Abstract
Whole gene duplications and triplications of alpha-synuclein (SNCA) can cause Parkinson’s disease (PD), and variation in the promoter region (Rep1) and 3′ region of SNCA has been reported to increase disease susceptibility. Within our cohort, one affected individual from each of 92 multiplex PD families showing the greatest evidence of linkage to the region around SNCA was screened for dosage alterations and sequence changes; no dosage or non-synonymous sequence changes were found. In addition, 737 individuals (from 450 multiplex PD families) that met strict diagnostic criteria for PD and did not harbor a known causative mutation, as well as 359 neurologically normal controls, were genotyped for the Rep1 polymorphism and four SNPs in the 3′ region of SNCA. The four SNPs were in high LD (r2 > 0.95) and were analyzed as a haplotype. The effects of the Rep1 genotype and the 3′ haplotype were evaluated using regression models employing only one individual per family. Cases had a 3% higher frequency of the Rep1 263 bp allele compared with controls (OR = 1.54; empirical P-value = 0.02). There was an inverse linear relationship between the number of 263 bp alleles and age of onset (empirical P-value = 0.0004). The 3′ haplotype was also associated with disease (OR = 1.29; empirical P-value = 0.01), but not age of onset (P = 0.40). These data suggest that dosage and sequence changes are a rare cause of PD, but variation in the promoter and 3′ region of SNCA convey an increased risk for PD.
Keywords: Parkinson’s disease, alpha-synuclein, dosage, Rep1, association
Mutations in five genes have been found to cause Parkinson’s disease (PD) in a small subset of patients with disease (<5% of cases). One of these genes, alpha-synuclein (SNCA), has been reported to act as both a causative and a susceptibility gene for PD. Missense mutations,1–3 as well as whole gene dosage changes,4–6 segregate with disease in an autosomal pattern of inheritance. Variation in the promoter region of SNCA, specifically the dinucleotide repeat polymorphism known as Rep1, has been reported to increase the risk for PD.7 A meta-analysis has further bolstered evidence that Rep1 is associated with a slight, but significant, increase in the risk of PD.8 Variation in the 3′ region of SNCA has also been associated with risk for PD.9
The two domains flanking the Rep1 repeat appear to interact with each other to enhance expression of SNCA, while Rep1 acts as a negative modulator.10 In addition, different alleles can vary the expression levels of SNCA in SH-SY5Y cells by up to threefold.10 There is also evidence from a genomewide study of gene expression that variation at the 3′ end of the gene can affect expression levels of SNCA.11 Because a triplication or a duplication of the normal gene can lead to a highly penetrant form of PD, and since a triplication (four copies of the gene) tends to lead to an earlier disease onset than a duplication (three copies of the gene), it is possible that even a subtle increase in expression could, over the course of many decades, predispose an individual to develop PD.
The purpose of this study is to test in familial PD whether: (1) dosage and coding changes in SNCA are a more frequent cause of PD than in sporadic cases; (2) Rep1 allele-length variability is associated with PD susceptibility or age of onset; and (3) variation in the 3′ region of SNCA is associated with PD susceptibility or age of onset.
SUBJECTS AND METHODS
Subjects
Multiplex PD families were ascertained through a pair of siblings reported to have PD. A total of 1,325 individuals with PD from 566 multiplex PD families were recruited. All available affected individuals were seen by a movement disorder specialist at one of 59 Parkinson Study Group sites. A uniform clinical assessment was completed that included the Unified Parkinson’s Disease Rating Scale (UPDRS)12 Parts II & III. A diagnostic checklist with inclusion criteria consisting of clinical features highly associated with autopsy-confirmed PD and exclusion criteria highly associated with non-PD pathological diagnoses was used to classify individuals as having either verified PD (65%) or non-verified PD (35%).13 Peripheral blood was obtained from all individuals after appropriate written informed consent approved by each institution’s review board was completed.
