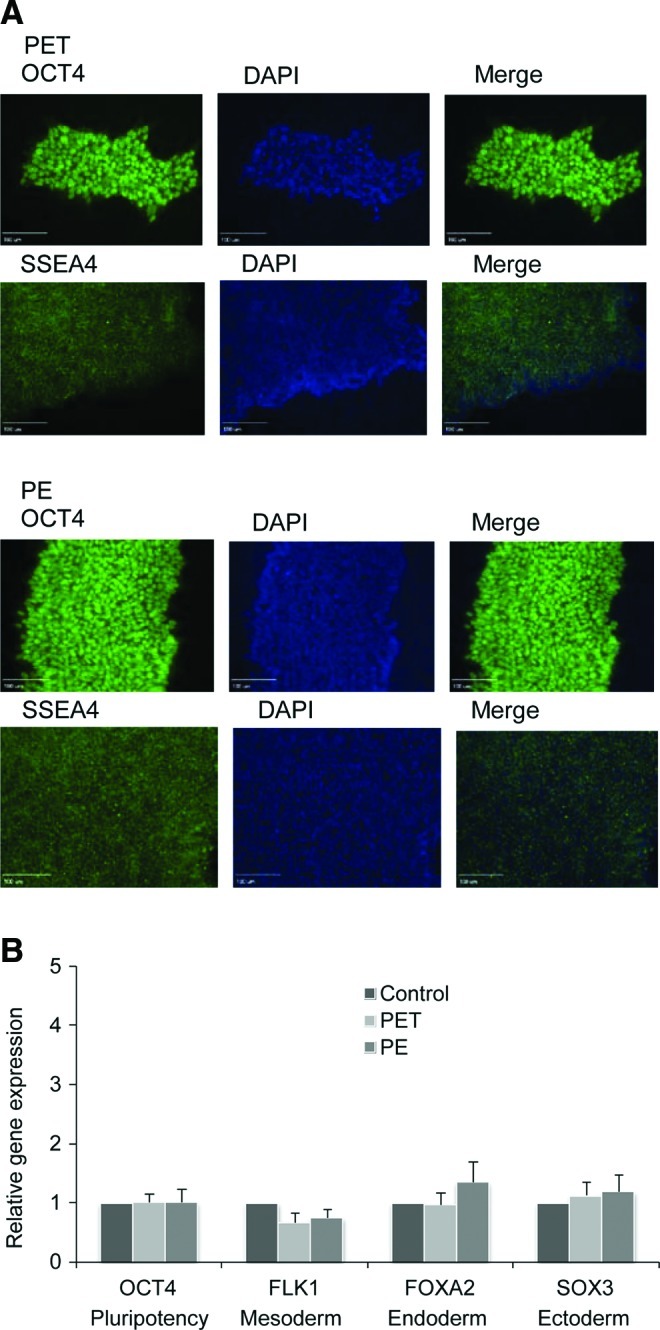
Detection of pluripotency of hESCs after five passages on PET and PE membranes. (A) Cells were stained with antibodies against OCT4 and SSEA4 marker proteins after five passages. Cells were also labeled with diaminophenylindole (DAPI) in order to localize the nucleus. Stained cells were observed under a fluorescence microscope (Olympus IX 71) equipped with a charge-coupled device (CCD) camera. Scar bar: 100 μm. (B) Gene marker of hESCs and markers of the three germ layers were detected in cells grown on membrane substrates using quantitative real-time–polymerase chain reaction (qRT-PCR) analysis. Control: hESC line H9 cultured on a TCP substrate in undifferentiated state. Color images available online at www.liebertonline.com/tea
Augmented lineage specification of hESCs by membrane substrates
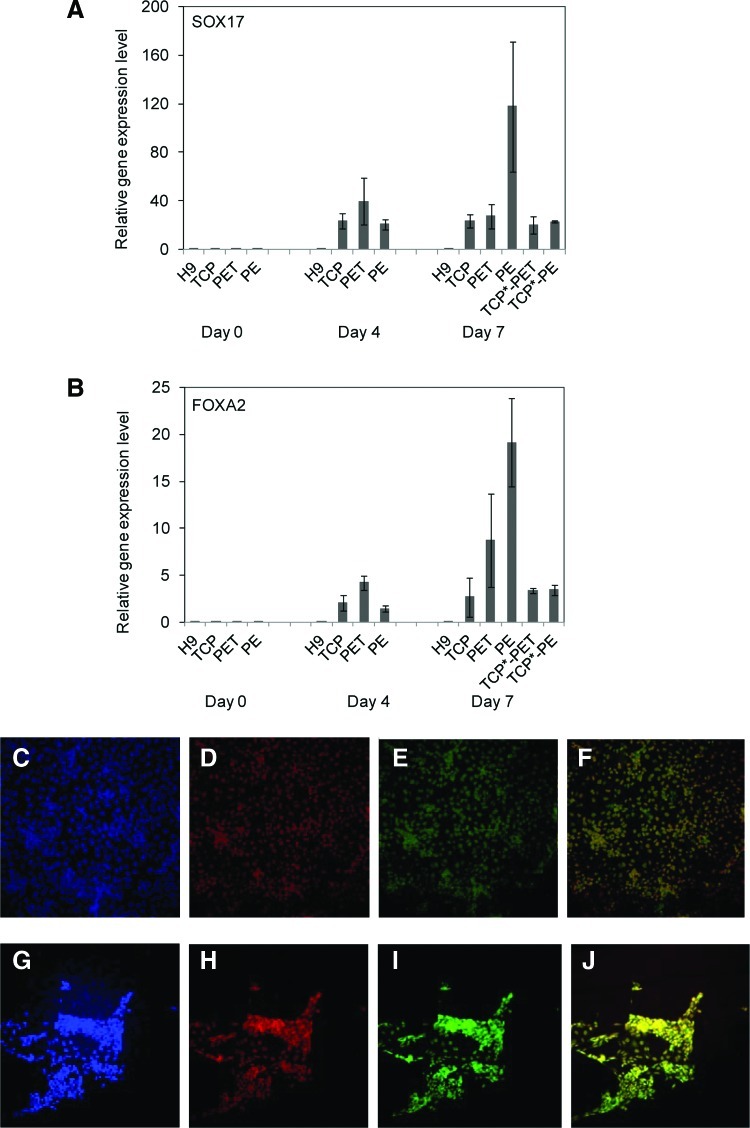
Having verified hESC attachment and proliferation, we next examined whether membrane substrates improve cell lineage specification. hESCs were differentiated into DE cells on either PET (pore size of 1 μm) or PE (pore size of 0.4 μm) membrane substrates. We analyzed DE marker genes, SOX17 and FOXA2, through TaqMan qRT-PCR assay and compared their expression levels with those on TCP cultures. As shown in Figure 4A and B, the expression of SOX17 and FOXA2 in cells differentiated on PE membrane substrates was 4.9- and 9.6-fold higher than those on TCP cultures, indicating significant enhancement of DE differentiation on porous membrane substrates. No SOX17 and FOXA2 expression could be detected in H9 undifferentiated cells. Interestingly, the increase in SOX17 and FOXA2 expressing cells differentiated on PET membrane substrates was marginal, although both PET and PE membrane substrates supported hESC attachment and proliferation. The immunofluorescence staining of SOX17 and FOXA2 in cells differentiated on membrane substrates was confirmed by the qRT-PCR assay. Moreover, we observed the formation of cell aggregates when hESCs were differentiated on PE membrane substrates, whereas no cell aggregates could be observed on PET membrane substrates (Fig. 4C–J). The formation of cell aggregates is an essential step for hESC differentiation toward DE tissues. Thus, PE membrane substrates offered better physicochemical cues for promoting hESC DE differentiation. To exclude the possible effect of chemical leakages from membranes on hESC DE differentiation, we performed hESC DE differentiation on a TCP substrate in the presence or absence of PET or PE membrane hanging inserts. As revealed by Figure 4A and B, no significant difference could be observed when cells were differentiated in the presence of PET or PE membranes, suggesting that any leakage of chemicals from PET or PE membrane hanging inserts does not influence the DE differentiation of hESCs. The effect of membrane hanging inserts on hESC DE differentiation is indeed due to the porous structure of a membrane substrate.
FIG. 4.
Definitive endoderm (DE) marker gene and protein expression in DE cells differentiated from hESCs on membrane substrates. Total RNA was extracted from DE cells on indicated days and subjected to TaqMan qRT-PCR analysis for detection of expression of SOX17 (A) and FOXA2 (B). Relative gene expression levels were normalized to their levels detected from adult pancreatic samples. Data were presented as mean±SD. (C–J) Immunofluorescence detection of DE marker proteins in DE lineage-specific differentiated hESCs on membrane substrates PET (C–F) and PE (G–J). Cells were immunostained with antibodies against SOX17 (E and I) and FOXA2 (D and H) at day 7 postdifferentiation. Cells were also counter-labeled with DAPI (C and G). (F) and (J) are overlay of SOX17 and FOXA2 fluorescence microscopy images. TCP* stands for the DE differentiation on a TCP surface in the presence of PET and PE membrane insert. Scar bar: 100 μm. Color images available online at www.liebertonline.com/tea
Upregulation of ECM and adhesion molecules in hESCs grown on membrane substrates
To understand the mechanisms underlying the enhancement of hESC attachment, proliferation, and differentiation on membrane substrates, we next performed global gene expression profiling analysis. We detected 92 genes that are considered to be involved in cell adhesion, cell–ECM, and cell–cell interactions at 48 h after seeding. We observed the upregulation of a number of ECM genes, including collagen type XI α1; Catenin α1, β1, and δ1; and laminin α3 and γ1, in hESCs grown on PET membrane substrates (Table 1). These genes, with the exception of laminin α3, were upregulated in cells grown on PE membrane substrates as well (Table 2). In addition, a number of integrin and collagen genes were upregulated considerably in cells grown on PE membrane substrates. In particular, the expression of integrin β1, αV, and β5; collagen type XI α1, type XII α1, and type XVI α1; and laminin γ1 increased from three- to ninefold in hESCs on PE membrane substrates. The increase in connective tissue growth factors and cadherin 1 expression was also detected in PE membrane cultures. Cadherin is a transmembrane protein that helps cells cling to each other, forming organized tissues. These results suggest that a PE membrane substrate, as compared with a PET membrane substrate, tends to encourage more cell–cell interactions. Interestingly, we observed an increase in the expression of matrix metallopeptidase (MMP) family proteins in hESCs grown on membrane substrates. The upregulation of CD44 expression in cells grown on membrane substrates was also observed. CD44 molecules are cell surface adhesion molecules involved in cell–cell and cell–ECM interactions.44 The upregulation of this gene expression indicates the enhancement of cell–cell and cell–ECM interactions in these cells when they were grown on the membrane substrates. The upregulation of neural cell adhesion molecule 1 gene in hESCs grown on PE membrane substrates further suggests the enhancement of cell–cell adhesion,45 which is essential for hESC proliferation.
Table 1.
Upregulation of Expression of Extracellular Matrix and Cell Adhesion Molecules in H9 Cells Grown on Polyethylene Terephthalate Membrane Substrate
| Gene title | Gene ID | Fold changea | p-Value |
|---|---|---|---|
| Collagen type XI, α1 | COL11A1 | 4.2 | 0.088 |
| Catenin α1 | CTNNA1 | 2.3 | 0.090 |
| Catenin β1 | CTNNB1 | 4.7 | 0.043 |
| Catenin δ1 | CTNND1 | 2.6 | 0.069 |
| Laminin α3 | LAMA3 | 2.6 | 0.046 |
| Laminin γ1 | LAMC1 | 2.4 | 0.063 |
| Matrix metallopeptidase 15 | MMP15 | 2.2 | 0.031 |
Genes expressed in cells grown on TCP substrates served as a control for determining the fold-change and up- or downregulation of relevant genes in cells grown on membrane substrates.
TCP, tissue culture polystyrene.
Table 2.
Upregulation of Expression of Extracellular Matrix and Cell Adhesion Molecules in H9 Cells Grown on Polyester Membrane Substrate
| Gene title | Gene ID | Fold changea | p-Value |
|---|---|---|---|
| CD44 molecule | CD44 | 5.0 | 0.039 |
| Cadherin 1, type I | CDH1 | 2.9 | 0.025 |
| Collagen type XI, α1 | COL11A1 | 6.8 | 0.016 |
| Collagen type XII, α1 | COL12A1 | 6.5 | 0.020 |
| Collagen type XVI, α1 | COL16A1 | 7.2 | 0.019 |
| Collagen type VII, α1 | COL7A1 | 7.2 | 0.023 |
| Connective tissue growth factor | CTGF | 4.9 | 0.026 |
| Catenin α1 | CTNNA1 | 3.4 | 0.021 |
| Catenin β1 | CTNNB1 | 5.9 | 0.027 |
| Catenin δ1 | CTNND1 | 5.2 | 0.029 |
| Integrin αV | ITGAV | 2.8 | 0.036 |
| Integrin β1 | ITGB1 | 9.3 | 0.011 |
| Integrin β5 | ITGB5 | 2.6 | 0.035 |
| Laminin γ1 | LAMC1 | 3.0 | 0.029 |
| Matrix metallopeptidase 15 | MMP15 | 2.0 | 0.039 |
| Neural cell adhesion molecule 1 | NCAM1 | 4.7 | 0.034 |
Genes expressed in cells grown on TCP substrates served as a control for determining the fold-change and up- or downregulation of relevant genes in cells grown on membrane substrates.