All samples were previously genotyped for the LRRK2 G2019S mutation,14 and a subset (43%) were screened for PRKN mutations.15 Individuals with either two PRKN mutations (n = 36) or at least one LRRK2 mutation (n = 42) were removed from all further analyses. Only samples from self-reported Caucasian, non-Hispanic subjects were included in statistical analyses. Data from a previously completed genome screen16 were used to identify the 92 families demonstrating the greatest evidence of linkage (NPL > 1.0) to the region containing SNCA. One individual from each of these were then screened for dosage and sequence changes in SNCA. The control sample consisted of 359 neurologically normal, non-Hispanic Caucasians who provided appropriate written informed consent (see Table 1).
TABLE 1.
Demographics
| Source | Type | N | Mean age at onset/exam (range) | Male (%) |
|---|---|---|---|---|
| PROGENI (samples reported to have PD) | Cases | 1,325 | 60.9 (18–89) | 58.0 |
| PROGENI (Caucasian Non-Hispanic with verified PD and no known causative mutation) | Cases | 737 | 61.8 (18–84) | 59.9 |
| PROGENI-CARES | Controls | 46 | 68.4 (55–82) | 19.6 |
| National Cell Repository for Alzheimer’s Disease | Controls | 44 | 76.9 (58–92) | 43.2 |
| NINDS Human Genetics Resource Center (Coriell) | Controls | 269 | 69.5 (55–88) | 48.0 |
Molecular Analysis
Multiplex ligation-dependent probe amplification (MLPA) was performed with 100 ng of genomic DNA according to manufacturer’s instructions using the P051 Salsa MLPA Parkinson probe set (MRC-Holland, Amsterdam, The Netherlands). This probe set includes probes for all SNCA exons except exon 2 and also contains a mutation-specific probe for the A30P mutation. Probe amplification products were run on an ABI 3730xl DNA Analyzer using GS500 size standard (Applied Biosystems, Foster City, CA). MLPA peak plots were visualized using Genemapper Software version 3.7 (Applied Biosystems, Foster City, CA) and analyzed as previously described.17 PCR and sequencing primers were designed using the chromosome 4 genomic contig sequence NC_000004.10 enabling PCR/sequencing of all coding exons and intron/exon boundaries of SNCA. PCR products were purified and sequenced as previously described.18
Screening of Rep1 and the four most significant SNPs identified by Mueller et al.9 was performed in the multiplex PD families and in the 359 controls. The SNCA Rep1 allele-length variants were genotyped as follows: 50 ng of genomic DNA was polymerase chain reaction (PCR)—amplified using fluorescently labeled forward and reverse primers Fam5′-GACTGGCCCAA GATTAACCA-3′ and 5′-CCTGGCATATTTGATTG CAA-3′. The resulting PCR products were evaluated on an ABI 3730xl DNA Analyzer. Allelic sizes were assessed using GeneMapper version 4.0 software (Applied Biosytems, Foster City, CA). TaqMan allelic-discrimination assays (Applied Biosystems, Foster City, CA) were developed to genotype four SNPs in the 3′ region of the gene (rs356219, rs356220, rs356165, rs356203) as previously described.18
Statistical Analysis
Rep1 has three predominant alleles, with sizes 259 bp, 261 bp, and 263 bp. As done previously,7,8 rare alleles (frequencies <0.1%) were excluded from analyses. Allele frequencies and genotype frequencies were weighted using one individual per family to create a representative sample. This distribution was then evaluated for deviations from Hardy-Weinberg equilibrium, which can indicate the presence of genotyping error.
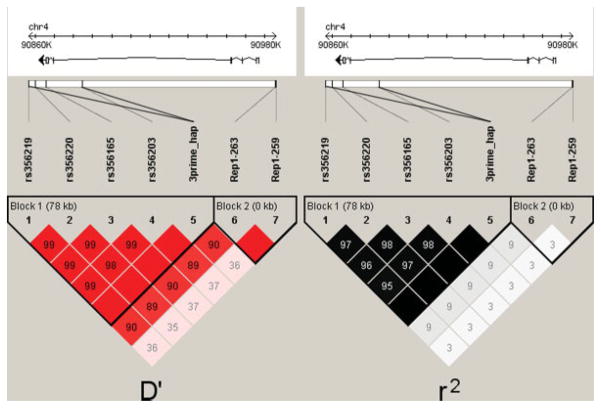
Linkage disequilibrium (LD) was assessed using Haploview (Fig. 1).19 Because of limitations in calculating pairwise LD, the Rep1 263 and 259 alleles were separated to create a pair of two-allele systems. The four SNPs (rs356219|rs356220|rs356165|rs356203) in the 3′ region of SNCA were in high linkage disequilibrium (0.95 < r2 < 0.99) and therefore, were analyzed as a haplotype.20 The two most common haplotypes (GTGC, ACAT) accounted for 99% of the observed haplotypes. Those individuals carrying any other haplotype(s) were removed from analysis.