Promoted β-catenin-mediated Wnt signaling in hESCs grown on membrane substrates
As shown earlier, we observed the upregulation of a number of ECM genes in hESCs grown on PET membrane substrates. No significant downregulation of ECM and cell-adhesion-related genes in these cells was detected. For cells grown on PE membrane substrates, 16 cell-adhesion-related genes were upregulated. Among these upregulated genes, a number of catenin family genes were elevated remarkably in both PET and PE membrane substrates. The expression of catenin α1, β1, and δ1 had 2.3-, 4.7-, and 2.6-fold increases in hESCs grown on PET membrane substrates, and 3.4-, 5.9-, and 5.2-fold increases were observed in hESCs grown on PE membrane substrates. We also noticed that the expression of a panel of integrin and collagen genes was upregulated considerably. For instance, the expression of integrins α1, αV, and β5 and collagen type VII, XIα1, XIIα1, and XVIα1 was increased from 2.6- to 7.2-fold on PE membrane substrates. These upregulations might suggest the activation of the Wnt signaling pathway by the topographical, texture, and biochemical properties of a membrane substrate. Wnt signals modulate β-catenin expression, which triggers a high-level expression of integrins.46 To investigate this possibility, we detected the nuclear translocation of β-catenin in hESCs grown on membrane substrates. Proteins in the cytoplasm and in the nucleus were extracted and detected through western blotting. β-Actin served as an internal control for the assay. As revealed in Figure 5, most β-catenin was translocated from cytoplasm to nucleus when cells were grown on membrane substrates. The ratio of β-catenin in nucleus to that in the cytoplasm was 0.21 in cells grown on TCP substrates. It was, however, 1.58 in cells grown on PET and 3.74 in cells grown on PE membrane substrates. The translocation of more β-catenin to the nucleus suggested the activation of β-catenin-mediated Wnt signaling pathway in hESCs grown on membrane substrates. The enhancement of Wnt signaling appeared to lead to the increase in collagen and integrin expression, which in turn improved hESC attachment, proliferation, and differentiation. This is in consistence with others work.47–49 The upregulation of MMP expression in these cells (Tables 1 and 2) further confirms this observation. As revealed by Doyle and Haas50 and Brabletz et al.,51 the activation of Wnt/β-catenin signaling activates MMP expression. We indeed observed the upregulation of MMP in cells grown on membrane substrates as mentioned previously. On the other hand, the upregulation of ECM and adhesion molecules in these cells could also be induced by the spontaneous differentiation of hESCs on these membrane substrates. To eliminate this possibility, we analyzed the expression of two ESC marker proteins, OCT4 and SSEA4, after growing cells on PET or PE membrane substrates for 2 days when the cells were collected for Wnt signaling analysis. As illustrated in Figure 5C, hESCs expressed a very high level of OCT4 and SSEA4. Thus, these cells remained in undifferentiated state when they were collected for Wnt signaling analysis. This undifferentiated state was maintained during passages. As shown in Figure 3B, hESC marker genes remained expressed at a high level after their five passages on the membrane substrates. Thus, we reasoned that the expression of ECM and adhesion molecules was upregulated by β-catenin/Wnt signaling in the cells.
FIG. 5.
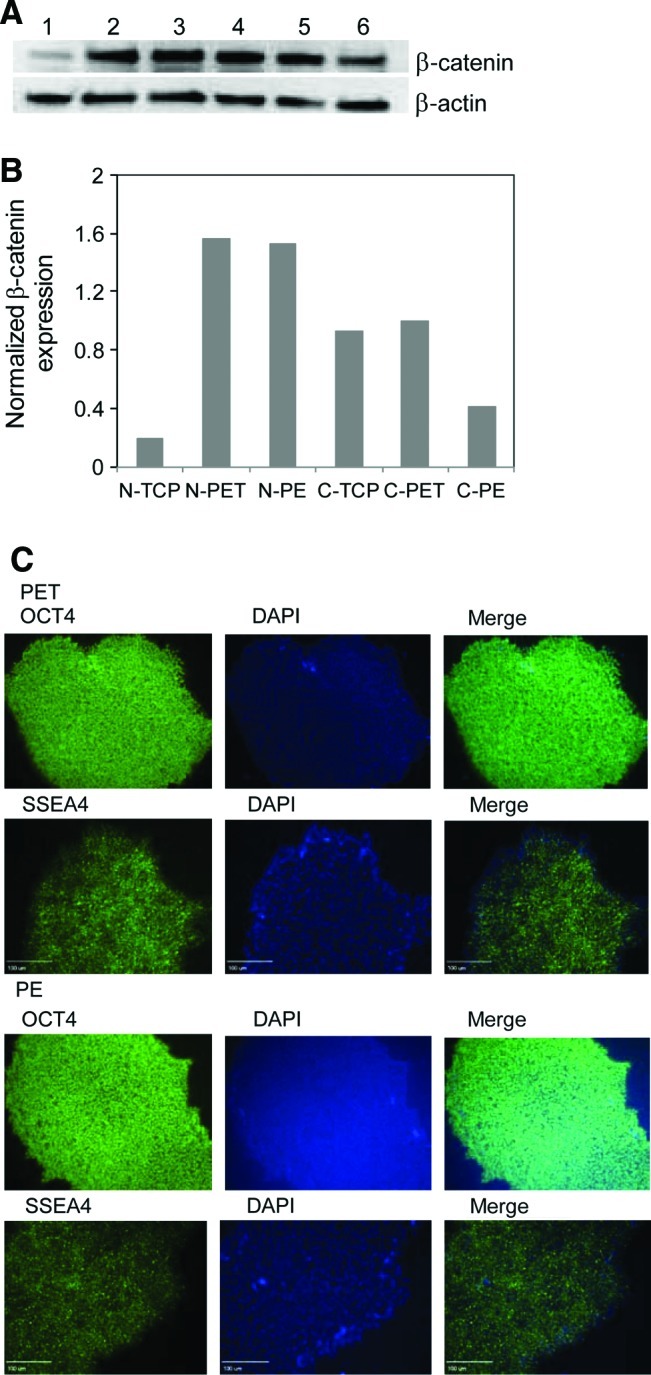
Translocation of β-catenin to nucleus of H9 cells grown on membrane substrates. (A) Western blotting analysis of β-catenin expression in nucleus (lanes 1–3) and cytoplasm (lanes 4–6) of hESCs grown on TCP substrates (lanes 1 and 4), PET (lanes 2 and 5), and PE (lanes 3 and 6) membrane substrates. (B) Semiquantification of β-catenin in nucleus (N) and cytoplasm (C) through western blotting assay. The Kodak 1D gel imaging analysis software was used to perform these semiquantification analyses. β-Actin served as a control for semiquantification. (C) Detection of pluripotency of hESCs after culturing cells on PET and PE membranes for 2 days. Cells were stained with antibodies against OCT4 and SSEA4 marker proteins and also labeled with DAPI. The labeled cells were examined under a fluorescence microscope (Olympus IX 71) equipped with a CCD camera. Scar bar: 100 μm. Color images available online at www.liebertonline.com/tea
Discussion
In this work, we provided evidence that a porous membrane substrate offers better niches for hESC attachment, expansion, and differentiation. Porous, thin, and permeable membrane substrates enable cells to take up and secrete molecules from both basal and apical surfaces. They act on cells more like basement membranes, allowing cells to grow into a polarized morphology and carry out metabolic activities in a more natural fashion. Their porous structure provides suitable stiffness and topographic cues to promote cell growth and differentiation. Our study supports this idea. We showed that the topographical structure of a membrane substrate significantly influences hESC behaviors. The detection of ECM and cell adhesion gene expression in hESCs grown on membrane substrates divulged a significant increase in β-catenin, collagen, integrin, and laminin gene expression in these cells. These noticeable upregulations suggested the activation of β-catenin-mediated Wnt signaling by membrane-substrate-associated physicochemical cues.