FIG. 1.
Linkage disequilibrium between Rep1 and the 3′ SNPs. The gene structure of SNCA is based on transcript NM_000345, isoform NACP140. The direction of transcription is indicated by an arrow, which explains why the promoter is on the right side and the 3′ end of the gene is on the left side. The size of the gene is indicated by the ruler at the top of the figure. The pairwise linkage disequilibrium (D′ on the left side; r2 on the right side) estimates for the genotyped SNPs are represented by the boxes. Darkly shaded boxes have strong evidence of LD, whereas lightly shaded boxes have lower evidence of LD. Boxes without numbers are in complete LD (100%). Note that the 263 and 259 alleles have been separated as two distinct alleles for the purpose of this diagram. Similarly, the two major haplotypes were incorporated as a two-allele system (which means that the information from these SNPs is presented twice – once individually and once collectively as a haplotype).
Statistical analyses were limited to the 737 PD non-Hispanic Caucasian cases meeting the strictest diagnostic criteria (verified PD) and that did not harbor a known causative PD mutation. These verified PD cases were distributed across 450 families. Two hypotheses were tested. First, odds ratios were calculated using logistic regression to test the hypothesis that particular alleles (263 bp or 259 bp for the Rep1 polymorphism; the GTGC haplotype for the 3′ region) influenced the risk for PD as compared with the common reference allele (261 bp for the Rep1 polymorphism; the ACAT haplotype for the 3′ region). Second, a linear regression model was employed to test the hypothesis that the age of PD onset was influenced by these alleles. All analyses were carried out using SAS software (release 9.13; SAS Institute, Cary, NC).
In the logistic models, affection status was the dependent variable and the independent variable(s) indicated the number of alleles harbored by each individual (0, 1, or 2). For the Rep1 model, two variables were used: one for the number of 263 bp allele and one for the number of 259 bp allele. Note that the common 261 bp allele was not excluded from the model, but rather was inferred from the other two states. Age at exam and gender were included as covariates in the initial models to see if they affected the odds ratios for the alleles of interest. To ensure statistical validity, a single individual was sampled at random from each of the families. This was repeated 50,000 times, and common resampling techniques (bootstrapping) were employed to obtain a representative value. The median t-statistic was determined for each variable, and the corresponding odds ratio and P-value are reported. This P-value, however, is conservative since information from 737 cases was used to derive the estimate but only 450 independent observations (one for each family) were used to determine the significance level. We therefore simulated the null distribution 10,000 times with the program Allegro21 using the given family structure and allele frequencies. For each replicate, the same permutation and bootstrapping methods were used to determine the representative t-statistic for that replicate. An empirical P-value was then calculated for each independent variable by determining the number of replicates with a t-statistic greater than or equal to the t-statistic observed for the real data and dividing by the total number of replicates.
To determine the effect of these alleles on age of disease onset, a linear regression model was fitted with age of onset as the dependent variable and the allele counts as independent variables (as with the logistic model, the common 261 bp allele for the Rep1 analyses was inferred, not excluded). Education, gender, and smoking were considered as possible covariates; however, those found to be nonsignificant were dropped from the final model. Similar to the approach used for the logistic regression, a single individual from each of the families was sampled at random, and 50,000 test statistics were generated. The median bootstrapped statistic was determined for each variable, and the corresponding P-values are reported. Empirical P-values were also determined for the age of onset methods by randomly permuting the genotypes between the families 10,000 times and determining the representative statistic for each replicate. Data were only swapped between families of the same size, thereby preserving the familiality of both the age of onset distribution and the familiality of the genotypes. Similar to the age of onset analyses, the association between other factors (i.e. disease duration, disease severity) and Rep1 genotype were also considered and empirical P-values were computed in the same fashion.
RESULTS
Detailed molecular screening was performed in a single PD case from each of the 92 families having the strongest evidence of linkage to the chromosomal region containing SNCA. No dosage alterations or non-synonymous sequence variants were found in the 92 individuals who were screened.
Rep1
Using an independent representative sample, no deviation from Hardy–Weinberg equilibrium was seen in the distributions of the Rep1 genotypes (Table 2A; χ2 = 1.87; df = 3; P = 0.60). When a logistic regression model was fitted to predict affection status, there was a significant increase in the risk of PD for each copy of the 263 bp allele (OR = 1.54; P = 0.02; empirical P-value = 0.02; Table 3A). The presence of a 259 bp allele did not have a significant protective or deleterious effect on the risk for PD (OR = 0.99; P = 0.78; empirical P-value = 0.94). Age and gender did not have an effect on the magnitude or significance of these alleles.
TABLE 2.
Genotypic counts for cases and controls permuted one per family
| A: SNCA Rep1 genotype | PD cases | Controls |
|---|---|---|
| 259/259 | 30.67 (6.65%) | 23.00 (6.34%) |
| 259/261 | 148.08 (32.12%) | 137.00 (37.74%) |
| 259/263 | 20.17 (4.37%) | 9.00 (2.48%) |
| 261/261 | 205.25 (44.52%) | 161.00 (44.35%) |
| 261/263 | 49.33 (10.70%) | 32.00 (8.82%) |
| 263/263 | 7.50 (1.63%) | 1.00 (0.28%) |
| Totals: | 461.00 (100.00%) | 363.00 (100.00%) |
| B: 3′ haplotype | PD cases | Controls |
|---|---|---|
| ACAT/ACAT | 142.25 (31.97%) | 121.000 (36.12%) |
| ACAT/GTGC | 211.75 (47.58%) | 172.000 (51.34%) |
| GTGC/GTGC | 91.00 (20.45%) | 42.000 (12.54%) |
| Totals: | 445.00 (100.00%) | 335.00 (100.00%) |
Table 3.
Logistic regression models for predicting affection status and linear regression models for predicting age of onset
| A: | Beta | Wald statistic | Odds ratio | Empirical P-value |
|---|---|---|---|---|
| Number of 263 bp alleles | 0.43 | 5.10 | 1.54 | 0.02 |
| Number of 259 bp alleles | −0.01 | 0.07 | 0.99 | 0.94 |
| Constant | 0.73 | |||
| Number of GTGC haplotypes | 0.25 | 5.80 | 1.29 | 0.01 |
| Constant | 0.55 |
| B: | Beta | T statistic | Empirical P-value |
|---|---|---|---|
| Number of 263 bp alleles | −3.58 | 3.17 | <0.001 |
| Number of 259 bp alleles | 0.67 | 0.87 | 0.32 |
| Constant | 62.32 | ||
| Number of GTGC haplotypes | 0.606 | 0.85 | 0.40 |
| Constant | 62.30 |
Subsequently, we tested the hypothesis that Rep1 genotype is associated with the age of disease onset. An inverse linear relationship between the number of 263 bp alleles and age of onset was observed (β = −3.76; P = 0.002; empirical P-value = 0.0004; Table 3B) predicting that the age of onset will be, on average, 3.76 years earlier in those with a single 263 bp allele and 7.52 years earlier in those with two 263 bp alleles. Gender, education, and smoking did not have a major effect on the magnitude or significance of these alleles. No association was detected with the 259 bp allele.
There were no other factors that were significantly associated with the 263 bp allele after applying a Bonferroni correction for multiple tests (14 tests would require an α of 0.004) (Table 4A). Nevertheless, individuals with a 263 allele tended to have more severe scores on the Geriatric Depression Scale and on the UPDRS Part II (Activities of Daily Living).
Table 4.