Interestingly, we observed upregulation of NCAM1 gene expression as well in hESCs grown on membrane substrates. NCAM1 is one of the protein markers associated with neural differentiation. However, a spontaneous differentiation of hESCs toward neuron lineages during their passages on membrane substrates was unlikely. As discussed previously, we detected expression of hESC pluripotency marker genes and proteins after five consecutive passages on membrane substrates (Fig. 3). If the cells started spontaneous differentiation toward neuron lineages, then the expression of these stem cell markers would be significantly suppressed and became undetectable. The fact of the high expression levels of stem cell markers in these cells indicated the pluripotency of the cells after passages on membrane substrates. Thus, the upregulation of NCAM1 might suggest other functions of this gene in regulating hESC attachment, proliferation, and differentiation. Indeed, a number of studies suggested that NCAM1 is also an important cell adhesion molecule that plays an essential role in cell division, migration, and differentiation.52 Its high-level expression is found during embryo development.52 Thus, we were not surprised by the upregulation of the NCAM1 in hESCs grown on membrane substrates. It might indicate that cues offered by a membrane substrate stimulated upregulation of adhesion molecules, leading to their enhanced attachment to membrane substrates, as suggested by our experimental data.
Another interesting phenomenon that we observed is the upregulation of Wnt pathway in hESCs grown on membrane substrates. Wnt/β-catenin signaling pathway plays a vital role in regulating cellular proliferation, cell fate decision, and organ development.53–55 It has been well understood that Wnt signals modulate β-catenin expression and activate a higher level expression of integrins.46 We observed an increase in expression levels of ECM and integrins in hESCs grown on membrane substrates. The upregulation of these gene expressions contributed to the improvement of cell attachment and proliferation. The role of Wnt/β-catenin signaling in regulating stem cell self-renewal has been revealed in many other studies.56,57 The Wnt/β-catenin pathway is found to be active in hematopoietic stem cells (HSCs), and is required for self-renewal of neural stem cells, limbal stem cells, and leukemia stem cells.58–60 Overexpression of β-catenin in HSCs enables cells to be expanded in long-term cultures.61
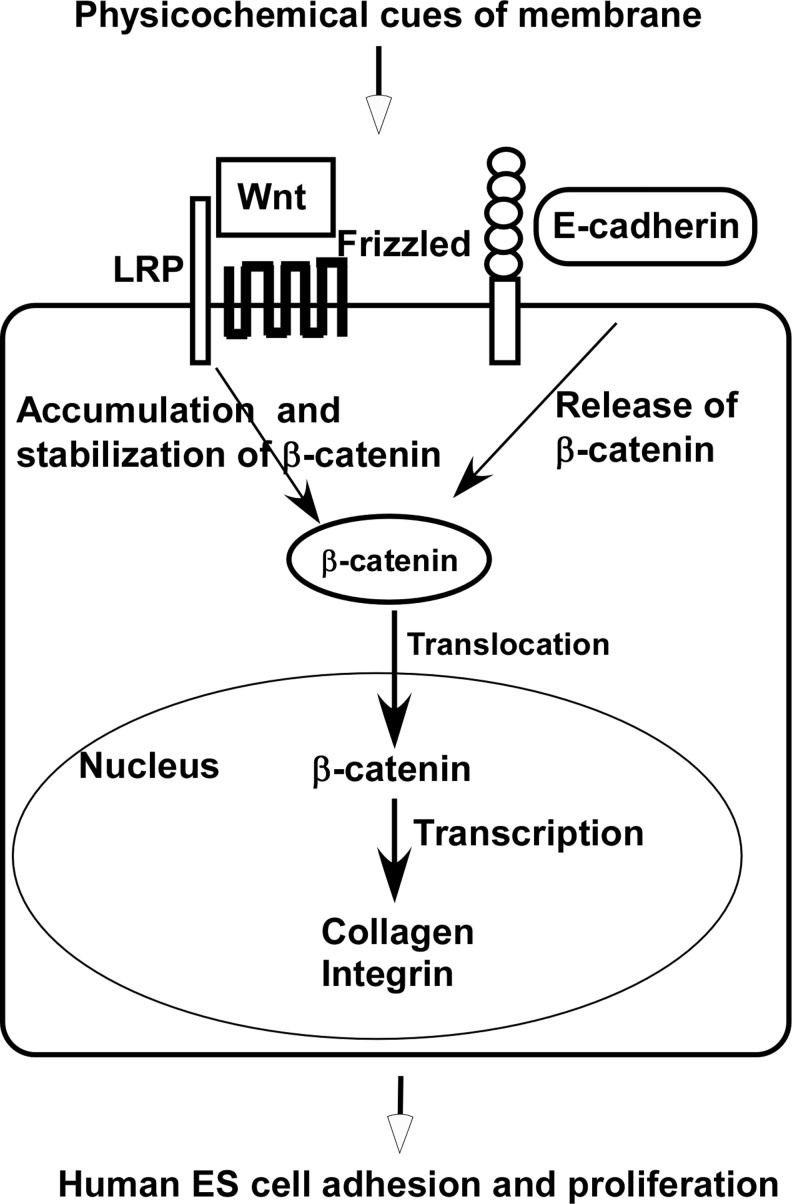
β-Catenin exists in both cytoplasm and nucleus. In the absence of Wnt signals, cytoplasmic β-catenin is phosphorylated and trapped in a multiprotein complex that includes casein kinase-1 α, glucogen synthesis kinase-3, and tumor suppressors adenomatous polyposis coli and AXIN.53,62 The phosphorylated β-catenin is rapidly degraded.63–65 In the presence of Wnt signals, β-catenin is translocated from the cytoplasm to the nucleus where it interacts with T-cell factor/lymphoid enhancer factor family transcription factors to activate the transcription of target genes for regulating cell growth and differentiation.66 A number of recent studies revealed that the Wnt signaling enhances hESC self-renewal and differentiation.67 For example, the Wnt signaling pathway has been proven to play a critical role in maintaining hESCs in undifferentiated state. It is shown that enhancing Wnt signaling results in a considerable increase in the number of undifferentiated hESC colonies formed on substrates.68 In another study, maintenance of pluripotent hESCs can be achieved by activating the Wnt signaling pathway using a GSK-3-specific inhibitor.69 It was also discovered that some small molecules such as activin A can upregulate the Wnt signaling pathway, promoting self-renewal of hESCs.70 In this study, we observed significant enhancement of β-catenin translocation from the cytoplasm to the nucleus in hESCs grown on membrane substrates. It appears that the enhancement of Wnt signaling resulted in more rapid cell growth and better DE differentiation. We demonstrated herein that a porous membrane structure of a membrane substrate can stimulate the canonical Wnt signaling pathway, which improves the hESC proliferation and differentiation. We speculate that the topographic structure of the porous membrane triggers biomechanical stimuli that activate Wnt signaling pathway in hESCs grown on membrane substrates. A hypothesized pathway regulation is shown in Figure 6. We reasoned that physicochemical cues offer by a membrane substrate activate the translocation