Distribution of PD-related factors (permuted one per family) based upon the presence of either the 263 bp allele or the GTGC haplotype
| A: | X/X | X/263 | 263/263 | P-value |
|---|---|---|---|---|
| Percentage of cases | 83.7% | 14.8% | 1.5% | |
| Age at exam | 71.0 | 68.7 | 65.9 | 0.008 |
| Age of onset | 62.7 | 59.8 | 50.9 | 0.0004 |
| Duration of disease | 8.3 | 8.9 | 15.1 | 0.02 |
| % with early onset (<60 years) | 38.6% | 53.2% | 73.3% | 0.0008 |
| % male | 59.6% | 54.2% | 46.7% | 0.22 |
| % with an affected parent | 28.8% | 30.6% | 42.9% | 0.43 |
| Geriatric depression scale | 8.9 | 10.6 | 9.6 | 0.03 |
| MMSE | 26.8 | 26.4 | 27.1 | 0.56 |
| Blessed functionality scale | 3.9 | 4.9 | 2.5 | 0.24 |
| Education (years) | 13.7 | 13.1 | 13.9 | 0.31 |
| Previous smoker | 42.2% | 38.9% | 46.7% | 0.73 |
| Hoehn and Yahr stage | 2.5 | 2.6 | 2.8 | 0.13 |
| UPDRS Part II (ADL) | 13.4 | 15.8 | 13.3 | 0.03 |
| UPDRS Part III (motor) | 27.4 | 31.3 | 23.8 | 0.11 |
| B: | ACAT/ACAT | ACAT/GTGC | GTGC/GTGC | P-value |
|---|---|---|---|---|
| Percentage of cases | 32.0% | 47.6% | 20.4% | |
| Age at exam | 70.9 | 70.5 | 70.0 | 0.47 |
| Age of onset | 62.4 | 62.2 | 61.1 | 0.40 |
| Duration of disease | 8.5 | 8.3 | 9.0 | 0.70 |
| % with early onset (<60 years) | 38.1% | 42.0% | 43.1% | 0.42 |
| % male | 57.9% | 60.5% | 56.0% | 0.86 |
| % with an affected parent | 39.8% | 30.7% | 36.8% | 0.46 |
| Geriatric depression scale | 8.8 | 9.2 | 9.4 | 0.45 |
| MMSE | 26.3 | 27.0 | 26.9 | 0.21 |
| Blessed functionality scale | 3.9 | 3.9 | 4.3 | 0.60 |
| Education (years) | 13.6 | 13.6 | 13.7 | 0.92 |
| Previous smoker | 41.5% | 44.9% | 35.6% | 0.50 |
| Hoehn and Yahr stage | 2.5 | 2.5 | 2.5 | 0.93 |
| UPDRS Part II (ADL) | 13.7 | 13.7 | 13.6 | 0.88 |
| UPDRS Part III (motor) | 27.6 | 27.5 | 28.5 | 0.66 |
X = Rep1 alleles other than the 263 bp allele.
3′ SNPs
Using the independent representative sample, no deviation from Hardy–Weinberg equilibrium was seen in the distributions of any of the four SNPs at the 3′ end of the gene (Table 2B; P > 0.30 for all four SNPs). There was a significant increase in the risk of PD for each copy of the deleterious haplotype (OR = 1.29; P = 0.02; empirical P-value = 0.01; Table 3B). Age and gender did not have an effect on the magnitude or significance of these alleles. The deleterious haplotype was not associated with age of onset (P > 0.40). There were no other factors that were significantly associated with the deleterious 3′ haplotype (Table 4B).
Linkage disequilibrium between the Rep1 alleles, the individual 3′ SNPs, and the 3′ haplotypes is displayed in Figure 1. There was evidence of strong LD (as measured by D′) between the Rep1 263 allele and the deleterious 3′ haplotype. However, the LD as measured by r2 is substantially lower. This is because of the substantial differences in minor allele frequencies in the two associated alleles (7% for the 263 allele and 42% for the GTGC haplotype).
DISCUSSION
We found that both the 263 bp allele in the promoter as well as a haplotype in the 3′ region of SNCA are both associated with the risk of PD. We also found the 263 bp allele was associated with an earlier age of onset (P < 0.001), with each copy of the allele predicted to lead to a decrease of 3.8 years in age of onset. A previous study by Hadjigeorgiou et al.22 also found an association of the 263 bp allele with an earlier age of onset. In contrast, a previous meta-analysis8 found the 263 bp allele to be significantly associated with disease risk but did not find this Rep I allele to be associated with an earlier age of onset. Several recent articles corroborate the association of both Rep1 and the 3′ region with disease status.23,24 Another recent manuscript found one of the 3′ SNPs studied here (rs356219) to be associated with SNCA mRNA levels in the substantia nigra and cerebellum,25 thereby providing a link between the observed association with PD and SNCA gene expression.
The reduction in age of onset associated with the 263 bp RepI allele is modest; however, any significant predictor of age of onset has potential clinical relevance for future pharmacological interventions designed to delay or prevent PD onset. Furthermore, by accounting for the variation because of Rep1 or the 3′ haplotype, the power of a study to identify additional susceptibility genes for PD is increased. Therefore, the identification of a common variant, even with only a small effect size, is likely to facilitate the identification of additional factors affecting the onset and progression of PD.
Those carrying a Rep1 263 bp allele tended to have a more severe disease phenotype, with higher UPDRS Part II subscores and higher depression scores. Depression has been associated with a more severe PD phenotype in many studies, including our own.26–28 The trend remained after controlling for the effects of PD duration and age at exam (UPDRS II (ADL) P = 0.05; UPDRS II (motor) P = 0.05). Although these findings are not statistically significant, they may be hypothesis-generating, and perhaps suggest that Rep1 is associated not only with PD susceptibility and an earlier age of onset, but also disease severity. If Rep1 genotype contributes to variation in the expression levels of SNCA, as has been hypothesized,8 and if overexpression of wild-type SNCA leads to increased aggregation and toxicity,29,30 then accelerated neurodegeneration may result in earlier onset and more severe clinical phenotype.
The linkage disequilibrium between the Rep1 263 allele and the deleterious 3′ haplotype (D′ = 0.90) is intriguing. It raises the possibility that the Rep1 263 allele is cotransmitted due to its ancestral origin with the deleterious 3′ background. If true, the extensive LD in both regions will make it difficult to discern whether there is variation in both the 5′ and 3′ regions of SNCA contributing to PD susceptibility or if the predisposing variation is located in only one of the two regions and the association observed in the other is simply because of LD between the two regions.
Advantages of our study include the use of strict diagnostic criteria that have been validated against autopsy confirmed PD and the uniform genotyping of a well-characterized sample by a single lab. One distinguishing characteristic of our study is the exclusive focus on familial PD. The meta-analysis found a similar odds ratio when they limited their analyses to the 15% of the sample that reported a family history of the disease; however, due to sample size, their finding was not statistically significant.8 Similarly, this subset of the meta-analysis sample did not show a significant relationship between Rep1 allele size and age of disease onset.8
The genotyping of our sample of PD families bolsters the evidence that variation in the promoter and/or 3′ region of SNCA is associated with risk for PD in a dose dependent fashion and that the Rep1 polymorphism is associated with age of PD onset. Although the effect sizes of these polymorphisms are small, it does fit with the common disease-common variant hypothesis, which states that genetic susceptibility to common diseases like hypertension, diabetes and PD are largely because of alleles that have moderate frequency in the population. In the future, it is likely that additional variants like Rep1 and the 3′ haplotype will be identified. While these known and yet unknown variants may individually contribute to a small proportion of disease susceptibility, collectively these variants could account for a significant fraction of disease risk.
Acknowledgments
This study was supported by the National Institutes of Health (R01 NS37167 and MO1 RR-00750), the NINDS Human Genetics Resource Center DNA and Cell Line Repository (http://ccr.coriell.org/ninds), and the National Cell Repository for Alzheimer’s Disease (U24 AG021886). We thank the subjects for their participation in this research study.
Footnotes
Potential conflict of interest: Nothing to report.