of β-catenin from cytoplasm to nucleus, thereby enhancing the Wnt signaling. This observation will have significant implications for hESC lineage differentiation. For example, Wnt signals have been shown to induce the expression of proglucagon gene, which cascades the glucagon-like peptide-1 expression.71,72 This cascaded induction promotes β-cell proliferation and insulin production, while inhibiting β-cell apoptosis in a mature pancreas. Wnt signaling can also stimulate islet β-cell proliferation. Knockdown Wnt protein decreases β-cell proliferation and mutation of genes associated to Wnt signaling increases the susceptibility of type 2 diabetes.73–75 Thus, the enhancement of Wnt signaling by membrane substrates can potentially improve hESC pancreatic differentiation for producing glucose-responsive, insulin-secreting β-cells.
FIG. 6.
Enhanced Wnt signaling in hESCs grown on a membrane substrate. Physicochemical cues of membrane substrates stimulate the binding of Wnt ligand to its receptor complex, resulting in stabilization and nuclear translocation of β-catenin, which triggers upregulation of collagen and integrin expression in hESCs grown on a membrane substrate, enhancing cell proliferation and differentiation.
In addition, we observed the upregulation of MMP family proteins in hESCs grown on membrane substrates. MMP proteins are involved in controlling the kinetics and the breakdown of ECM in many physiological processes, such as embryonic development and tissue remodeling. The upregulation of these protein expressions contributes to the improvement of hESC attachment and spreading on membrane substrates. In our experiments, we revealed the formation of large colonies when hESCs were grown on membrane substrates. It has been well documented that hESCs proliferate better when they form colonies. The characterization of the mechanical properties of membrane substrates revealed the different stiffness of two membrane substrates tested in this work. The Young's moduli of the PE and PET membrane substrates with pore size of 0.4 μm were 1.280±0.033 GPa and 0.700±0.061 GPa (Table 3), respectively. Indeed, induced hESC differentiation can be greatly promoted on PE membrane substrates, suggesting the effect of membrane stiffness on hESC differentiation. It is noteworthy to point out that three-pore-size (3, 1, and 0.4 μm) membrane substrates were tested in this work due to the availability of membrane substrates. However, the data gained from these experiments clearly demonstrated the advantage of membrane substrates over the conventional TCP substrate for improving hESC attachment, proliferation, and lineage-specific differentiation.
Table 3.
Comparison of Mechanical Properties of Polyethylene Terephthalate and Polyester Membrane Substratesa
| PET2 (pore size: 0.4 μm) | PE (pore size: 0.4 μm) | |
|---|---|---|
| Hardness (MPa) | 115.3±8.95 | 273.6±15.12 |
| Modulus (GPa) | 0.700±0.061 | 1.280±0.033 |
The indentation tests were performed six times for every sample.
PE, polyester; PET, polyethylene terephthalate.
Another advantage of using a membrane substrate for hESC growth and differentiation is its coculture capability. The coculture of hESCs with other signaling cells can potentially elevate the maturity of cells differentiated from hESCs. For example, the coculture of mouse embryoid bodies (EBs) with human microvascular endothelial cells (hMECs) shows a significant increase in the expression level of pancreatic marker genes, such as Pdx1, Ngn3, Nkx6, proinsulin, GLUT-2, and Pft1a, at the interfaces between the EBs and hMECs.76 The communication between islet cells and paracrine signals exchanged between endocrine and nonendocrine cells has been found to be crucial to β-cell differentiation, maturation, and homeostasis.77,78 Such cell–cell interaction can readily be achieved in a membrane culture system. Both hESCs and signaling cells can be cultured on two sides of the membrane, or one on the membrane and the other on a solid plate that supports the membrane. The transportable and permeable capabilities of a membrane substrate facilitate cell–cell interaction and promote cell lineage differentiation and maturation.
Acknowledgments
This work was partly supported by the NSF grant CBET 0756455, Juvenile Diabetes Research Foundation (JDRF) grant 5-2009-381, Arkansas Biosciences Institute grant 0152-27504-231108, and NIH funding 1P30RR031154-01 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH). Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NCRR or NIH.
Disclosure Statement
No competing financial interests exist.
References
- 1.Bershadsky A.D. Balaban N.Q. Geiger B. Adhesion-dependent cell mechanosensitivity. Annu Rev Cell Dev Biol. 2003;19:677. doi: 10.1146/annurev.cellbio.19.111301.153011. [DOI] [PubMed] [Google Scholar]
- 2.Katsumi A. Orr A.W. Tzima E. Schwartz M.A. Integrins in mechanotransduction. J Biol Chem. 2004;279:12001. doi: 10.1074/jbc.R300038200. [DOI] [PubMed] [Google Scholar]
- 3.Stupack D.G. The biology of integrins. Oncology (Williston Park) 2007;21:6. [PubMed] [Google Scholar]
- 4.Geiger B. Bershadsky A. Assembly and mechanosensory function of focal contacts. Curr Opin Cell Biol. 2001;13:584. doi: 10.1016/s0955-0674(00)00255-6. [DOI] [PubMed] [Google Scholar]
- 5.Balaban N.Q. Schwarz U.S. Riveline D. Goichberg P. Tzur G. Sabanay I., et al. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat Cell Biol. 2001;3:466. doi: 10.1038/35074532. [DOI] [PubMed] [Google Scholar]
- 6.Geiger B. Bershadsky A. Pankov R. Yamada K.M. Transmembrane crosstalk between the extracellular matrix—cytoskeleton crosstalk. Nat Rev Mol Cell Biol. 2001;2:793. doi: 10.1038/35099066. [DOI] [PubMed] [Google Scholar]
- 7.Zamir E. Geiger B. Molecular complexity and dynamics of cell-matrix adhesions. J Cell Sci. 2001;114:3583. doi: 10.1242/jcs.114.20.3583. [DOI] [PubMed] [Google Scholar]
- 8.von Wichert G. Jiang G. Kostic A. De Vos K. Sap J. Sheetz M.P. RPTP-alpha acts as a transducer of mechanical force on alphav/beta3-integrin-cytoskeleton linkages. J Cell Biol. 2003;161:143. doi: 10.1083/jcb.200211061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sakai T. Li S. Docheva D. Grashoff C. Sakai K. Kostka G., et al. Integrin-linked kinase (ILK) is required for polarizing the epiblast, cell adhesion, and controlling actin accumulation. Genes Dev. 2003;17:926. doi: 10.1101/gad.255603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gillespie P.G. Walker R.G. Molecular basis of mechanosensory transduction. Nature. 2001;413:194. doi: 10.1038/35093011. [DOI] [PubMed] [Google Scholar]
- 11.Hamill O.P. Martinac B. Molecular basis of mechanotransduction in living cells. Physiol Rev. 2001;81:685. doi: 10.1152/physrev.2001.81.2.685. [DOI] [PubMed] [Google Scholar]
- 12.Yeung T. Georges P.C. Flanagan L.A. Marg B. Ortiz M. Funaki M., et al. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil Cytoskeleton. 2005;60:24. doi: 10.1002/cm.20041. [DOI] [PubMed] [Google Scholar]
- 13.Dalby M.J. Riehle M.O. Johnstone H. Affrossman S. Curtis A.S. In vitro reaction of endothelial cells to polymer demixed nanotopography. Biomaterials. 2002;23:2945. doi: 10.1016/s0142-9612(01)00424-0. [DOI] [PubMed] [Google Scholar]
- 14.Markert L.D. Lovmand J. Foss M. Lauridsen R.H. Lovmand M. Fuchtbauer E.M., et al. Identification of distinct topographical surface microstructures favoring either undifferentiated expansion or differentiation of murine embryonic stem cells. Stem Cells Dev. 2009;18:1331. doi: 10.1089/scd.2009.0114. [DOI] [PubMed] [Google Scholar]
- 15.Kantawong F. Burgess K.E. Jayawardena K. Hart A. Riehle M.O. Oreffo R.O., et al. Effects of a surface topography composite with puerariae radix on human STRO-1-positive stem cells. Acta Biomater. 2010;6:3694. doi: 10.1016/j.actbio.2010.02.038. [DOI] [PubMed] [Google Scholar]
- 16.Berry C.C. Campbell G. Spadiccino A. Robertson M. Curtis A.S. The influence of microscale topography on fibroblast attachment and motility. Biomaterials. 2004;25:5781. doi: 10.1016/j.biomaterials.2004.01.029. [DOI] [PubMed] [Google Scholar]
- 17.McNamara L.E. McMurray R.J. Biggs M.J.P. Kantawong F. Oreffo R.O.C. Dalby M.J. Nanotopographical control of stem cell differentiation. J Tissue Eng. 2010;2010:1. doi: 10.4061/2010/120623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.You M.H. Kwak M.K. Kim D.H. Kim K. Levchenko A. Kim D.Y., et al. Synergistically enhanced osteogenic differentiation of human mesenchymal stem cells by culture on nanostructured surfaces with induction media. Biomacromolecules. 2010;11:1856. doi: 10.1021/bm100374n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steedman M.R. Tao S.L. Klassen H. Desai T.A. Enhanced differentiation of retinal progenitor cells using microfabricated topographical cues. Biomed Microdevices. 2010;12:363. doi: 10.1007/s10544-009-9392-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chua K.N. Chai C. Lee P.C. Tang Y.N. Ramakrishna S. Leong K.W., et al. Surface-aminated electrospun nanofibers enhance adhesion and expansion of human umbilical cord blood hematopoietic stem/progenitor cells. Biomaterials. 2006;27:6043. doi: 10.1016/j.biomaterials.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 21.Anselme K. Bigerelle M. Noel B. Dufresne E. Judas D. Iost A., et al. Qualitative and quantitative study of human osteoblast adhesion on materials with various surface roughnesses. J Biomed Mater Res. 2000;49:155. doi: 10.1002/(sici)1097-4636(200002)49:2<155::aid-jbm2>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 22.Zhu X. Chen J. Scheideler L. Reichl R. Geis-Gerstorfer J. Effects of topography and composition of titanium surface oxides on osteoblast responses. Biomaterials. 2004;25:4087. doi: 10.1016/j.biomaterials.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 23.Ahmad M. Gawronski D. Blum J. Goldberg J. Gronowicz G. Differential response of human osteoblast-like cells to commercially pure (cp) titanium grades 1 and 4. J Biomed Mater Res. 1999;46:121. doi: 10.1002/(sici)1097-4636(199907)46:1<121::aid-jbm14>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 24.Webster T.J. Siegel R.W. Bizios R. Osteoblast adhesion on nanophase ceramics. Biomaterials. 1999;20:1221. doi: 10.1016/s0142-9612(99)00020-4. [DOI] [PubMed] [Google Scholar]
- 25.Fan Y.W. Cui F.Z. Hou S.P. Xu Q.Y. Chen L.N. Lee I.S. Culture of neural cells on silicon wafers with nano-scale surface topograph. J Neurosci Methods. 2002;120:17. doi: 10.1016/s0165-0270(02)00181-4. [DOI] [PubMed] [Google Scholar]