Author Roles: Nathan Pankratz contributed to the conception and design of the study, statistical analysis, drafting, editing, and revising the text. William C. Nichols contributed to the conception and design of the study, data acquisition, editing, and revising of the text. Veronika E. Elsaesser, Michael W. Pauciulo, Diane K. Marek, Cheryl A. Halter, and Joanne Wojcieszek contributed to the data acquisition and revising of the text. Alice Rudolph, Ronald F. Pfeiffer, and Tatiana Foroud contributed to the conception and design, editing and revising of the text.
References
- 1.Polymeropoulos MH, Lavedan C, Leroy E, et al. Mutation in the α-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 2.Kruger R, Kuhn W, Muller T, et al. Ala30Pro mutation in the gene encoding α-synuclein in Parkinson’s disease. Nat Genet. 1998;18:106–108. doi: 10.1038/ng0298-106. [DOI] [PubMed] [Google Scholar]
- 3.Zarranz JJ, Alegre J, Gomez-Esteban JC, et al. The new mutation, E46K, of α-synuclein causes Parkinson and Lewy body dementia. Ann Neurol. 2004;55:164–173. doi: 10.1002/ana.10795. [DOI] [PubMed] [Google Scholar]
- 4.Singleton AB, Farrer M, Johnson J, et al. α-synuclein locus triplication causes Parkinson’s disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 5.Chartier-Harlin MC, Kachergus J, Roumier C, et al. α-synuclein locus duplication as a cause of familial Parkinson’s disease. Lancet. 2004;364:1167–1169. doi: 10.1016/S0140-6736(04)17103-1. [DOI] [PubMed] [Google Scholar]
- 6.Ibanez P, Bonnet AM, Debarges B, et al. Causal relation between α-synuclein gene duplication and familial Parkinson’s disease. Lancet. 2004;364:1169–1171. doi: 10.1016/S0140-6736(04)17104-3. [DOI] [PubMed] [Google Scholar]
- 7.Kruger R, Vieira-Saecker AM, Kuhn W, et al. Increased susceptibility to sporadic Parkinson’s disease by a certain combined α-synuclein/apolipoprotein E genotype. Ann Neurol. 1999;45:611–617. doi: 10.1002/1531-8249(199905)45:5<611::aid-ana9>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 8.Maraganore DM, de Andrade M, Elbaz A, et al. Collaborative analysis of α-synuclein gene promoter variability and Parkinson disease. JAMA. 2006;296:661–670. doi: 10.1001/jama.296.6.661. [DOI] [PubMed] [Google Scholar]
- 9.Mueller JC, Fuchs J, Hofer A, et al. Multiple regions of α-synuclein are associated with Parkinson’s disease. Ann Neurol. 2005;57:535–541. doi: 10.1002/ana.20438. [DOI] [PubMed] [Google Scholar]
- 10.Chiba-Falek O, Kowalak JA, Smulson ME, Nussbaum RL. Regulation of α-synuclein expression by poly (ADP ribose) polymerase-1 (PARP-1) binding to the NACP-Rep1 polymorphic site upstream of the SNCA gene. Am J Hum Genet. 2005;76:478–492. doi: 10.1086/428655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dixon AL, Liang L, Moffatt MF, et al. A genome-wide association study of global gene expression. Nature Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 12.Fahn S, Elton RL. UPDRS Development Committee. Unified Parkinson’s disease rating scale. In: Fahn S, Marsden CD, Goldstein M, editors. Recent developments in Parkinson’s disease. New York: Macmillan Healthcare Information; 1987. pp. 153–163. [Google Scholar]
- 13.Pankratz N, Nichols WC, Uniacke SK, et al. Genome screen to identify susceptibility genes for Parkinson disease in a sample without Parkin mutations. Am J Hum Genet. 2002;71:124–135. doi: 10.1086/341282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nichols WC, Pankratz N, Hernandez D, et al. Genetic screening for a single common LRRK2 mutation in familial Parkinson’s disease. Lancet. 2005;365:410–412. doi: 10.1016/S0140-6736(05)17828-3. [DOI] [PubMed] [Google Scholar]