- 26.D'Angelo F. Armentano I. Mattioli S. Crispoltoni L. Tiribuzi R. Cerulli G.G., et al. Micropatterned hydrogenated amorphous carbon guides mesenchymal stem cells towards neuronal differentiation. Eur Cell Mater. 2010;20:231. doi: 10.22203/ecm.v020a19. [DOI] [PubMed] [Google Scholar]
- 27.Dickinson L.E. Kusuma S. Gerecht S. Reconstructing the differentiation niche of embryonic stem cells using biomaterials. Macromol Biosci. 2011;11:36. doi: 10.1002/mabi.201000245. [DOI] [PubMed] [Google Scholar]
- 28.Gerecht S. Bettinger C.J. Zhang Z. Borenstein J.T. Vunjak-Novakovic G. Langer R. The effect of actin disrupting agents on contact guidance of human embryonic stem cells. Biomaterials. 2007;28:4068. doi: 10.1016/j.biomaterials.2007.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McBeath R. Pirone D.M. Nelson C.M. Bhadriraju K. Chen C.S. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6:483. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 30.Park J.Y. Takayama S. Lee S.H. Regulating microenvironmental stimuli for stem cells and cancer cells using microsystems. Integr Biol (Camb) 2010;2:229. doi: 10.1039/c000442a. [DOI] [PubMed] [Google Scholar]
- 31.Luo W. Jones S.R. Yousaf M.N. Geometric control of stem cell differentiation rate on surfaces. Langmuir. 2008;24:12129. doi: 10.1021/la802836g. [DOI] [PubMed] [Google Scholar]
- 32.Ruiz S.A. Chen C.S. Emergence of patterned stem cell differentiation within multicellular structures. Stem Cells. 2008;26:2921. doi: 10.1634/stemcells.2008-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teo B.K. Ankam S. Chan L.Y. Yim E.K. Nanotopography/mechanical induction of stem-cell differentiation. Methods Cell Biol. 2010;98:241. doi: 10.1016/S0091-679X(10)98011-4. [DOI] [PubMed] [Google Scholar]
- 34.Abrams G.A. Goodman S.L. Nealey P.F. Franco M. Murphy C.J. Nanoscale topography of the basement membrane underlying the corneal epithelium of the rhesus macaque. Cell Tissue Res. 2000;299:39. doi: 10.1007/s004419900074. [DOI] [PubMed] [Google Scholar]
- 35.Abrams G.A. Schaus S.S. Goodman S.L. Nealey P.F. Murphy C.J. Nanoscale topography of the corneal epithelial basement membrane and Descemet's membrane of the human. Cornea. 2000;19:57. doi: 10.1097/00003226-200001000-00012. [DOI] [PubMed] [Google Scholar]
- 36.Ma T. Li Y. Yang S.T. Kniss D.A. Effects of pore size in 3-D fibrous matrix on human trophoblast tissue development. Biotechnol Bioeng. 2000;70:606. doi: 10.1002/1097-0290(20001220)70:6<606::aid-bit2>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 37.Idelson M. Alper R. Obolensky A. Ben-Shushan E. Hemo I. Yachimovich-Cohen N., et al. Directed differentiation of human embryonic stem cells into functional retinal pigment epithelium cells. Cell Stem Cell. 2009;5:396. doi: 10.1016/j.stem.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 38.Metallo C.M. Vodyanik M.A. de Pablo J.J. Slukvin II. Palecek S.P. The response of human embryonic stem cell-derived endothelial cells to shear stress. Biotechnol Bioeng. 2008;100:830. doi: 10.1002/bit.21809. [DOI] [PubMed] [Google Scholar]
- 39.Grobstein C. Morphogenetic interaction between embryonic mouse tissues separated by a membrane filter. Nature. 1953;172:869. doi: 10.1038/172869a0. [DOI] [PubMed] [Google Scholar]
- 40.Jiang J. Au M. Lu K. Eshpeter A. Korbutt G. Fisk G., et al. Generation of insulin-producing islet-like clusters from human embryonic stem cells. Stem Cells. 2007;25:1940. doi: 10.1634/stemcells.2006-0761. [DOI] [PubMed] [Google Scholar]
- 41.Jin S. Chen C. Montelaro R.C. Equine infectious anemia virus Gag p9 function in early steps of virus infection and provirus production. J Virol. 2005;79:8793. doi: 10.1128/JVI.79.14.8793-8801.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu Y. Dong Z. Wejinya U.C. Jin S. Ye K. Nanoindentation and Mechanical Properties Investigation of Soft Tissue Scaffolds by Atomic Force Microscopy. J Biomech. 2011;44:2356. doi: 10.1016/j.jbiomech.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 43.Jin S. Issel C.J. Montelaro R.C. Serological method using recombinant S2 protein to differentiate equine infectious anemia virus (EIAV)-infected and EIAV-vaccinated horses. Clin Diagn Lab Immunol. 2004;11:1120. doi: 10.1128/CDLI.11.6.1120-1129.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goodison S. Urquidi V. Tarin D. CD44 cell adhesion molecules. Mol Pathol. 1999;52:189. doi: 10.1136/mp.52.4.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sachlos E. Auguste D.T. Embryoid body morphology influences diffusive transport of inductive biochemicals: a strategy for stem cell differentiation. Biomaterials. 2008;29:4471. doi: 10.1016/j.biomaterials.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 46.Gradl D. Kuhl M. Wedlich D. The Wnt/Wg signal transducer beta-catenin controls fibronectin expression. Mol Cell Biol. 1999;19:5576. doi: 10.1128/mcb.19.8.5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He X. Semenov M. Tamai K. Zeng X. LDL receptor-related proteins 5 and 6 in Wnt/beta-catenin signaling: arrows point the way. Development. 2004;131:1663. doi: 10.1242/dev.01117. [DOI] [PubMed] [Google Scholar]
- 48.De Langhe S.P. Sala F.G. Del Moral P.M. Fairbanks T.J. Yamada K.M. Warburton D., et al. Dickkopf-1 (DKK1) reveals that fibronectin is a major target of Wnt signaling in branching morphogenesis of the mouse embryonic lung. Dev Biol. 2005;277:316. doi: 10.1016/j.ydbio.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 49.Baarsma H.A. Spanjer A.I. Haitsma G. Engelbertink L.H. Meurs H. Jonker M.R., et al. Activation of WNT/beta-catenin signaling in pulmonary fibroblasts by TGF-beta is increased in chronic obstructive pulmonary disease. PLoS One. 2011;6:e25450. doi: 10.1371/journal.pone.0025450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Doyle J.L. Haas T.L. Differential role of beta-catenin in VEGF and histamine-induced MMP-2 production in microvascular endothelial cells. J Cell Biochem. 2009;107:272. doi: 10.1002/jcb.22123. [DOI] [PubMed] [Google Scholar]
- 51.Brabletz T. Jung A. Dag S. Hlubek F. Kirchner T. beta-catenin regulates the expression of the matrix metalloproteinase-7 in human colorectal cancer. Am J Pathol. 1999;155:1033. doi: 10.1016/s0002-9440(10)65204-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crossin K.L. Krushel L.A. Cellular signaling by neural cell adhesion molecules of the immunoglobulin superfamily. Dev Dyn. 2000;218:260. doi: 10.1002/(SICI)1097-0177(200006)218:2<260::AID-DVDY3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 53.Wang H.X. Tekpetey F.R. Kidder G.M. Identification of WNT/beta-CATENIN signaling pathway components in human cumulus cells. Mol Hum Reprod. 2009;15:11. doi: 10.1093/molehr/gan070. [DOI] [PubMed] [Google Scholar]
- 54.Hoppler S. Kavanagh C.L. Wnt signalling: variety at the core. J Cell Sci. 2007;120:385. doi: 10.1242/jcs.03363. [DOI] [PubMed] [Google Scholar]
- 55.Wodarz A. Nusse R. Mechanisms of Wnt signaling in development. Annu Rev Cell Dev Biol. 1998;14:59. doi: 10.1146/annurev.cellbio.14.1.59. [DOI] [PubMed] [Google Scholar]
- 56.Reya T. Regulation of hematopoietic stem cell self-renewal. Recent Prog Horm Res. 2003;58:283. doi: 10.1210/rp.58.1.283. [DOI] [PubMed] [Google Scholar]
- 57.Nusse R. Wnt signaling and stem cell control. Cell Res. 2008;18:523. doi: 10.1038/cr.2008.47. [DOI] [PubMed] [Google Scholar]
- 58.Kalani M.Y. Cheshier S.H. Cord B.J. Bababeygy S.R. Vogel H. Weissman I.L., et al. Wnt-mediated self-renewal of neural stem/progenitor cells. Proc Natl Acad Sci U S A. 2008;105:16970. doi: 10.1073/pnas.0808616105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakatsu M.N. Ding Z. Ng M.Y. Truong T.T. Yu F. Deng S.X. Wnt/{beta}-catenin signaling regulates proliferation of human cornea epithelial stem/progenitor cells. Invest Ophthalmol Vis Sci. 2011;52:4734. doi: 10.1167/iovs.10-6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Y. Krivtsov A.V. Sinha A.U. North T.E. Goessling W. Feng Z., et al. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010;327:1650. doi: 10.1126/science.1186624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reya T. Duncan A.W. Ailles L. Domen J. Scherer D.C. Willert K., et al. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003;423:409. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- 62.Hendriksen J. Fagotto F. van der Velde H. van Schie M. Noordermeer J. Fornerod M. RanBP3 enhances nuclear export of active (beta)-catenin independently of CRM1. J Cell Biol. 2005;171:785. doi: 10.1083/jcb.200502141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sakanaka C. Weiss J.B. Williams L.T. Bridging of beta-catenin and glycogen synthase kinase-3beta by axin and inhibition of beta-catenin-mediated transcription. Proc Natl Acad Sci U S A. 1998;95:3020. doi: 10.1073/pnas.95.6.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gordon M.D. Nusse R. Wnt signaling: multiple pathways, multiple receptors, and multiple transcription factors. J Biol Chem. 2006;281:22429. doi: 10.1074/jbc.R600015200. [DOI] [PubMed] [Google Scholar]
- 65.Habas R. Dawid I.B. Dishevelled and Wnt signaling: is the nucleus the final frontier? J Biol. 2005;4:2. doi: 10.1186/jbiol22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Katoh M. WNT signaling pathway and stem cell signaling network. Clin Cancer Res. 2007;13:4042. doi: 10.1158/1078-0432.CCR-06-2316. [DOI] [PubMed] [Google Scholar]
- 67.Kikuchi A. Yamamoto H. Sato A. Matsumoto S. New insights into the mechanism of Wnt signaling pathway activation. Int Rev Cell Mol Biol. 2011;291:21. doi: 10.1016/B978-0-12-386035-4.00002-1. [DOI] [PubMed] [Google Scholar]
- 68.Villa-Diaz L.G. Garcia-Perez J.L. Krebsbach P.H. Enhanced transfection efficiency of human embryonic stem cells by the incorporation of DNA liposomes in extracellular matrix. Stem Cells Dev. 2010;19:1949. doi: 10.1089/scd.2009.0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sato N. Meijer L. Skaltsounis L. Greengard P. Brivanlou A.H. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 70.Xiao L. Yuan X. Sharkis S.J. Activin A maintains self-renewal and regulates fibroblast growth factor, Wnt, and bone morphogenic protein pathways in human embryonic stem cells. Stem Cells. 2006;24:1476. doi: 10.1634/stemcells.2005-0299. [DOI] [PubMed] [Google Scholar]
- 71.Nostro M.C. Sarangi F. Ogawa S. Holtzinger A. Corneo B. Li X., et al. Stage-specific signaling through TGF{beta} family members and WNT regulates patterning and pancreatic specification of human pluripotent stem cells. Development. 2010;138:861. doi: 10.1242/dev.055236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu Z. Habener J.F. Glucagon-like peptide-1 activation of TCF7L2-dependent Wnt signaling enhances pancreatic beta cell proliferation. J Biol Chem. 2008;283:8723. doi: 10.1074/jbc.M706105200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bordonaro M. Role of Wnt signaling in the development of type 2 diabetes. Vitam Horm. 2009;80:563. doi: 10.1016/S0083-6729(08)00619-5. [DOI] [PubMed] [Google Scholar]
- 74.Rulifson I.C. Karnik S.K. Heiser P.W. ten Berge D. Chen H. Gu X., et al. Wnt signaling regulates pancreatic beta cell proliferation. Proc Natl Acad Sci U S A. 2007;104:6247. doi: 10.1073/pnas.0701509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu Z. Habener J.F. Wnt signaling in pancreatic islets. Adv Exp Med Biol. 2010;654:391. doi: 10.1007/978-90-481-3271-3_17. [DOI] [PubMed] [Google Scholar]
- 76.Talavera-Adame D. Wu G. He Y. Ng T.T. Gupta A. Kurtovic S., et al. Endothelial cells in co-culture enhance embryonic stem cell differentiation to pancreatic progenitors and insulin-producing cells through BMP signaling. Stem Cell Rev. 2011;7:532. doi: 10.1007/s12015-011-9232-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Eberhard D. Lammert E. The pancreatic beta-cell in the islet and organ community. Curr Opin Genet Dev. 2009;19:469. doi: 10.1016/j.gde.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 78.Zhao Y. Xu J. Wei J. Li J. Cai J. Miao G. Preservation of islet survival by upregulating alpha3 integrin signaling: the importance of 3-dimensional islet culture in basement membrane extract. Transplant Proc. 2010;42:4638. doi: 10.1016/j.transproceed.2010.09.154. [DOI] [PubMed] [Google Scholar]