- 15.Foroud T, Uniacke SK, Liu L, et al. Heterozygosity for a mutation in the parkin gene leads to later onset Parkinson disease. Neurology. 2003;60:796–801. doi: 10.1212/01.wnl.0000049470.00180.07. [DOI] [PubMed] [Google Scholar]
- 16.Pankratz N, Nichols WC, Uniacke SK, et al. Genome-wide linkage analysis and evidence of gene-by-gene interactions in a sample of 362 multiplex Parkinson disease families. Hum Mol Genet. 2003;12:2599–2608. doi: 10.1093/hmg/ddg270. [DOI] [PubMed] [Google Scholar]
- 17.Cogan JD, Pauciulo MW, Batchman AP, et al. High frequency of BMPR2 exonic deletions/duplications in familial pulmonary arterial hypertension. Am J Respir Crit Care Med. 2006;174:590–598. doi: 10.1164/rccm.200602-165OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nichols WC, Elsaesser VE, Pankratz N, et al. LRRK2 mutation analysis in Parkinson disease families with evidence of linkage to PARK8. Neurology. 2007;69:1737–1744. doi: 10.1212/01.wnl.0000278115.50741.4e. [DOI] [PubMed] [Google Scholar]
- 19.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 20.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gudbjartsson DF, Jonasson K, Frigge ML, Kong A. Allegro, a new computer program for multipoint linkage analysis. Nature Genet. 2000;25:12–13. doi: 10.1038/75514. [DOI] [PubMed] [Google Scholar]
- 22.Hadjigeorgiou GM, Xiromerisiou G, Gourbali V, et al. Association of α-synuclein Rep1 polymorphism and Parkinson’s disease: influence of Rep1 on age at onset. Mov Disord. 2006;21:534–539. doi: 10.1002/mds.20752. [DOI] [PubMed] [Google Scholar]
- 23.Myhre R, Toft M, Kachergus J, et al. Multiple α-synuclein gene polymorphisms are associated with Parkinson’s disease in a Norwegian population. Acta Neurol Scand. 2008;118:320–327. doi: 10.1111/j.1600-0404.2008.01019.x. [DOI] [PubMed] [Google Scholar]
- 24.Kay DM, Factor SA, Samii A, et al. Genetic association between α-synuclein and idiopathic Parkinson’s disease. Am J Med Genet B Neuropsychiatr Genet B. 2008;147:1222–1230. doi: 10.1002/ajmg.b.30758. [DOI] [PubMed] [Google Scholar]
- 25.Fuchs J, Tichopad A, Golub Y, et al. Genetic variability in the SNCA gene influences α-synuclein levels in the blood and brain. FASEB J. 2008;22:1327–1334. doi: 10.1096/fj.07-9348com. [DOI] [PubMed] [Google Scholar]
- 26.Starkstein SE, Mayberg HS, Leiguarda R, Preziosi TJ, Robinson RG. A prospective longitudinal study of depression, cognitive decline, and physical impairments in patients with Parkinson’s disease. J Neurol Neurosurg Psychiatry. 1992;55:377–382. doi: 10.1136/jnnp.55.5.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cubo E, Bernard B, Leurgans S, Raman R. Cognitive and motor function in patients with Parkinson’s disease with and without depression. Clin Neuropharmacol. 2000;23:331–334. doi: 10.1097/00002826-200011000-00006. [DOI] [PubMed] [Google Scholar]
- 28.Pankratz N, Marder KS, Halter CA, et al. Clinical correlates of depressive symptoms in familial Parkinson disease. Mov Disord. 2008;23:2216–2223. doi: 10.1002/mds.22285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirik D, Rosenblad C, Burger C, et al. Parkinson-like neurodegeneration induced by targeted overexpression of α-synuclein in the nigrostriatal system. J Neurosci. 2002;22:2780–2791. doi: 10.1523/JNEUROSCI.22-07-02780.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou W, Schaack J, Zawada WM, Freed CR. Overexpression of human α-synuclein causes dopamine neuron death in primary human mesencephalic culture. Brain Res. 2002;926:42–50. doi: 10.1016/s0006-8993(01)03292-9. [DOI] [PubMed] [Google Scholar]